Simple Summary
Long noncoding RNAs (lncRNAs) are important regulators of cell progression or regression in gastric cancer (GC). The lncRNA LOC441461 was downregulated in TNM stage IV GC. The downregulation of LOC441461 promoted the proliferation, cell cycle progression, and metastasis of GC cells in vitro. We confirmed, with predictable targets, that LOC441461 interacts with transcription factors and promotes tumor progression. Our study suggests LOC441461 as a potential biomarker, which could also lead to a therapeutic target in patients with advanced TNM stage gastric cancer.
Abstract
Gastric cancer is a common tumor, with a high mortality rate. The severity of gastric cancer is assessed by TNM staging. Long noncoding RNAs (lncRNAs) play a role in cancer treatment; investigating the clinical significance of novel biomarkers associated with TNM staging, such as lncRNAs, is important. In this study, we investigated the association between the expression of the lncRNA LOC441461 and gastric cancer stage. LOC441461 expression was lower in stage IV than in stages I, II, and III. The depletion of LOC441461 promoted cell proliferation, cell cycle progression, apoptosis, cell motility, and invasiveness. LOC441461 downregulation increased the epithelial-to-mesenchymal transition, as indicated by increased TRAIL signaling and decreased RUNX1 interactions. The interaction of the transcription factors RELA, IRF1, ESR1, AR, POU5F1, TRIM28, and GATA1 with LOC441461 affected the degree of the malignancy of gastric cancer by modulating gene transcription. The present study identified LOC441461 and seven transcription factors as potential biomarkers and therapeutic targets for the treatment of gastric cancer.
1. Introduction
Gastric cancer is a common malignancy, with a high mortality rate worldwide, and shows substantial incidence in East Asia, Eastern Europe, and South America [1]. The two most important risk factors for gastric cancer are Helicobacter pylori infection, which can induce atrophic gastritis, as well as intestinal metaplasia, and salted food intake [1]. H. pylori infection is the most prevalent, infecting more than 50% of all gastric cancers [2]. Common symptoms of gastric cancer include indigestion, anorexia, weight loss, and abdominal pain [1]. Gastric cancer with the persistence of symptoms before diagnosis can be incurable or in an advanced stage [1]. The diagnosis of gastric cancer is determined using the tumor, node, and metastasis (TNM) classification system, according to the American Joint Committee on Cancer [3]. Clinical staging is determined according to physical examination, biopsy, radiological imaging, and the results of an endoscopy [4,5,6]. In patients who undergo surgical resection, pathological staging can be performed, according to the analysis of surgical specimens [4,5,6]. The clinical and pathological staging of gastric cancer is determined by various combinations of the T, N, and M categories [7,8]. Although only 20% of patients survive more than 5 years after diagnosis, there is no universal standard first-line therapy [2,9]. For early gastric cancer with moderate differentiation or without invasion, upfront surgery is the preferred means of management. Perioperative or postoperative chemotherapy with chemoradiation is recommended in the current guidelines [10]. For the most advanced gastric cancer, a fluoropyrimidine chemotherapy and platinum-based doublet are typically the backbone regimen [9,10].
Long noncoding RNAs (lncRNAs) are longer than 200 nucleotides and transcribed from noncoding regions [11]. Although previous studies have focused on the role of mRNAs in gastric cancer, recent studies have highlighted the involvement of lncRNAs [12,13]. Due to this nature, lncRNAs have their own structural characteristics that play various roles in epigenetic regulation, transcriptional regulation, and post-transcriptional regulation [14]. LncRNAs regulate histone modifications at the chromatin level through histone methylation and acetylation, DNA methylation, and transcriptional activation, and function together with various transcription factors and post-transcriptional modifications [12]. The function of lncRNAs as a decoy molecule blocks the function of chromosome-folding proteins or transcription regulators [14]. In addition, lncRNAs associate with the regulation of gene expression through sponging miRNAs [14]. As guide molecules, lncRNAs lead specific molecules to target locations [14]. If the target molecules are transcription factors, lncRNAs have a role in cisregulation [14,15]. Alternatively, other lncRNAs play a role in transregulation with histone methylation [14,16]. LncRNAs also act as a scaffold that can recruit numerous molecules [14]. For example, a well-known scaffold lncRNA is an X-inactive specific transcript (Xist) that suppresses the expression of the X chromosome in females, and as a result promotes the assembly of polycomb complexes 1 and 2 [14,16]. From the perspective of mRNA processing, lncRNAs modulate splicing factors through regulating the phosphorylation of splicing factors, and hijacking them [16,17]. Based on the molecular function of lncRNAs, lncRNAs play different roles in development and disease, and are involved in the progression or regression of cancer, suggesting their potential as novel therapeutic targets [18,19]. For example, the expression of HOX transcript antisense RNA, which recruits chromatin modifiers, is associated with metastasis and a poor prognosis in lung cancer [20]. Metastasis-associated lung adenocarcinoma transcript 1 is a novel therapeutic target for decreasing resistance in prostate cancer [21].
