Human Cytomegalovirus Seropositivity and Viral DNA in Breast Tumors Are Associated with Poor Patient Prognosis
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Human Specimens and Patient Information
2.3. Positive and Negative Controls for HCMV Infection
2.4. Extraction of DNA and RNA
2.5. LightCycler (LC) PCR
2.6. Nested PCR Amplification and Visualization
2.7. Sequencing of PCR Product
2.8. qPCR for mRNA Expression
2.9. HCMV IgG Detection
2.10. Statistical Analysis
3. Results
3.1. Comparison of HCMV gB DNA Detection Using LC-PCR versus Nested PCR
3.2. Detection of HCMV gB DNA in Tissues and HCMV IgG in Serum
3.3. Association of HCMV gB DNA-Positive Breast Tumors or HCMV IgG Positivity with Patient Characteristics
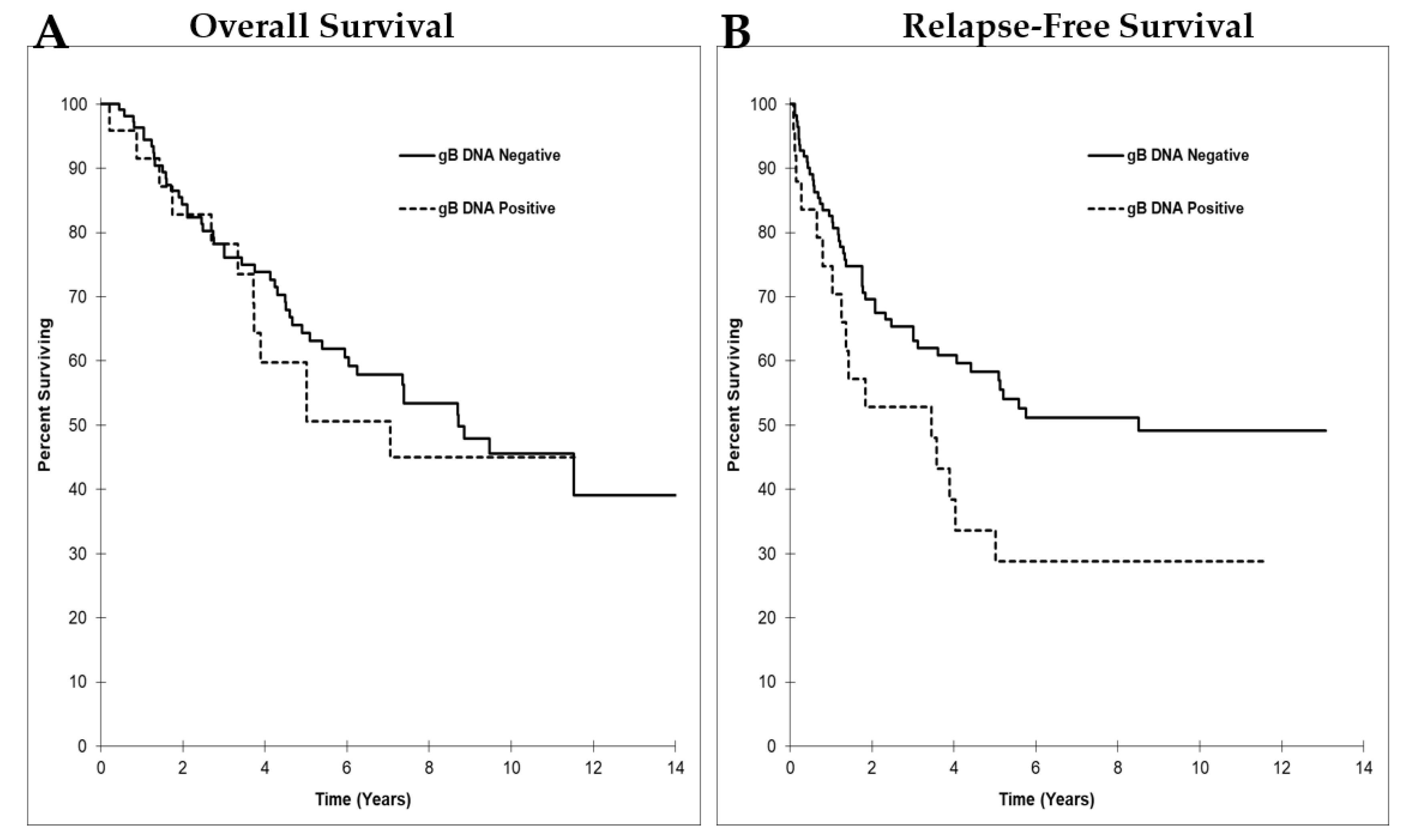
3.4. HCMV gB DNA Positivity in Breast Tumors was Associated with Reduced Relapse-Free Survival
3.5. mRNA Expression of HCMV IE1 Was Not Detected in any Human Breast Tumors or Normal Breast Tissue
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ER | estrogen receptor-1 |
| gB | glycoprotein B |
| HCMV | human cytomegalovirus |
| HER2 | human epidermal growth factor receptor 2 |
| IE | immediate early |
| IgG | immunoglobulin G (IgG) |
| IL | interleukin |
| mCMV | mouse cytomegalovirus |
| PR | progesterone receptor |
| PDGFRα | platelet-derived growth factor receptor alpha |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Akram, M.; Iqbal, M.; Daniyal, M.; Khan, A.U. Awareness and current knowledge of breast cancer. Biol. Res. 2017, 50, 33. [Google Scholar] [CrossRef] [PubMed]
- Brewer, H.R.; Jones, M.E.; Schoemaker, M.J.; Ashworth, A.; Swerdlow, A.J. Family history and risk of breast cancer: An analysis accounting for family structure. Breast Cancer Res. Treat. 2017, 165, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Endogenous, H.; Breast Cancer Collaborative, G.; Key, T.J.; Appleby, P.N.; Reeves, G.K.; Travis, R.C.; Alberg, A.J.; Barricarte, A.; Berrino, F.; Krogh, V.; et al. Sex hormones and risk of breast cancer in premenopausal women: A collaborative reanalysis of individual participant data from seven prospective studies. Lancet Oncol. 2013, 14, 1009–1019. [Google Scholar] [CrossRef]
- Makarem, N.; Chandran, U.; Bandera, E.V.; Parekh, N. Dietary fat in breast cancer survival. Annu. Rev. Nutr. 2013, 33, 319–348. [Google Scholar] [CrossRef]
- Jung, S.; Wang, M.; Anderson, K.; Baglietto, L.; Bergkvist, L.; Bernstein, L.; van den Brandt, P.A.; Brinton, L.; Buring, J.E.; Eliassen, A.H.; et al. Alcohol consumption and breast cancer risk by estrogen receptor status: In a pooled analysis of 20 studies. Int. J. Epidemiol. 2016, 45, 916–928. [Google Scholar] [CrossRef]
- Plummer, M.; de Martel, C.; Vignat, J.; Ferlay, J.; Bray, F.; Franceschi, S. Global burden of cancers attributable to infections in 2012: A synthetic analysis. Lancet Glob. Health 2016, 4, e609–e616. [Google Scholar] [CrossRef]
- Epstein, M.A.; Achong, B.G.; Barr, Y.M. Virus Particles in Cultured Lymphoblasts from Burkitt’s Lymphoma. Lancet 1964, 1, 702–703. [Google Scholar] [CrossRef]
- de-The, G.; Geser, A.; Day, N.E.; Tukei, P.M.; Williams, E.H.; Beri, D.P.; Smith, P.G.; Dean, A.G.; Bronkamm, G.W.; Feorino, P.; et al. Epidemiological evidence for causal relationship between Epstein-Barr virus and Burkitt’s lymphoma from Ugandan prospective study. Nature 1978, 274, 756–761. [Google Scholar] [CrossRef]
- zur Hausen, H. Human Papillomaviruses and Their Possible Role in Squamous Cell Carcinomas; Current Topics in Microbiology and Immunology; Springer: New York, NY, USA, 1977; Volume 78, pp. 1–30. [Google Scholar] [CrossRef]
