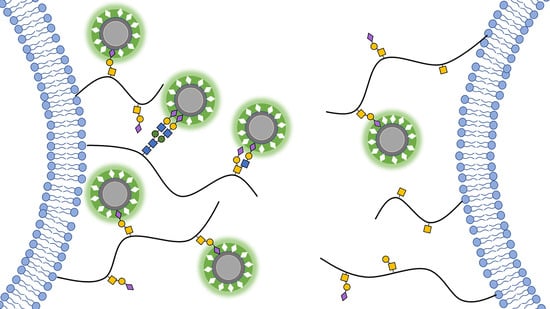
Fluorescent Molecularly Imprinted Polymer Layers against Sialic Acid on Silica-Coated Polystyrene Cores—Assessment of the Binding Behavior to Cancer Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Cell Culture
2.2. SA-MIP Synthesis
2.3. Flow Cytometry Assay for Lectin Staining
2.4. Flow Cytometry Assay for MIPs Staining
2.5. Pre-Treatment of SA-MIPs with Pentavalent SA Conjugates
2.6. Confocal Fluorescence Microscopy Analysis
3. Results
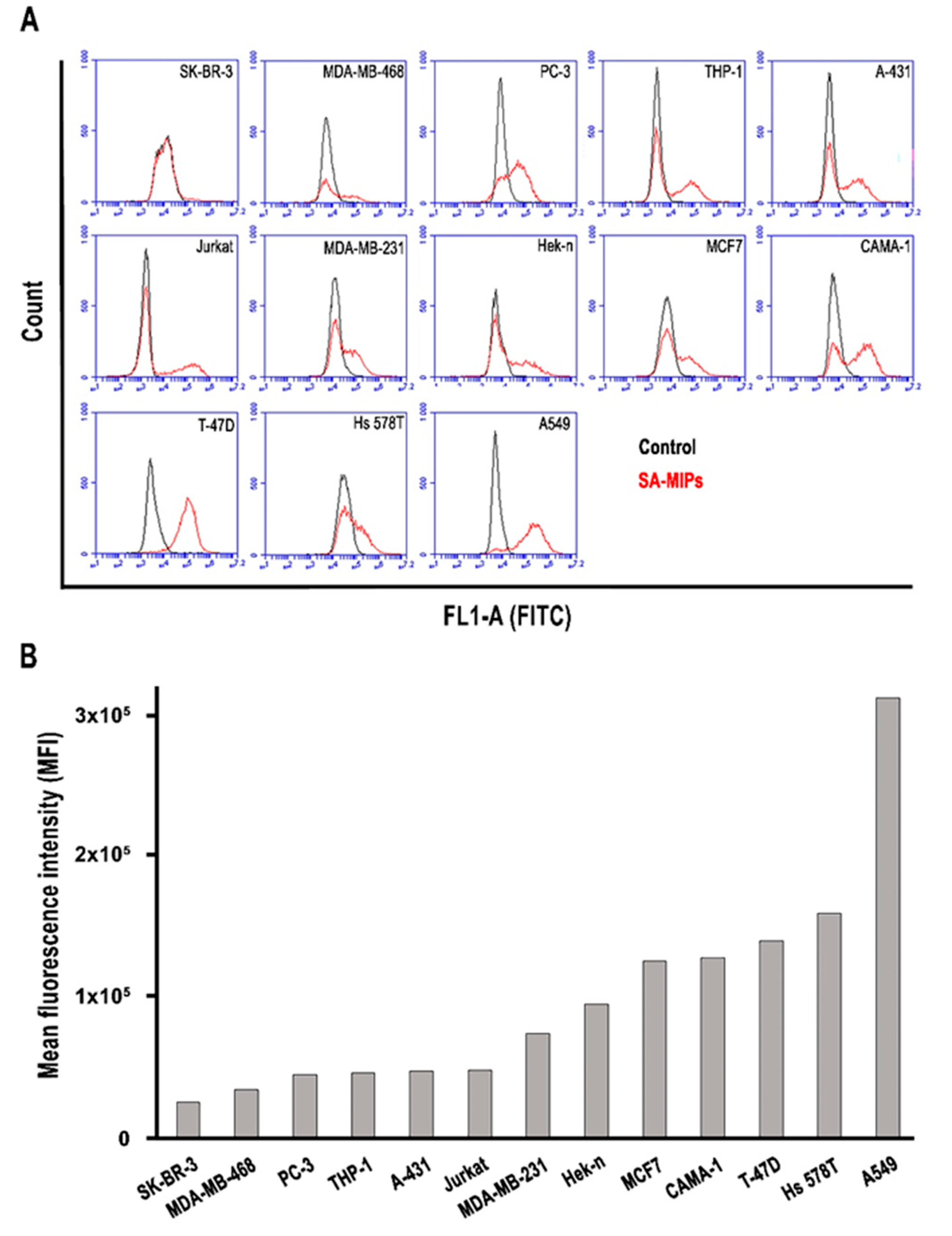
3.1. SA Staining with Lectins MAL I and SNA of a Variety of Cancer Cell Lines
3.2. The Binding Patterns of SA-MIPs to Several Cancer Cell Lines
3.3. Pre-Treatment of SA-MIPs with SA Conjugates to Reduce Binding to Cancer Cell Lines
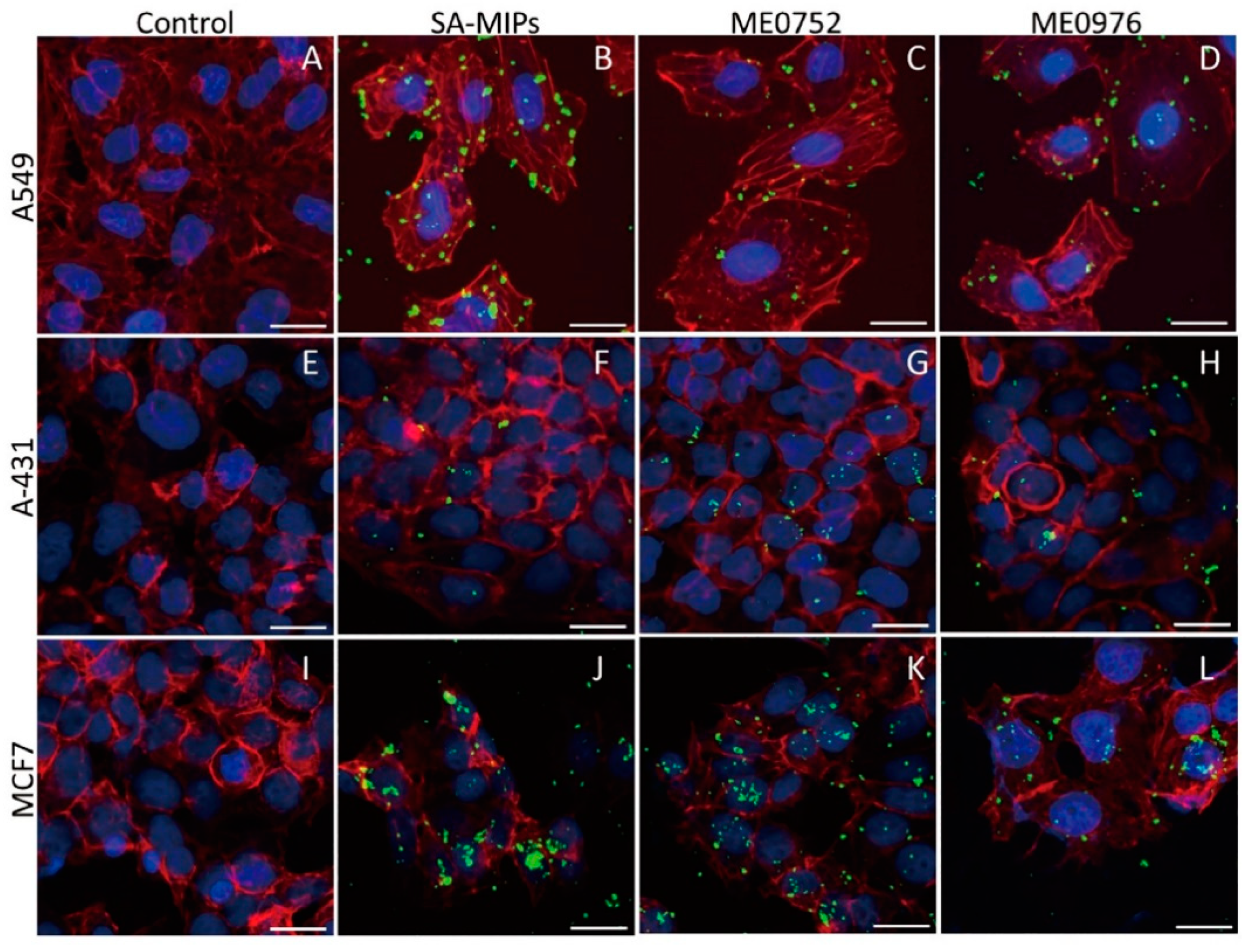
3.4. SA-MIPs Staining Patterns Visualized by Confocal Fluorescence Microscopy
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Varki, A. Sialic acids in human health and disease. Trends Mol. Med. 2008, 14, 351–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varki, A. Biological roles of glycans. Glycobiology 2017, 27, 3–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues, E.; Macauley, M.S. Hypersialylation in cancer: Modulation of inflammation and therapeutic opportunities. Cancers 2018, 10, 207. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Yang, G.; Guan, F. Biological functions and analytical strategies of sialic acids in tumor. Cells 2020, 9, 273. [Google Scholar] [CrossRef] [Green Version]
- Bogenrieder, T.; Herlyn, M. Axis of evil: Molecular mechanisms of cancer metastasis. Oncogene 2003, 22, 6524–6536. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, F.; Tiralongo, E.; Tiralongo, J. Sialic acid-specific lectins: Occurrence, specificity and function. Cell. Mol. Life Sci. 2006, 63, 1331–1354. [Google Scholar] [CrossRef]