In this study, we show that the lncRNA LOC441461 is downregulated in clinical stage IV gastric cancer. The function of LOC441461 has not been studied in detail, except in colorectal cancer, in which LOC441461 expression is associated with cell growth and motility [22]. The role of LOC441461 in modulating various cellular functions was investigated in human gastric cancer cell lines. The downregulation of LOC441461 expression increased the aggressiveness of gastric cancer cells, suggesting that decreased LOC441461 is associated with a poor prognosis.
2. Materials and Methods
2.1. Expression Data from TCGA
Clinical data, survival data, and RNA-seq data of STAD were downloaded from UCSC XENA (https://xena.ucsc.edu/ accessed on 10 December 2021). A total of 379 gastric cancer cases were analyzed for clinical TNM staging and RNA-seq data. The unit of RNA-seq data was log2(RPKM + 1).
2.2. Information Gain (IG)
Clinical TNM staging was performed for each individual. IG was assessed using the clinical staging of each individual, in addition to gene-level expression data using FSelector package v. 0.31 (a package in R, a language and environment for statistical computing, R Core Team, R Foundation for Statistical Computing, Vienna, Austria) [23,24]. Genes with an IG > 0 were used as the feature selection threshold for the clinical stage.
2.3. Survival Analysis
A survival analysis was conducted to investigate the difference in prognoses between groups, which were composed of a combination of the T and M categories. A logrank test was conducted with Survival v. 3.2 (https://cran.r-project.org/web/packages/survival/index.html accessed on 8 January 2022). The Kaplan–Meier plot was visualized with Survminer v 0.4.8 (https://cran.r-project.org/web/packages/survminer/index.html accessed on 8 January 2022).
2.4. Gastric Cancer Cell Lines and Culture
All human gastric cancer cell lines (AGS, SNU16, SNU216, SNU638, MKN45, and MKN74) were purchased from the Korean Cell Line Bank (KCLB; Seoul, Republic of Korea) and cultured in RPMI-1640 (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin (Gibco). Cells were grown at 37 °C in a 5% CO2 incubator, under humidified conditions.
2.5. siRNA Transfection
The LOC441461 siRNA oligonucleotide (si-LOC441461; BIONEER, Daejeon, Republic of Korea) and negative control scrambled siRNA (N.C, BIONEER) were transfected into MKN74 and SNU216 cells using Lipofectamine™ RNAiMAX (Invitrogen, Carlsbad, CA, USA) and Opti-MEM (Gibco) at a final concentration of 100 nM. After 24 or 48 h, cells were harvested for subsequent experiments. The oligonucleotide sequences of the siRNAs are listed in Table S1.
2.6. Subcellular Fractionation and RNA Isolation
The cytoplasmic or nuclear location of LOC441461 was assessed in MKN74 and SNU216 cells using a PARIS kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol.
2.7. qRT-PCR
Total RNA was extracted from cells using an RNeasy mini kit (Qiagen, Hilden, Germany). The purity and concentration of total RNA were measured using a NanoDrop 1000 spectrophotometer (ND-1000, Thermo Fisher Scientific). One microgram of RNA was used for cDNA synthesis with an AccuPower CycleScript RT Premix (BIONEER) according to the manufacturer’s instructions. qPCR was performed on a CFX96 Touch real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA, USA) using an iQ™ SYBR Green Supermix (Bio-Rad Laboratories) and the indicated primer sets (Table S2). The following PCR reactions were performed: initial denaturation (95 °C for 10 min) followed by 45 cycles of denaturation (95 °C for 10 s) and annealing (55 °C for 30 s). Data were analyzed using the 2−ΔΔCt method and normalized to GAPDH or U6 as an endogenous internal control.
2.8. Proliferation Assay
Living cells (5000 living cells/well) transfected with N.C or si-LOC441461 were seeded in 96-well culture plates. After 6 h of transfection (or incubation) at 37 °C in 5% CO2 conditions, the medium was replaced with a fresh culture medium. Cell Counting Kit 8 solution (CCK8; Abcam, Cambridge, UK) was added directly to the cells. Cell growth was determined by measuring the absorbance at 450 nm every 24 h.