- Durst, M.; Gissmann, L.; Ikenberg, H.; zur Hausen, H. A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proc. Natl. Acad. Sci. USA 1983, 80, 3812–3815. [Google Scholar] [CrossRef]
- Haidar Ahmad, S.; Al Moussawi, F.; El Baba, R.; Nehme, Z.; Pasquereau, S.; Kumar, A.; Molimard, C.; Monnien, F.; Algros, M.P.; Karaky, R.; et al. Identification of UL69 Gene and Protein in Cytomegalovirus-Transformed Human Mammary Epithelial Cells. Front. Oncol. 2021, 11, 627866. [Google Scholar] [CrossRef] [PubMed]
- Moussawi, F.A.; Kumar, A.; Pasquereau, S.; Tripathy, M.K.; Karam, W.; Diab-Assaf, M.; Herbein, G. The transcriptome of human mammary epithelial cells infected with the HCMV-DB strain displays oncogenic traits. Sci. Rep. 2018, 8, 12574. [Google Scholar] [CrossRef] [PubMed]
- Nehme, Z.; Pasquereau, S.; Haidar Ahmad, S.; Coaquette, A.; Molimard, C.; Monnien, F.; Algros, M.P.; Adotevi, O.; Diab Assaf, M.; Feugeas, J.P.; et al. Polyploid giant cancer cells, stemness and epithelial-mesenchymal plasticity elicited by human cytomegalovirus. Oncogene 2021, 40, 3030–3046. [Google Scholar] [CrossRef] [PubMed]
- Bai, B.; Wang, X.; Chen, E.; Zhu, H. Human cytomegalovirus infection and colorectal cancer risk: A meta-analysis. Oncotarget 2016, 7, 76735–76742. [Google Scholar] [CrossRef] [PubMed]
- Samanta, M.; Harkins, L.; Klemm, K.; Britt, W.J.; Cobbs, C.S. High prevalence of human cytomegalovirus in prostatic intraepithelial neoplasia and prostatic carcinoma. J. Urol. 2003, 170, 998–1002. [Google Scholar] [CrossRef]
- Radestad, A.F.; Estekizadeh, A.; Cui, H.L.; Kostopoulou, O.N.; Davoudi, B.; Hirschberg, A.L.; Carlson, J.; Rahbar, A.; Soderberg-Naucler, C. Impact of Human Cytomegalovirus Infection and its Immune Response on Survival of Patients with Ovarian Cancer. Transl. Oncol. 2018, 11, 1292–1300. [Google Scholar] [CrossRef]
- Dziurzynski, K.; Chang, S.M.; Heimberger, A.B.; Kalejta, R.F.; McGregor Dallas, S.R.; Smit, M.; Soroceanu, L.; Cobbs, C.S.; Hcmv; Gliomas, S. Consensus on the role of human cytomegalovirus in glioblastoma. Neuro-Oncology 2012, 14, 246–255. [Google Scholar] [CrossRef]
- Harkins, L.E.; Matlaf, L.A.; Soroceanu, L.; Klemm, K.; Britt, W.J.; Wang, W.; Bland, K.I.; Cobbs, C.S. Detection of human cytomegalovirus in normal and neoplastic breast epithelium. Herpesviridae 2010, 1, 8. [Google Scholar] [CrossRef]
- Richardson, A.K.; Walker, L.C.; Cox, B.; Rollag, H.; Robinson, B.A.; Morrin, H.; Pearson, J.F.; Potter, J.D.; Paterson, M.; Surcel, H.M.; et al. Breast cancer and cytomegalovirus. Clin. Transl. Oncol. 2020, 22, 585–602. [Google Scholar] [CrossRef]
- Geisler, J.; Touma, J.; Rahbar, A.; Soderberg-Naucler, C.; Vetvik, K. A Review of the Potential Role of Human Cytomegalovirus (HCMV) Infections in Breast Cancer Carcinogenesis and Abnormal Immunity. Cancers 2019, 11, 1842. [Google Scholar] [CrossRef]
- Michaelis, M.; Doerr, H.W.; Cinatl, J. The story of human cytomegalovirus and cancer: Increasing evidence and open questions. Neoplasia 2009, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Soroceanu, L.; Cobbs, C.S. Is HCMV a tumor promoter? Virus Res. 2011, 157, 193–203. [Google Scholar] [CrossRef]
- Davison, A.J.; Eberle, R.; Ehlers, B.; Hayward, G.S.; McGeoch, D.J.; Minson, A.C.; Pellett, P.E.; Roizman, B.; Studdert, M.J.; Thiry, E. The order Herpesvirales. Arch. Virol. 2009, 154, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Goodrum, F.; Caviness, K.; Zagallo, P. Human cytomegalovirus persistence. Cell Microbiol. 2012, 14, 644–655. [Google Scholar] [CrossRef] [PubMed]
- Bate, S.L.; Dollard, S.C.; Cannon, M.J. Cytomegalovirus seroprevalence in the United States: The national health and nutrition examination surveys, 1988-2004. Clin. Infect. Dis. 2010, 50, 1439–1447. [Google Scholar] [CrossRef]