- Loureiro, L.R.; Carrascal, M.A.; Barbas, A.; Ramalho, J.S.; Novo, C.; Delannoy, P.; Videira, P.A. Challenges in antibody development against Tn and Sialyl-Tn antigens. Biomolecules 2015, 5, 1783–1809. [Google Scholar] [CrossRef] [Green Version]
- Xing, R.; Wang, S.; Bie, Z.; He, H.; Liu, Z. Preparation of molecularly imprinted polymers specific to glycoproteins, glycans and monosaccharides via boronate affinity controllable–oriented surface imprinting. Nat. Protoc. 2017, 12, 964–987. [Google Scholar] [CrossRef]
- Sellergren, B. Molecularly Imprinted Polymers: Man-Made Mimics of Antibodies and Their Application in Analytical Chemistry; Elsevier: Amsterdam, The Netherlands, 2000. [Google Scholar]
- El-Schich, Z.; Zhang, Y.; Feith, M.; Beyer, S.; Sternbæk, L.; Ohlsson, L.; Stollenwerk, M.; Wingren, A.G. Molecularly imprinted polymers in biological applications. Biotechniques 2020, 69, 406–419. [Google Scholar] [CrossRef]
- Hillberg, A.; Brain, K.; Allender, C. Molecular imprinted polymer sensors: Implications for therapeutics. Adv. Drug Del. Rev. 2005, 57, 1875–1889. [Google Scholar] [CrossRef]
- Mavliutova, L.; Verduci, E.; Shinde, S.A.; Sellergren, B. Combinatorial Design of a Sialic Acid-Imprinted Binding Site. ACS Omega 2021, 6, 12229–12237. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Dong, Y.; Xu, S.; Wang, L.; Liu, Z. Molecularly Imprinted Polymer-Based Smart Prodrug Delivery System for Specific Targeting, Prolonged Retention, and Tumor Microenvironment-Triggered Release. Angew. Chem. 2021, 60, 2663–2667. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.; Li, X.; Ma, Y.; Liu, Z. Targeted cancer imaging and photothermal therapy via monosaccharide-imprinted gold nanorods. Chem. Commun. 2017, 53, 6716–6719. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wen, Y.; Wang, Y.; Ma, Y.; Liu, Z. Pattern Recognition of Cells via Multiplexed Imaging with Monosaccharide-Imprinted Quantum Dots. Anal. Chem. 2017, 89, 5646–5652. [Google Scholar] [CrossRef]