2.9. Colony Formation Assay
N.C- or si-LOC441461-transfected MKN74 cells were seeded into 6-well plates at a density of 2500 cells per well. After 2 weeks of incubation at 37 °C in 5% CO2, cells were fixed with 3.7% formaldehyde for 10 min and stained with 0.01% crystal violet solution in 10% ethanol for 20 min. Colony formation was quantified using 33% acetic acid and measured on a VICROR3 Multilabel Plate Reader (PerkinElmer, Waltham, MA, USA) at a wavelength of 590 nm.
2.10. Cell Cycle Assay
A total of 1 × 106 cells were harvested and fixed with 70% ethanol added dropwise to the pellet. After 12 h of incubation at −20 °C, cells were stained with PI/RNase Staining Buffer (BD Biosciences, San Jose, CA, USA) for 15 min at RT and detected using flow cytometry (Beckman Coulter, Brea, CA, USA). Cell cycle progression was analyzed using FlowJo v. 10.8.1 (FlowJo LLC, Ashland, OR, USA) with the Watson (Pragmatic) model.
2.11. Apoptosis Assay
N.C- or si-LOC441461-transfected MKN74 and SNU216 cells were seeded into 6-well plates at a density of 3 × 105 cells per well and incubated in the presence of 5-FU (2.5 μg/mL). After 24 h, apoptotic cells were detected with an Annexin V Apoptosis Detection Kit (BD Biosciences), following the manufacturer’s instructions using flow cytometry (Beckman Coulter).
2.12. Migration and Invasion Assays
Migration and invasion assays were performed using Transwell plates (Corning, NY, USA). N.C- or si-LOC441461-transfected MKN74 and SNU216 cells (7.5 × 104) were seeded into the upper chamber of Transwell plates in a serum-free medium. For invasion assays the upper chamber was precoated with Matrigel Matrix (Corning). After incubation under culture conditions for 24 h, the lower chamber of the Transwell plates was fixed with 10% formaldehyde solution, and cells on the upper chamber were removed with cotton swabs. A 0.01% crystal violet solution was used to stain cells, to detect migration and invasion. After air-drying, the stained solution was eluted using 33% acetic acid, and the absorbance of the eluted solution was measured using a VICROR3 Multilabel Plate Reader (PerkinElmer) at a wavelength of 590 nm.
2.13. Wound Healing Assay
For the wound healing assay, a 24-well culture insert dish (ibid GmbH, Münster, Germany) was used to determine the migration ability of MKN74 cells. N.C- and si-LOC441461-transfected MKN74 cells were seeded onto the culture inserts and incubated for 12 h after a confluent monolayer was formed. The inserts were gently removed, and a complete growth medium was added. Wound closure at 24 and 48 h was photographed under a microscope. The wound area was measured and quantified using ImageJ v. 1.53k (National Institutes of Health, Bethesda, MD, USA).
2.14. RNA-Seq Analysis
The total RNA of N.C- or si-LOC441461-transfected MKN74 cells was extracted with Trizol (Invitrogen), and the concentration was calculated using Quant-IT RiboGreen (Invitrogen). The integrity of the total RNA was measured on TapeStation RNA ScreenTape (Agilent Technologies, Santa Clara, CA, USA), and an RNA integrity number >7.0 was used for RNA library construction.
Total RNA (0.5 µg) was used to construct a library using an Illumina TruSeq Stranded Total RNA Library Prep Gold Kit (Illumina, Inc., San Diego, CA, USA). After removing the rRNA, fragmentation was performed with mRNA under elevated temperatures. First-strand cDNA was synthesized using SuperScript II Reverse Transcriptase (Invitrogen) and random primers. Second-strand cDNA synthesis was performed using DNA polymerase I, RNase H, and dUTP. These cDNA fragments went through an end repair process mediated by the addition of a single ‘A’ base, as well as the ligation of the adapters. The products were purified and enriched by PCR to create the final cDNA library.
The libraries were quantified using KAPA Library Quantification Kits for Illumina sequencing platforms according to the qPCR Quantification Protocol Guide (KAPA BIOSYSTEMS, #KK4854) and qualified using the TapeStation D1000 ScreenTape (Agilent Technologies, #5067-5582). Indexed libraries were then submitted to Illumina NovaSeq (Illumina), and paired-end (2 × 100 bp) sequencing was performed by Macrogen Incorporated (Seoul, Republic of Korea). Data have been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/ accessed on 8 January 2022), under the accession number GSE193700.