- Pawelec, G.; McElhaney, J.E.; Aiello, A.E.; Derhovanessian, E. The impact of CMV infection on survival in older humans. Curr. Opin. Immunol. 2012, 24, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Adland, E.; Klenerman, P.; Goulder, P.; Matthews, P.C. Ongoing burden of disease and mortality from HIV/CMV coinfection in Africa in the antiretroviral therapy era. Front. Microbiol. 2015, 6, 1016. [Google Scholar] [CrossRef]
- Asanuma, H.; Numazaki, K.; Nagata, N.; Hotsubo, T.; Horino, K.; Chiba, S. Role of milk whey in the transmission of human cytomegalovirus infection by breast milk. Microbiol. Immunol. 1996, 40, 201–204. [Google Scholar] [CrossRef]
- Hayes, K.; Danks, D.M.; Gibas, H.; Jack, I. Cytomegalovirus in human milk. N. Engl. J. Med. 1972, 287, 177–178. [Google Scholar] [CrossRef]
- Griffiths, P.; Baraniak, I.; Reeves, M. The pathogenesis of human cytomegalovirus. J. Pathol. 2015, 235, 288–297. [Google Scholar] [CrossRef]
- Wreghitt, T.G.; Teare, E.L.; Sule, O.; Devi, R.; Rice, P. Cytomegalovirus infection in immunocompetent patients. Clin. Infect. Dis. 2003, 37, 1603–1606. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Sasaki, Y.; Maeda, T.; Komatsu, F.; Suzuki, T.; Urita, Y. Clinical differentiation of infectious mononucleosis that is caused by Epstein-Barr virus or cytomegalovirus: A single-center case-control study in Japan. J. Infect. Chemother. 2019, 25, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Kocak, E.D.; Sherwin, J.C.; Hall, A.J. Cytomegalovirus disease in immunocompetent adults. Med. J. Aust. 2015, 202, 419. [Google Scholar] [CrossRef]
- Cheng, S.; Caviness, K.; Buehler, J.; Smithey, M.; Nikolich-Zugich, J.; Goodrum, F. Transcriptome-wide characterization of human cytomegalovirus in natural infection and experimental latency. Proc. Natl. Acad. Sci. USA 2017, 114, E10586–E10595. [Google Scholar] [CrossRef] [PubMed]
- Rahbar, A.; Bostrom, L.; Lagerstedt, U.; Magnusson, I.; Soderberg-Naucler, C.; Sundqvist, V.A. Evidence of active cytomegalovirus infection and increased production of IL-6 in tissue specimens obtained from patients with inflammatory bowel diseases. Inflamm. Bowel Dis. 2003, 9, 154–161. [Google Scholar] [CrossRef]
- Frantzeskaki, F.G.; Karampi, E.S.; Kottaridi, C.; Alepaki, M.; Routsi, C.; Tzanela, M.; Vassiliadi, D.A.; Douka, E.; Tsaousi, S.; Gennimata, V.; et al. Cytomegalovirus reactivation in a general, nonimmunosuppressed intensive care unit population: Incidence, risk factors, associations with organ dysfunction, and inflammatory biomarkers. J. Crit. Care 2015, 30, 276–281. [Google Scholar] [CrossRef]
- Tu, W.; Rao, S. Mechanisms Underlying T Cell Immunosenescence: Aging and Cytomegalovirus Infection. Front. Microbiol. 2016, 7, 2111. [Google Scholar] [CrossRef]
- McDevitt, L.M. Etiology and impact of cytomegalovirus disease on solid organ transplant recipients. Am. J. Health Syst. Pharm. 2006, 63, S3–S9. [Google Scholar] [CrossRef]
- Bodeus, M.; Feyder, S.; Goubau, P. Avidity of IgG antibodies distinguishes primary from non-primary cytomegalovirus infection in pregnant women. Clin. Diagn. Virol. 1998, 9, 9–16. [Google Scholar] [CrossRef]
- Taher, C.; de Boniface, J.; Mohammad, A.A.; Religa, P.; Hartman, J.; Yaiw, K.C.; Frisell, J.; Rahbar, A.; Soderberg-Naucler, C. High prevalence of human cytomegalovirus proteins and nucleic acids in primary breast cancer and metastatic sentinel lymph nodes. PLoS ONE 2013, 8, e56795. [Google Scholar] [CrossRef]