- Mavliutova, L.; Aldeguer, B.M.; Wiklander, J.; Wierzbicka, C.; Sellergren, B. Discrimination between sialic acid linkage modes using sialyllactose-imprinted polymers. RSC Adv. 2021, 11, 22409–22418. [Google Scholar] [CrossRef]
- Shinde, S.; El-Schich, Z.; Malakpour, A.; Wan, W.; Dizeyi, N.; Mohammadi, R.; Rurack, K.; Gjörloff Wingren, A.; Sellergren, B.r. Sialic acid-imprinted fluorescent core–shell particles for selective labeling of cell surface glycans. J. Am. Chem. Soc. 2015, 137, 13908–13912. [Google Scholar] [CrossRef]
- Bie, Z.; Chen, Y.; Ye, J.; Wang, S.; Liu, Z. Boronate-affinity glycan-oriented surface imprinting: A new strategy to mimic lectins for the recognition of an intact glycoprotein and its characteristic fragments. Angew. Chem. 2015, 54, 10211–10215. [Google Scholar] [CrossRef]
- Wang, S.; Yin, D.; Wang, W.; Shen, X.; Zhu, J.-J.; Chen, H.-Y.; Liu, Z. Targeting and imaging of cancer cells via monosaccharide-imprinted fluorescent nanoparticles. Sci. Rep. 2016, 6, 22757. [Google Scholar] [CrossRef] [Green Version]
- Panagiotopoulou, M.; Salinas, Y.; Beyazit, S.; Kunath, S.; Duma, L.; Prost, E.; Mayes, A.G.; Resmini, M.; Tse Sum Bui, B.; Haupt, K. Molecularly imprinted polymer coated quantum dots for multiplexed cell targeting and imaging. Angew. Chem. 2016, 128, 8384–8388. [Google Scholar] [CrossRef]
- Panagiotopoulou, M.; Kunath, S.; Medina-Rangel, P.X.; Haupt, K.; Bui, B.T.S. Fluorescent molecularly imprinted polymers as plastic antibodies for selective labeling and imaging of hyaluronan and sialic acid on fixed and living cells. Biosens. Bioelectron. 2017, 88, 85–93. [Google Scholar] [CrossRef]
- Kimani, M.; Beyer, S.; El-Schich, Z.; Gawlitza, K.; Gjörloff-Wingren, A.; Rurack, K. Imprinted Particles for Direct Fluorescence Detection of Sialic Acid in Polar Media and on Cancer Cells with Enhanced Control of Nonspecific Binding. ACS Appl. Polym. Mater. 2021, 3, 2363–2373. [Google Scholar] [CrossRef]