2.15. Differentially Expressed Gene Analysis
Differentially expressed genes between N.C and si-LOC441461 groups were analyzed using the DESeq2 package, which is included in R [25]. Genes with an absolute value of log2 fold change > 1 and a Benjamini–Hochberg-adjusted p-value < 0.05 were included.
2.16. Pathway Enrichment Analysis
Reactome pathway (https://reactome.org/ accessed on 18 November 2021) enrichment analysis was performed using ShinyGO software, and significant pathways with an FDR correction < 0.05 were extracted in ShinyGO v. 0.741 [26]. The p-values of the enriched pathways are presented as −log10 (p-value).
2.17. Transcription Factor Enrichment Analysis Using the LncRNA-TF Interactome
Molecules interacting with LOC441461 were predicted using RNAinter v. 4.0 [27]. Transcription factor enrichment analysis was performed to identify factors affected by LOC441461 using ChEA (https://www.encodeproject.org/chip-seq/ accessed on 18 November 2021) and ENCODE ChIP-seq databases (https://maayanlab.cloud/chea3/ accessed on 18 November 2021), and the differentially expressed gene set between N.C and LOC441461 knockdown samples using ShinyGO [26]. The common transcription factors were visualized using Venny v. 2.1.0 (https://bioinfogp.cnb.csic.es/tools/venny/ accessed on 20 November 2021).
2.18. Transcription Factor Target Gene Extraction
The target genes of each transcription factor were extracted according to cotarget genes from ENCODE ChIP-seq data, based on the target gene database [28] and the ChEA ChIP-X target gene database [29].
2.19. Hierarchical Clustering
Hierarchical clustering was performed to examine the differences in the expression of target genes between the si-LOC441461 and N.C groups. The expression level of genes was normalized to transcripts per million. ComplexHeatmap v. 2.8.0, a package of R, was used to visualize the heatmap with annotation group information [30].
2.20. Statistical Analysis
Significant differences between treatment means were determined using Student’s t-test. All qRT-PCR, cell proliferation, colony formation, cell cycle, apoptosis, migration, invasion, wound healing, and RNA-seq assays were performed in triplicate. Error bars indicate standard deviation. One asterisk (*) denotes p < 0.05; two asterisks (**) denote p < 0.01; three asterisks (***) denote p < 0.001; and four asterisks (****) denote p < 0.0001.
3. Results
3.1. Comparison of Gene Expression between Stage I and Stage IV Gastric Cancer
The result of IG feature selection between TNM stage I and IV gastric cancer patients using RNA-seq data identified 59 genes with an IG value > 0. The 59 genes were subjected to a t-test comparing TNM stage I and stage IV, and seven statistically significant genes were identified after Bonferroni correction. The gene names, including IG scores and p-values determined via a t-test, are presented in Table 1. The IG value results are presented in Table S3. The p-values of genes with an IG >0 determined by t-test are presented in Table S4.

Table 1.
Selected genes with IG and p-values according to t-tests based on XENA TCGA STAD.
3.2. Comparison of LOC441461 Expression and Prognosis between Different Gastric Cancer Stages
The expression of LOC441461 was assessed in samples from different TNM stages and compared between tumor and normal tissues. LOC441461 expression was lower in stage IV than in stages I, II, and III, whereas its expression was similar between stages I, II, and III (Figure 1A). LOC441461 expression did not differ between gastric cancer and adjacent normal tissues (Figure 1B). As LOC441461 expression differed between stage IV and stage I, II, and III gastric cancer, a logistic regression analysis was performed according to the T, N, and M categories. T4 tumors showed lower LOC441461 expression than T1, T2, and T3 tumors, whereas there was no difference between the different N categories. In the M category, LOC441461 expression was lower in M1 than in M0 tumors (Table 2). Based on the difference in expression of LOC441461 in the T and M categories, a survival analysis was conducted with overall survival (OS) and progression-free interval (PFI). The combination of the T category and the M category was divided into six groups. The difference in the prognosis in these six groups was significant in both OS and PFI (Figure 1C,D). The prognoses of T4 with M0 and M1 with T4 were significantly worse than T1 with M0 and M0 with T4, respectively, in OS (Table S5). Otherwise, only T4 with M0 showed a worse prognosis than T1 with M0 in PFI (Table S5). The entire results of the logrank test are described in Table S5.