- Rahbar, A.; Touma, J.; Costa, H.; Davoudi, B.; Bukholm, I.R.; Sauer, T.; Vetvik, K.; Geisler, J.; Soderberg-Naucler, C. Low Expression of Estrogen Receptor-alpha and Progesterone Receptor in Human Breast Cancer Tissues Is Associated with High-Grade Human Cytomegalovirus Protein Expression. Clin. Breast Cancer 2017, 17, 526–535.e1. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Wang, Q.; Wang, H.B.; Wang, B.; Li, L. Protein and DNA evidences of HCMV infection in primary breast cancer tissues and metastatic sentinel lymph nodes. Cancer Biomark. 2018, 21, 769–780. [Google Scholar] [CrossRef] [PubMed]
- Al-Nuaimi, B.N.; Al-Azzawi, R.H.; Naji, R.Z. Association of human cytomegalovirus with HER2 proto-oncogene overexpression in Iraqi breast cancer patients. Biochem. Cell. Arch. 2019, 19, 1691–1698. [Google Scholar]
- El Shazly, D.F.; Bahnassey, A.A.; Omar, O.S.; Elsayed, E.T.; Al-Hindawi, A.; El-Desouky, E.; Youssef, H.; Zekri, A.N. Detection of Human Cytomegalovirus in Malignant and Benign Breast Tumors in Egyptian Women. Clin. Breast Cancer 2018, 18, e629–e642. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, R.A.; Yussif, S.M. Immunohistochemical detection of human cytomegalovirus, Epstein-Barr virus and human papillomavirus in invasive breast carcinoma in Egyptian women: A tissue microarray study. J. Solid Tumors 2016, 6, 8–16. [Google Scholar] [CrossRef]
- Mohammadizadeh, F.; Mahmudi, F. Evaluation of human cytomegalovirus antigen expression in invasive breast carcinoma in a population of Iranian patients. Infect. Agent Cancer 2017, 12, 39. [Google Scholar] [CrossRef]
- Tsai, J.H.; Tsai, C.H.; Cheng, M.H.; Lin, S.J.; Xu, F.L.; Yang, C.C. Association of viral factors with non-familial breast cancer in Taiwan by comparison with non-cancerous, fibroadenoma, and thyroid tumor tissues. J. Med. Virol. 2005, 75, 276–281. [Google Scholar] [CrossRef]
- Sepahvand, P.; Makvandi, M.; Samarbafzadeh, A.; Talaei-Zadeh, A.; Ranjbari, N.; Nisi, N.; Azaran, A.; Jalilian, S.; Pirmoradi, R.; Makvandi, K.; et al. Human Cytomegalovirus DNA among Women with Breast Cancer. Asian Pac. J. Cancer Prev. 2019, 20, 2275–2279. [Google Scholar] [CrossRef]
- El-Shinawi, M.; Mohamed, H.T.; Abdel-Fattah, H.H.; Ibrahim, S.A.; El-Halawany, M.S.; Nouh, M.A.; Schneider, R.J.; Mohamed, M.M. Inflammatory and Non-inflammatory Breast Cancer: A Potential Role for Detection of Multiple Viral DNAs in Disease Progression. Ann. Surg. Oncol. 2016, 23, 494–502. [Google Scholar] [CrossRef]
- Antonsson, A.; Bialasiewicz, S.; Rockett, R.J.; Jacob, K.; Bennett, I.C.; Sloots, T.P. Exploring the prevalence of ten polyomaviruses and two herpes viruses in breast cancer. PLoS ONE 2012, 7, e39842. [Google Scholar] [CrossRef]
- Richardson, A.K.; Currie, M.J.; Robinson, B.A.; Morrin, H.; Phung, Y.; Pearson, J.F.; Anderson, T.P.; Potter, J.D.; Walker, L.C. Cytomegalovirus and Epstein-Barr virus in breast cancer. PLoS ONE 2015, 10, e0118989. [Google Scholar] [CrossRef] [PubMed]
- Utrera-Barillas, D.; Valdez-Salazar, H.A.; Gomez-Rangel, D.; Alvarado-Cabrero, I.; Aguilera, P.; Gomez-Delgado, A.; Ruiz-Tachiquin, M.E. Is human cytomegalovirus associated with breast cancer progression? Infect. Agent Cancer 2013, 8, 12. [Google Scholar] [CrossRef] [PubMed]
- Bakhtiyrizadeh, S.; Hosseini, S.Y.; Yaghobi, R.; Safaei, A.; Sarvari, J. Almost Complete Lack of Human Cytomegalovirus and Human papillomaviruses Genome in Benign and Malignant Breast Lesions in Shiraz, Southwest of Iran. Asian Pac. J. Cancer Prev. 2017, 18, 3319–3324. [Google Scholar] [CrossRef]
- Golrokh Mofrad, M.; Sadigh, Z.A.; Ainechi, S.; Faghihloo, E. Detection of human papillomavirus genotypes, herpes simplex, varicella zoster and cytomegalovirus in breast cancer patients. Virol. J. 2021, 18, 25. [Google Scholar] [CrossRef] [PubMed]