- El-Schich, Z.; Abdullah, M.; Shinde, S.; Dizeyi, N.; Rosén, A.; Sellergren, B.; Wingren, A.G. Different expression levels of glycans on leukemic cells—A novel screening method with molecularly imprinted polymers (MIP) targeting sialic acid. Tumor Biol. 2016, 37, 13763–13768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sternbæk, L.; Kimani Wamaitha, M.; Gawlitza, K.; Janicke, B.; Alm, K.; Wingren Gjörloff, A. Digital holographic microscopy: Macrophage uptake of nanoprobes. Imaging Microsc. 2019, 1, 21–23. [Google Scholar]
- Patel, M.; Feith, M.; Janicke, B.; Alm, K.; El-Schich, Z. Evaluation of the impact of imprinted polymer particles on morphology and motility of breast cancer cells by using digital holographic cytometry. Appl. Sci. 2020, 10, 750. [Google Scholar] [CrossRef] [Green Version]
- Kimoto, S.; Dick, W.D.; Hunt, B.; Szymanski, W.W.; McMurry, P.H.; Roberts, D.L.; Pui, D.Y.J.A.S. Characterization of nanosized silica size standards. Technology 2017, 51, 936–945. [Google Scholar] [CrossRef] [Green Version]
- Johansson, E.; Caraballo, R.; Mistry, N.; Zocher, G.; Qian, W.; Andersson, C.D.; Hurdiss, D.L.; Chandra, N.; Thompson, R.; Frängsmyr, L. Pentavalent sialic acid conjugates block coxsackievirus A24 variant and human adenovirus type 37–viruses that cause highly contagious eye infections. ACS Chem. Biol. 2020, 15, 2683–2691. [Google Scholar] [CrossRef]
- Xie, Y.; Sheng, Y.; Li, Q.; Ju, S.; Reyes, J.; Lebrilla, C.B. Determination of the glycoprotein specificity of lectins on cell membranes through oxidative proteomics. Chem. Sci. 2020, 11, 9501–9512. [Google Scholar] [CrossRef]
- Caraballo, R.; Saleeb, M.; Bauer, J.; Liaci, A.M.; Chandra, N.; Storm, R.J.; Frängsmyr, L.; Qian, W.; Stehle, T.; Arnberg, N. Triazole linker-based trivalent sialic acid inhibitors of adenovirus type 37 infection of human corneal epithelial cells. Org. Biomol. Chem. 2015, 13, 9194–9205. [Google Scholar] [CrossRef] [Green Version]