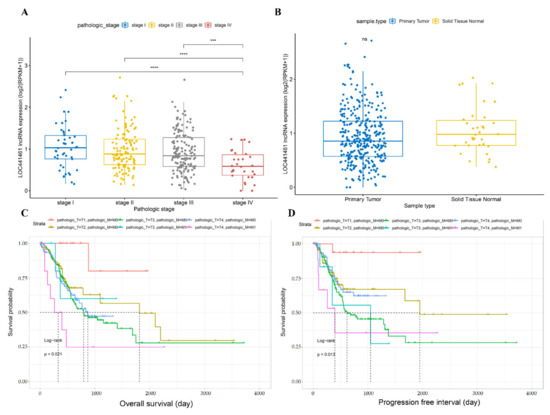
Figure 1.
Comparison of LOC441461 expression and survival analysis using TCGA STAD data. (A) The expression of LOC441461 in each TNM stage and statistical analysis. The statistical analysis was conducted via a t-test. **** p < 0.0001, *** p < 0.001. (B) LOC441461 expression in primary tumors and adjacent normal tissues. ns, not significant. (C,D) Comparison of prognoses between different combinations of T and M. The statistical method was a logrank test. Pathologic_T is the T category of TNM staging. Pathologic_M is the M category of TNM staging.

Table 2.
Results of the logistic regression analysis of TNM scores based on XENA TCGA STAD data.
3.3. Selection of Gastric Cancer Cell Lines and si-LOC441461 Treatment
The effect of LOC441461 on gastric cancer cells was investigated based on the TCGA STAD data. LOC441461 expression was first measured in different gastric cancer cell lines, to select the most adequate cells. The MKN74 and SNU216 cell lines showed high expression of LOC441461 and were, thus, selected for further experiments (Figure 2A). MKN74 and SNU216 cells were transfected with si-LOC441461, and depletion of LOC441461 was detected (Figure 2B). LOC441461 expression was higher in the nucleus than in the cytoplasm in both the SNU216 and MKN74 cell lines (Figure 2C).
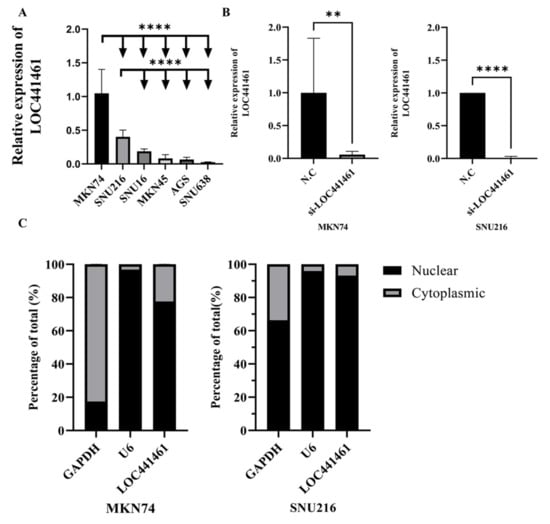
Figure 2.
Relative expression of LOC441461 and localization in human gastric cancer cell lines. (A) Expression levels of LOC441461 were measured in six gastric cancer cell lines (MKN74, SNU216, SNU16, MKN45, AGS, and SNU638) by qRT-PCR. (B) Knockdown of LOC441461 in MKN74 and SNU216 cells via transfection with small interfering RNA (siRNA) targeting LOC441461 (si-LOC441461) compared with a scrambled negative control (N.C). qRT-PCR was performed to quantify the relative expression levels of LOC441461 normalized to GAPDH; SNU638 cells were compared as a reference sample. (C) Subcellular fractionation was performed using a PARIS kit to separate the nuclear and cytoplasmic fractions of MKN74 and SNU216 cells. The localization of LOC441461 in the nucleus (normalized to U6) and cytoplasm (normalized to GAPDH) was compared via qRT-PCR using the 2−ΔΔCt method. All experiments were performed in triplicate, and data are expressed as the mean ± standard deviation (** p < 0.01, **** p < 0.0001; Student’s t-test).
3.4. Downregulation of LOC441461 Induces Proliferation and Accelerates Cell Cycle Progression
Lower LOC441461 expression was correlated with increased tumor volume, suggesting that negative LOC441461 expression affects the proliferation ability of MKN74 and SNU216 cells. The results of the CCK8 assay showed that LOC441461 significantly inhibited the proliferation of MKN74 and SNU216 cells, suggesting that LOC441461 promotes gastric cancer cell proliferation in vitro (Figure 3A and Figure S1). LOC441461 knockdown induced cell cycle progression; specifically, the G1/S transition was accelerated (Figure 3B). However, the colony formation ability, which indicates the ability of a single cell to grow into a colony, was significantly decreased in response to si-LOC441461 in both MKN74 and SNU216 cells (Figure 3C).