- Ghaffari, H.; Tavakoli, A.; Nafissi, N.; Farahmand, M.; Ghorbani, S.; Moochani, S.S.; Hashemi-Bahremani, M.; Alebouyeh, M.R.; Monavari, S.H. Human cytomegalovirus and Epstein-Barr virus infections in breast cancer: A molecular study on Iranian women. Breast Dis. 2021, 40, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Nakhaie, M.; Charostad, J.; Azaran, A.; Arabzadeh, S.A.M.; Motamedfar, A.; Iranparast, S.; Ahmadpour, F.; Talaeizadeh, A.; Makvandi, M. Molecular and Serological Prevalence of HCMV in Iranian Patients with Breast Cancer. Asian Pac. J. Cancer Prev. 2021, 22, 2011–2016. [Google Scholar] [CrossRef] [PubMed]
- El-Shinawi, M.; Mohamed, H.T.; El-Ghonaimy, E.A.; Tantawy, M.; Younis, A.; Schneider, R.J.; Mohamed, M.M. Human cytomegalovirus infection enhances NF-kappaB/p65 signaling in inflammatory breast cancer patients. PLoS ONE 2013, 8, e55755. [Google Scholar] [CrossRef]
- Yang, Z.; Tang, X.; McMullen, T.P.W.; Brindley, D.N.; Hemmings, D.G. PDGFRalpha Enhanced Infection of Breast Cancer Cells with Human Cytomegalovirus but Infection of Fibroblasts Increased Prometastatic Inflammation Involving Lysophosphatidate Signaling. Int. J. Mol. Sci. 2021, 22, 9817. [Google Scholar] [CrossRef]
- Touma, J.; Liu, Y.; Rahbar, A.; Pantalone, M.R.; Almazan, N.M.; Vetvik, K.; Soderberg-Naucler, C.; Geisler, J.; Sauer, T. Detection of Human Cytomegalovirus Proteins in Paraffin-Embedded Breast Cancer Tissue Specimens-A Novel, Automated Immunohistochemical Staining Protocol. Microorganisms 2021, 9, 1059. [Google Scholar] [CrossRef]
- Tsai, J.H.; Hsu, C.S.; Tsai, C.H.; Su, J.M.; Liu, Y.T.; Cheng, M.H.; Wei, J.C.; Chen, F.L.; Yang, C.C. Relationship between viral factors, axillary lymph node status and survival in breast cancer. J. Cancer Res. Clin. Oncol. 2007, 133, 13–21. [Google Scholar] [CrossRef]
- Taher, C.; Frisk, G.; Fuentes, S.; Religa, P.; Costa, H.; Assinger, A.; Vetvik, K.K.; Bukholm, I.R.; Yaiw, K.C.; Smedby, K.E.; et al. High prevalence of human cytomegalovirus in brain metastases of patients with primary breast and colorectal cancers. Transl. Oncol. 2014, 7, 732–740. [Google Scholar] [CrossRef]
- Yang, Z.; Tang, X.; Meng, G.; Benesch, M.G.K.; Mackova, M.; Belon, A.P.; Serrano-Lomelin, J.; Goping, I.S.; Brindley, D.N.; Hemmings, D.G. Latent Cytomegalovirus Infection in Female Mice Increases Breast Cancer Metastasis. Cancers 2019, 11, 447. [Google Scholar] [CrossRef]
- Davey, A.; Eastman, L.; Hansraj, P.; Hemmings, D.G. Human cytomegalovirus is protected from inactivation by reversible binding to villous trophoblasts. Biol. Reprod. 2011, 85, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.L.; Chui, L.; Fenton, J.; LeBlanc, B.; Preiksaitis, J.K. Comparison of LightCycler-based PCR, COBAS amplicor CMV monitor, and pp65 antigenemia assays for quantitative measurement of cytomegalovirus viral load in peripheral blood specimens from patients after solid organ transplantation. J. Clin. Microbiol. 2003, 41, 3167–3174. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nicoll, S.; Brass, A.; Cubie, H.A. Detection of herpes viruses in clinical samples using real-time PCR. J. Virol. Methods 2001, 96, 25–31. [Google Scholar] [CrossRef]
- Distefano, A.L.; Alonso, A.; Martin, F.; Pardon, F. Human cytomegalovirus: Detection of congenital and perinatal infection in Argentina. BMC Pediatr. 2004, 4, 11. [Google Scholar] [CrossRef]
- Renzette, N.; Bhattacharjee, B.; Jensen, J.D.; Gibson, L.; Kowalik, T.F. Extensive genome-wide variability of human cytomegalovirus in congenitally infected infants. PLoS Pathog. 2011, 7, e1001344. [Google Scholar] [CrossRef] [PubMed]
- Gibson, L.; Piccinini, G.; Lilleri, D.; Revello, M.G.; Wang, Z.; Markel, S.; Diamond, D.J.; Luzuriaga, K. Human cytomegalovirus proteins pp65 and immediate early protein 1 are common targets for CD8+ T cell responses in children with congenital or postnatal human cytomegalovirus infection. J. Immunol. 2004, 172, 2256–2264. [Google Scholar] [CrossRef]