- El-Schich, Z.; Zhang, Y.; Göransson, T.; Dizeyi, N.; Persson, J.L.; Johansson, E.; Caraballo, R.; Elofsson, M.; Shinde, S.; Sellergren, B. Sialic Acid as a Biomarker Studied in Breast Cancer Cell Lines In Vitro Using Fluorescent Molecularly Imprinted Polymers. Appl. Sci. 2021, 11, 3256. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beyer, S.; Kimani, M.; Zhang, Y.; Verhassel, A.; Sternbæk, L.; Wang, T.; Persson, J.L.; Härkönen, P.; Johansson, E.; Caraballo, R.; et al. Fluorescent Molecularly Imprinted Polymer Layers against Sialic Acid on Silica-Coated Polystyrene Cores—Assessment of the Binding Behavior to Cancer Cells. Cancers 2022, 14, 1875. https://doi.org/10.3390/cancers14081875
Beyer S, Kimani M, Zhang Y, Verhassel A, Sternbæk L, Wang T, Persson JL, Härkönen P, Johansson E, Caraballo R, et al. Fluorescent Molecularly Imprinted Polymer Layers against Sialic Acid on Silica-Coated Polystyrene Cores—Assessment of the Binding Behavior to Cancer Cells. Cancers. 2022; 14(8):1875. https://doi.org/10.3390/cancers14081875
Chicago/Turabian StyleBeyer, Sarah, Martha Kimani, Yuecheng Zhang, Alejandra Verhassel, Louise Sternbæk, Tianyan Wang, Jenny L. Persson, Pirkko Härkönen, Emil Johansson, Remi Caraballo, and et al. 2022. "Fluorescent Molecularly Imprinted Polymer Layers against Sialic Acid on Silica-Coated Polystyrene Cores—Assessment of the Binding Behavior to Cancer Cells" Cancers 14, no. 8: 1875. https://doi.org/10.3390/cancers14081875
APA StyleBeyer, S., Kimani, M., Zhang, Y., Verhassel, A., Sternbæk, L., Wang, T., Persson, J. L., Härkönen, P., Johansson, E., Caraballo, R., Elofsson, M., Gawlitza, K., Rurack, K., Ohlsson, L., El-Schich, Z., Wingren, A. G., & Stollenwerk, M. M. (2022). Fluorescent Molecularly Imprinted Polymer Layers against Sialic Acid on Silica-Coated Polystyrene Cores—Assessment of the Binding Behavior to Cancer Cells. Cancers, 14(8), 1875. https://doi.org/10.3390/cancers14081875