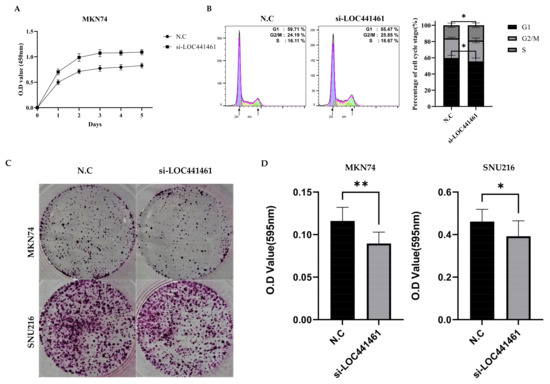
Figure 3.
LOC441461 suppresses cell proliferation by promoting G1/S transition in human gastric cancer cells. (A) Knockdown of LOC441461 by transfection with si-LOC441461 or a scrambled negative control (N.C) in MKN74 cells. The proliferation ability of N.C- and si-LOC441461-transfected cells was examined using the CCK-8 solution at every 24 h. (B) The cell cycle progression of N.C- and si-LOC441461-transfected cells was examined using flow cytometry with the PI/RNase solution. Each cell cycle phase was analyzed with FlowJo v. 10.8.1. Graph of each quantified phase. (C) MKN74 and SNU216 cells were transfected with N.C or si-LOC441461 and stained with 0.01% crystal violet solution after 14 days. (D) Graph of colony formation ability after elution with 33% acetic acid. All experiments were performed in triplicate, and data are expressed as the mean ± standard deviation (* p < 0.05, ** p < 0.01; Student’s t-test).
3.5. LOC441461 Knockdown Promotes 5-FU-Induced Gastric Cancer Cell Apoptosis
The present results indicated that LOC441461 knockdown induced gastric cancer cell apoptosis in vitro. As shown in Figure 4, LOC441461 knockdown promoted apoptosis in MKN74 and SNU216 cells treated with 5-FU (Figure 4A,B). We, thus, examined the effect of LOC441461 on apoptosis and drug resistance in MKN74 and SNU216 cells treated with 5-FU. The results showed that the expression of LOC441461 correlated negatively with the drug sensitivity of gastric cancer (Figure 4A); whereas, DMSO treatment decreased the rate of apoptosis of LOC441461 knockdown cells (Figure 4A,B).
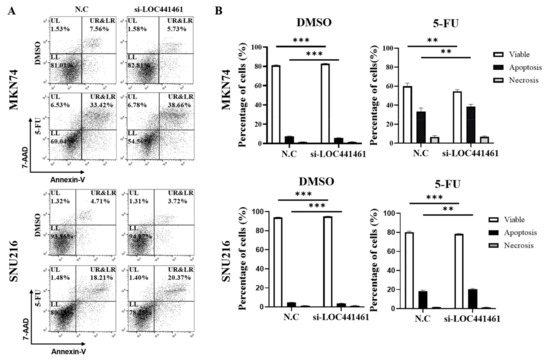
Figure 4.
Sensitivity to 5-FU was increased in gastric cancer cells with LOC441461 knockdown. MKN74 and SNU216 cells were transfected with a scrambled negative control (N.C) or si-LOC441461. After exposure to 5-FU (2 μg/mL), apoptotic cells were measured using an annexin V/7-AAD kit. (A) Representative dot plot of the apoptosis assay using flow cytometry. The upper-left (UL) quadrant indicates necrotic cells, the lower-left (LL) quadrant shows viable cells, the lower-right (LR) quadrant indicates early apoptotic cells, and the upper-right (UR) quadrant shows the late stage of apoptosis. (B) Graph showing the percentage of the cells in each stage. Experiments were performed in triplicate, and data are expressed as the mean ± standard deviation (** p < 0.01; *** p < 0.001; Student’s t-test).
3.6. LOC441461 Knockdown Increases Gastric Cancer Cell Motility and Invasiveness
Analysis of the migration and invasion of gastric cancer cells using the Transwell assay showed that SNU216 cells had a higher rate of migration and invasiveness than MKN74 cells (Figure S2). The knockdown of LOC441461 increased the migration and invasion capability of SNU216 cells (Figure 5A). Additionally, the migration and invasion abilities of SNU216 cells were also significantly increased in the absence of LOC441461 (Figure 5B). The results of the wound healing assay showed that wound closure was significantly higher in LOC441461-silenced MKN74 cells than in the negative control group (Figure 5C). The decrease in the wound area was statistically significant after 48 h, but not at 24 h.