- Pagano, J.S.; Blaser, M.; Buendia, M.A.; Damania, B.; Khalili, K.; Raab-Traub, N.; Roizman, B. Infectious agents and cancer: Criteria for a causal relation. Semin. Cancer Biol. 2004, 14, 453–471. [Google Scholar] [CrossRef]
- Gombos, R.B.; Brown, J.C.; Teefy, J.; Gibeault, R.L.; Conn, K.L.; Schang, L.M.; Hemmings, D.G. Vascular dysfunction in young, mid-aged and aged mice with latent cytomegalovirus infections. Am. J. Physiol. Heart Circ. Physiol. 2013, 304, H183–H194. [Google Scholar] [CrossRef]
- Benesch, M.G.; Tang, X.; Dewald, J.; Dong, W.F.; Mackey, J.R.; Hemmings, D.G.; McMullen, T.P.; Brindley, D.N. Tumor-induced inflammation in mammary adipose tissue stimulates a vicious cycle of autotaxin expression and breast cancer progression. FASEB J. 2015, 29, 3990–4000. [Google Scholar] [CrossRef] [PubMed]
- Hahn, G.; Jores, R.; Mocarski, E.S. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc. Natl. Acad. Sci. USA 1998, 95, 3937–3942. [Google Scholar] [CrossRef] [PubMed]
- Soderberg-Naucler, C.; Streblow, D.N.; Fish, K.N.; Allan-Yorke, J.; Smith, P.P.; Nelson, J.A. Reactivation of latent human cytomegalovirus in CD14(+) monocytes is differentiation dependent. J. Virol. 2001, 75, 7543–7554. [Google Scholar] [CrossRef] [PubMed]
- Reeves, M.B.; MacAry, P.A.; Lehner, P.J.; Sissons, J.G.; Sinclair, J.H. Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc. Natl. Acad. Sci. USA 2005, 102, 4140–4145. [Google Scholar] [CrossRef]
- Surendran, A.; Chisthi, M.M. Breast Cancer Association with Cytomegalo Virus-A Tertiary Center Case-Control Study. J. Investig. Surg. 2019, 32, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Richardson, A.K.; Cox, B.; McCredie, M.R.; Dite, G.S.; Chang, J.H.; Gertig, D.M.; Southey, M.C.; Giles, G.G.; Hopper, J.L. Cytomegalovirus, Epstein-Barr virus and risk of breast cancer before age 40 years: A case-control study. Br. J. Cancer 2004, 90, 2149–2152. [Google Scholar] [CrossRef]
- Soroceanu, L.; Akhavan, A.; Cobbs, C.S. Platelet-derived growth factor-alpha receptor activation is required for human cytomegalovirus infection. Nature 2008, 455, 391–395. [Google Scholar] [CrossRef]


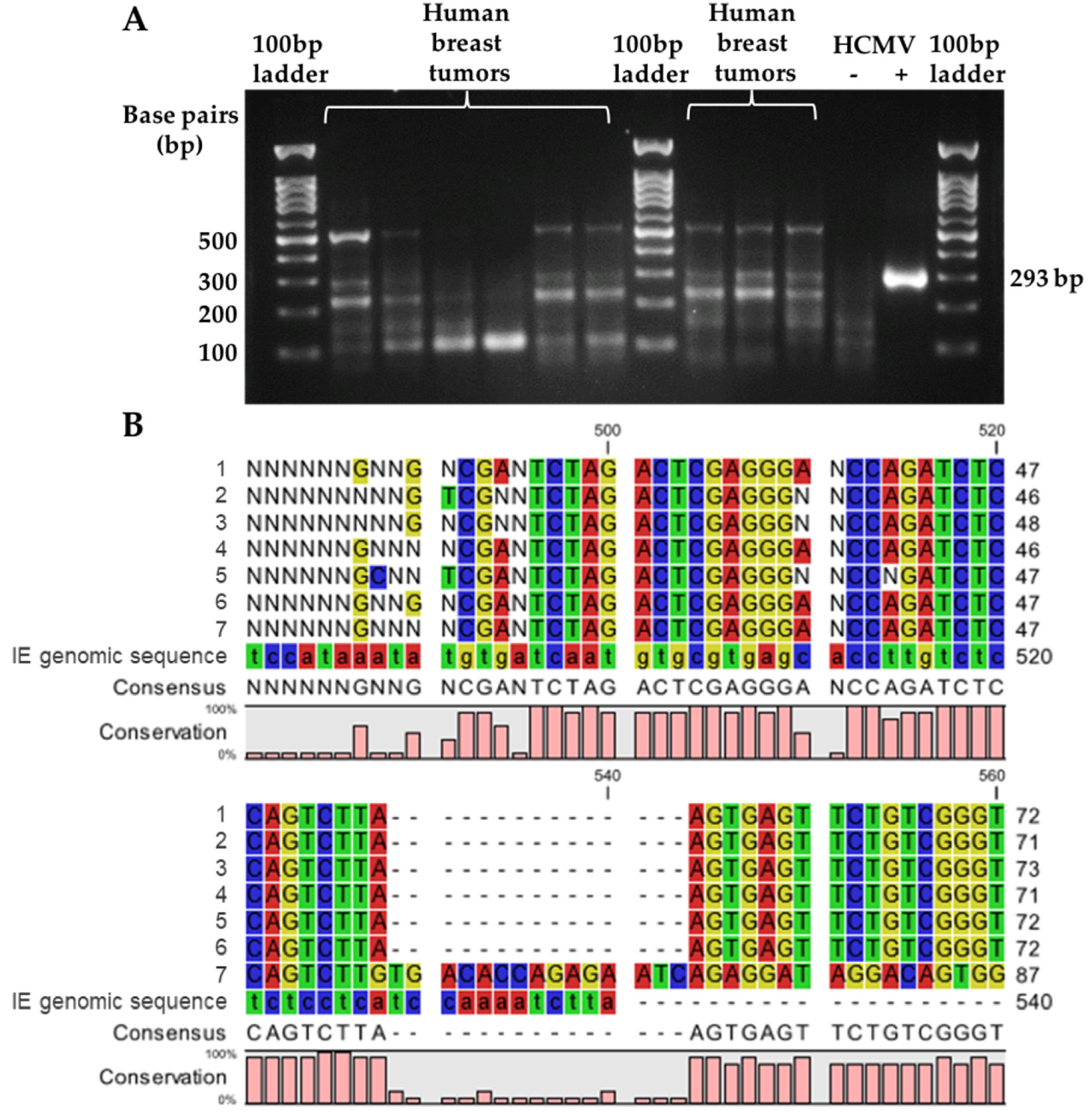
| Sample | PCR Cycle Number | Expected Concentration (Copies/PCR Reaction) | Calculated Concentration (Copies/PCR Reaction) |
|---|---|---|---|
| Standard 1800 copies | 29.72 | 1800 | 1480 |
| Standard 180,000 copies | 22.95 | 180,000 | 304,000 |
| HCMV original | 32.26 | Unknown | 200 |
| HCMV 1:10 | 33.10 | Unknown | 104 |
| HCMV 1:102 | 38.18 | Unknown | 2 |
| HCMV 1:103 | Undetermined | Unknown | NA |
| HCMV 1:104 | Undetermined | Unknown | NA |
| HCMV 1:105 | Undetermined | Unknown | NA |
| Negative control | Undetermined | Unknown | NA |
| Breast Tissues | All Breast Tumors | Serum | Breast Tumors from HCMV Seropositive Women | |
|---|---|---|---|---|
| HCMV gB, n (%) | ||||
| Negative | 9 (90) | 111 (81.6) | NA | 34 (57.6) |
| Positive | 1 (10) | 25 (18.4) | 25 (42.4) * | |
| HCMV IgG, n (%) | ||||
| Negative | NA | NA | 35 (37.2) | NA |
| Positive | 59 (62.8) * |
| Characteristics | HCMV gB− n = 111 | HCMV gB+ n = 25 | p-Value |
|---|---|---|---|
| Age, year (mean ± SD) | 56 ± 13 | 57 ± 13 | 0.596 |
| Menopause status, n (%) | 0.482 | ||
| Pre, n = 38 | 32 (29.9) | 6 (24.0) | |
| Post, n = 85 | 69 (64.5) | 16 (64.0) | |
| Peri, n = 9 | 6 (5.6) | 3 (12.0) | |
| Body mass index (mean ± SD) | 29.0 ± 6.8 | 31.0 ± 8.8 | 0.223 |
| Deceased, n (%) | 0.665 | ||
| No, n = 76 | 63 (56.8) | 13 (52.0) | |
| Yes, n = 60 | 48 (43.2) | 12 (48.0) | |
| Recurrence, n (%) | 0.060 | ||
| No, n = 72 | 63 (56.8) | 9 (36.0) | |
| Yes, n = 64 | 48 (43.2) | 16 (64.0) | |
| Time to recurrence event, days from diagnosis (median and IQR) | 1132 (41–4774) | 859 (54–4216) | 0.424 |
| Time to recurrence event, days from surgery (median and IQR) | 1132 (40–4774) | 673 (34–4216) | 0.237 |
| Tumor grade, n (%) | 0.616 | ||
| Low, n = 45 | 36 (33.6) | 9 (39.1) | |
| High, n = 85 | 71 (66.4) | 14 (60.9) | |
| Positive lymph nodes, n (mean ± SD) | 4 ± 5 | 4 ± 6 | 0.619 |
| Size of largest lymph nodes, cm(mean ± SD) | 1.58 ± 0.83 | 1.57 ± 0.93 | 0.959 |
| Tumor stage, n (%) | 0.007 | ||
| I–III, n = 116 | 99 (89.2) | 17 (68.0) | |
| IV = metastasis, n = 20 | 12 (10.8) | 8 (32.0) | |
| ER, n (%) | 0.745 | ||
| Negative, n = 18 | 14 (12.8) | 4 (16.0) | |
| Positive, n = 116 | 95 (87.2) | 21 (84.0) | |
| PR, n (%) | 0.745 | ||
| Negative, n = 47 | 37 (33.9) | 10 (40.0) | |
| Positive, n = 87 | 72 (66.1) | 15 (60.0) | |
| HER2, n (%) | 0.745 | ||
| Negative, n = 103 | 85 (76.6) | 18 (75.0) | |
| Positive, n = 32 | 26 (23.4) | 6 (25.0) | |
| Tumor subtypes, n (%) | 0.998 | ||
| Triple negative, n = 5 | 4 (3.6) | 1 (4.2) | |
| Luminal A, n = 97 | 80 (72.7) | 17 (70.8) | |
| Luminal B, n = 21 | 17 (15.5) | 4 (16.7) | |
| HER2 Enriched, n = 11 | 9 (8.2) | 2 (8.3) | |
| Vascular invasion, n (%) | 0.225 | ||
| No, n = 53 | 46 (42.6) | 7 (29.2) | |
| Yes, n = 79 | 62 (57.4) | 17 (70.8) | |
| Tumor PDGFRα mRNA (mean ± SD) | 7.92 ± 6.90 | 6.53 ± 5.15 | 0.374 |
| Characteristics | IgG− (n = 35) | IgG+ * (n = 59) | p-Value |
|---|---|---|---|
| Age, year (mean ± SD) | 51 ± 13 | 57 ± 12 | 0.017 |
| Menopause status, n (%) | 0.012 | ||
| Pre, n = 29 | 17 (50.0) | 12 (21.1) | |
| Post, n = 55 | 16 (47.1) | 39 (68.4) | |
| Peri, n = 7 | 1 (2.9) | 6 (10.5) | |
| Body mass index (mean ± SD) | 27.5 ± 6.0 | 30.0 ± 7.4 | 0.093 |
| Deceased, n (%) | 0.617 | ||
| No, n = 56 | 22 (62.9) | 34 (57.6) | |
| Yes, n = 38 | 13 (37.1) | 25 (42.4) | |
| Recurrence, n (%) | 0.148 | ||
| No, n = 50 | 22 (62.9) | 28 (47.5) | |
| Yes, n = 44 | 13 (37.1) | 31 (52.5) | |
| Time to recurrence event, days from diagnosis (median and IQR) | 1509 (126–4774) | 1128 (54–4216) | 0.375 |
| Time to recurrence event, days from surgery (median and IQR) | 1509 (126–4774) | 1007 (34–4216) | 0.290 |
| Tumor grade, n (%) | 0.343 | ||
| Low, n = 32 | 10 (29.4) | 22 (39.3) | |
| High, n = 58 | 24 (70.6) | 34 (60.7) | |
| Positive lymph nodes, n (mean ± SD) | 3 ± 4 | 4 ± 5 | 0.255 |
| Size of largest lymph nodes, cm (mean ± SD) | 1.52 ± 0.74 | 1.50 ± 0.79 | 0.941 |
| Tumor stage, n (%) | 0.157 | ||
| I–III, n = 79 | 32 (91.4) | 47 (79.7) | |
| IV = metastasis, n = 15 | 3 (8.6) | 12 (20.3) | |
| ER, n (%) | 0.741 | ||
| Negative, n = 11 | 3 (8.8) | 8 (13.8) | |
| Positive, n = 81 | 31 (91.2) | 50 (86.2) | |
| PR, n (%) | 0.937 | ||
| Negative, n = 32 | 12 (35.3) | 20 (34.5) | |
| Positive, n = 60 | 22 (64.7) | 38 (65.5) | |
| HER2, n (%) | 0.644 | ||
| Negative, n = 72 | 28 (80.0) | 44 (75.9) | |
| Positive, n = 21 | 7 (20.0) | 14 (24.1) | |
| Tumor subtypes, n (%) | 0.220 | ||
| Triple negative, n = 4 | 2 (5.9) | 2 (3.4) | |
| Luminal A, n = 67 | 25 (73.5) | 42 (72.4) | |
| Luminal B, n = 15 | 7 (20.6) | 8 (13.8) | |
| HER2 Enriched, n = 6 | 0 (0) | 6 (10.3) | |
| Vascular invasion, n (%) | 0.981 | ||
| No, n = 40 | 15 (44.1) | 25 (43.9) | |
| Yes, n = 51 | 19 (55.9) | 32 (56.1) | |
| Tumor PDGFRα mRNA (mean ± SD) | 7.8 ± 6.62 | 7.08 ± 5.90 | 0.591 |
| Characteristics | IgG+/gB− n = 34 | IgG+/gB+ * n = 25 | p-Value |
|---|---|---|---|
| Age, year (mean ± SD) | 57 ± 12 | 57 ± 13 | 0.985 |
| Menopause status, n (%) | 0.818 | ||