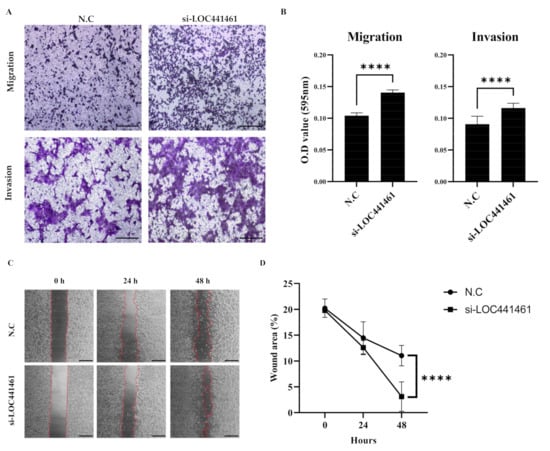
Figure 5.
LOC441461 knockdown promotes gastric cancer cell migration and invasion. (A) Representative microscopic images of the Transwell migration and invasion assays in SNU216 cells (stained with crystal violet). Scale bar = 200 μm. (B) Relative migration and invasion abilities were quantified after elution with 33% acetic acid. (C) Representative images of wound healing assays using culture preinserts in MKN74 cells with LOC441461 knockdown. Scale bar = 500 μm. (D) Wound closure was assessed by ImageJ v. 1.53k from 0 to 48 h. All experiments were performed in triplicate, and data were expressed as the mean ± standard deviation (**** p < 0.0001; Student’s t-test).
3.7. Effect of LOC441461 Knockdown on the Transcriptomic Landscape of Gastric Cancer Cells
The transcriptomic landscape of LOC441461 knockdown gastric cancer cells was analyzed by RNA sequencing. Three replicates were, respectively, included in two groups, including negative controls and the knockdown of LOC441461. Differentially expressed gene analysis was performed using expression data obtained by RNA-seq. A total of 2503 genes with an absolute log2 fold change >1 and an adjusted p < 0.05 were selected for pathway enrichment analysis. Of these, 1202 upregulated genes and 1301 downregulated genes in the knockdown group were selected. The genes are listed in Table S6. The selected genes were subjected to pathway enrichment analysis using the Reactome pathway database. Apoptosis- and cell-cycle-progression-associated terms were significantly enriched in genes that were upregulated in the LOC441461 knockdown group (Figure 6A). The detailed results are presented in Table S7. The mRNA expression of cyclin D1, which is involved in the G1/S transition of the cell cycle, and TRAIL, which is associated with apoptosis and the epithelial-to-mesenchymal transition (EMT), were higher in the LOC441461 knockdown group (Figure 6B).
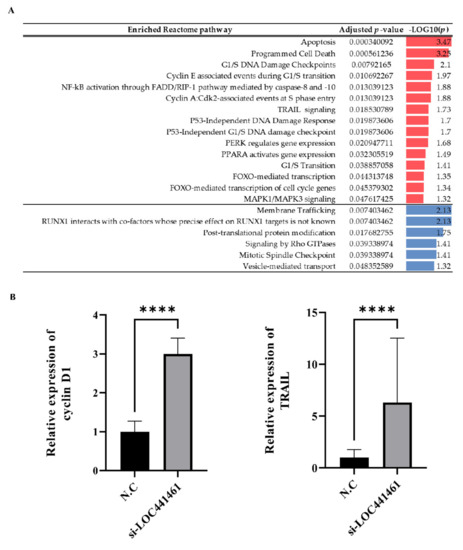
Figure 6.
Transcriptomic landscape after the depletion of LOC441461 in gastric cancer cell lines. (A) Results of pathway enrichment analysis. Red indicates upregulated pathways in LOC441461 knockdown groups. Blue indicates downregulated pathways in LOC441461 knockdown groups. (B) mRNA expression of cyclin D1 and TRAIL determined by qRT-PCR in MKN74 cells transfected with N.C or si-LOC441461. The data were normalized to GAPDH. All experiments were performed in triplicate, and data are expressed as the mean ± standard deviation (**** p < 0.0001; Student’s t-test).