| Pre, n = 12 | 6 (18.8) | 6 (24.0) | |
| Post, n = 39 | 23 (71.9) | 16 (64.0) | |
| Peri, n = 6 | 3 (9.4) | 3 (12.0) | |
| Body mass index (mean ± SD) | 29.3 ± 6.2 | 31.0 ± 8.8 | 0.406 |
| Deceased, n (%) | 0.453 | ||
| No, n = 34 | 21 (61.8) | 13 (52.0) | |
| Yes, n = 25 | 13 (38.2) | 12 (48.0) | |
| Recurrence, n (%) | 0.131 | ||
| No, n = 28 | 19 (55.9) | 9 (36.0) | |
| Yes, n = 31 | 15 (44.1) | 16 (64.0) | |
| Time to recurrence event, days from diagnosis (median and IQR) | 1130 (175–4205) | 859 (54–4216) | 0.453 |
| Time to recurrence event, days from surgery (median and IQR) | 1115 (87–4205) | 673 (34–4216) | 0.276 |
| Tumor grade, n (%) | 0.984 | ||
| Low, n = 22 | 13 (39.4) | 9 (39.1) | |
| High, n = 34 | 20 (60.6) | 14 (60.9) | |
| Positive lymph nodes, n (mean ± SD) | 4 ± 5 | 4 ± 6 | 0.671 |
| Size of largest lymph nodes, cm (mean ± SD) | 1.45 ± 0.68 | 1.57 ± 0.93 | 0.612 |
| Tumor stage, n (%) | 0.0995 | ||
| I–III, n = 47 | 30 (88.2) | 17 (68.0) | |
| IV = metastasis, n = 12 | 4 (11.8) | 8 (32.0) | |
| ER, n (%) | 0.715 | ||
| Negative, n = 8 | 4 (12.1) | 4 (16.0) | |
| Positive, n = 50 | 29 (87.9) | 21 (84.0) | |
| PR, n (%) | 0.442 | ||
| Negative, n = 20 | 10 (30.3) | 10 (40.0) | |
| Positive, n = 38 | 23 (69.7) | 15 (60.0) | |
| HER2, n (%) | 0.897 | ||
| Negative, n = 72 | 26 (76.5) | 18 (75.0) | |
| Positive, n = 21 | 8 (23.5) | 6 (25.0) | |
| Tumor subtypes, n (%) | 0.923 | ||
| Triple negative, n = 2 | 1 (2.9) | 1 (3.4) | |
| Luminal A, n = 42 | 25 (73.5) | 17 (72.4) | |
| Luminal B, n = 8 | 4 (11.8) | 4 (13.8) | |
| HER2 Enriched, n = 6 | 4 (11.8) | 2 (10.3) | |
| Vascular invasion, n (%) | 0.057 | ||
| No, n = 25 | 18 (54.5) | 7 (29.2) | |
| Yes, n = 32 | 15 (45.5) | 17 (70.8) | |
| Tumor PDGFRα mRNA (mean ± SD) | 7.45 ± 6.42 | 6.53 ± 5.15 | 0.579 |
| Variable | OR | 95% CI | p-Value | |
|---|---|---|---|---|
| gB status (n = 136) | Stage I–III Stage IV | 5.27 | 1.77–15.67 | 0.003 |
| Pre-menopause | ||||
| Post-menopause | 1.31 | 0.45–3.84 | 0.621 | |
| Peri-menopause | 3.95 | 0.73–21.44 | 0.112 | |
| IgG * status (n = 94) | Stage I–III Stage IV | 5.61 | 1.09–28.75 | 0.039 |
| Pre-menopause | ||||
| Post-menopause | 3.83 | 1.43–10.29 | 0.008 | |
| Peri-menopause | 11.42 | 1.18–110.31 | 0.035 | |
| IgG/gB* status (n = 57) | Stage I–III Stage IV | 3.48 | 0.88–13.78 | 0.076 |
| Pre-menopause | ||||
| Post-menopause | 0.80 | 0.21–3.11 | 0.748 | |
| Peri-menopause | 1.50 | 0.20–11.42 | 0.697 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Z.; Tang, X.; Hasing, M.E.; Pang, X.; Ghosh, S.; McMullen, T.P.W.; Brindley, D.N.; Hemmings, D.G. Human Cytomegalovirus Seropositivity and Viral DNA in Breast Tumors Are Associated with Poor Patient Prognosis. Cancers 2022, 14, 1148. https://doi.org/10.3390/cancers14051148
Yang Z, Tang X, Hasing ME, Pang X, Ghosh S, McMullen TPW, Brindley DN, Hemmings DG. Human Cytomegalovirus Seropositivity and Viral DNA in Breast Tumors Are Associated with Poor Patient Prognosis. Cancers. 2022; 14(5):1148. https://doi.org/10.3390/cancers14051148
Chicago/Turabian StyleYang, Zelei, Xiaoyun Tang, Maria Eloisa Hasing, Xiaoli Pang, Sunita Ghosh, Todd P. W. McMullen, David N. Brindley, and Denise G. Hemmings. 2022. "Human Cytomegalovirus Seropositivity and Viral DNA in Breast Tumors Are Associated with Poor Patient Prognosis" Cancers 14, no. 5: 1148. https://doi.org/10.3390/cancers14051148
APA StyleYang, Z., Tang, X., Hasing, M. E., Pang, X., Ghosh, S., McMullen, T. P. W., Brindley, D. N., & Hemmings, D. G. (2022). Human Cytomegalovirus Seropositivity and Viral DNA in Breast Tumors Are Associated with Poor Patient Prognosis. Cancers, 14(5), 1148. https://doi.org/10.3390/cancers14051148