3.8. LOC441461 Changes the Nature of Gastric Cancer Cells by Modulating Transcription Factor Activity
Analysis of the RNAinter database showed that most of the molecules interacting with LOC441461 were transcription factors (Table S8). To identify the transcription factors modulated by LOC441461, a transcription factor enrichment analysis was performed according to the ENCODE and ChEA consensus transcription factors obtained from the ChIP-seq database, as well as the differentially expressed genes from RNA-seq data (Table S9). RELA and IRF1 interacted with LOC441461 and were significantly enriched among the genes upregulated by LOC441461 knockdown, whereas ESR1, AR, POU5F1, YY1, FOXM1, TRIM28, SMAD4, E2F4, and GATA1 interacted with LOC441461 and were significantly enriched in the downregulated gene set (Figure 7A). Hierarchical clustering was performed based on the target gene expression of significantly enriched transcription factors from RNA-seq data. Transcription factors significantly affected by LOC441461 were identified by hierarchical clustering, which showed that the expression of the RELA, IRF1, ESR1, AR, POU5F1, TRIM28, and GATA1 target genes differed significantly between the knockdown and negative control groups (Figure 7B). The detailed results of hierarchical clustering and the expression of each target gene are presented in Figures S3 and S4. Target genes were extracted by the cotarget genes in the ENCODE ChIP-seq database and ChEA ChIP-X database, except AR and SMAD4, which were extracted from the ChEA ChIP-X database. The target genes of each transcription factor are listed in Table S10.
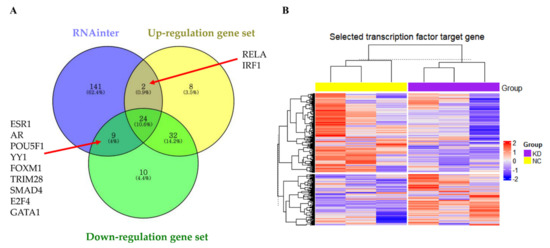
Figure 7.
Investigation of the role of LOC441461 in modulating transcription factor activity. (A) Enriched transcription factors in the up- or downregulated gene sets in interacting with LOC441461. (B) Hierarchical clustering based on the target gene expression of 11 transcription factors with RNA-seq data of LOC441461 knockdown. KD, knockdown of LOC441461. N.C, negative control.
4. Discussion
Recent reports on the different functions of lncRNAs have highlighted their involvement in tumor development and progression [18,31]. Moreover, the investigation of the molecular and cellular functions of lncRNAs may lead to the identification of therapeutic targets and the design of strategies for the treatment of gastric cancer [18,31,32,33]. By examining the association between lncRNA LOC441461 expression level and TNM stage, we identified biological processes in gastric cancer. In this study, LOC441461 expression was lower in stage IV than in stage I, II, and III gastric cancer samples from TCGA STAD data. The association between the degree of the malignancy of gastric cancer and higher stages, as assessed by the TNM staging system, supports the effect of LOC441461 downregulation on increasing the severity of the disease by promoting tumor growth, cell motility, and worse prognoses. Furthermore, the localization and local interactions of lncRNAs are key to predicting their function [34]. Overall, we suggest that LOC441461 modulates gene transcription, inducing the malignancy of gastric cancer by interacting with RELA, IRF1, ESR1, AR, POU5F1, TRIM28, and GATA1.
LncRNAs are involved in diverse biological processes, such as proliferation, motility, and the EMT in different aspects of regulation [11,35]. The latest research shows that LOC441461 upregulation promotes growth and motility in colon cancer cell lines [22]. In the present study we found that the different effects of LOC441461 may be attributed to the use of gastric cancer, and several experimental validations were conducted to investigate the function of LOC441461 in gastric cancer cells. The results of cell experiments and RNA-seq analysis showed that the downregulation of the lncRNA LOC441461 could promote the growth and metastasis of gastric cancer cells in vitro and in silico. Furthermore, lncRNAs modulate the cell cycle by regulating the expression of cyclins and cyclin-dependent kinases [36,37], and a similar mechanism may underlie the regulation of the cell cycle by LOC441461. The FOXO-mediated transcription of cell cycle genes was significantly increased in LOC441461 knockdown cells. However, the clonogenic ability showed a positive correlation with the expression of LOC441461.
5-FU treatment, a fluoropyrimidine chemotherapy, is the first treatment of choice of gastric cancer [9]. Previous studies suggest that 5-FU sensitivity increases the status of DNA damage [38,39]. Ultimately, the p53-independent DNA damage response was significantly enriched in LOC441461 knockdown cells, suggesting that the DNA damage response was promoted by the effect of LOC441461 knockdown, in inducing the 5-FU-induced apoptosis of gastric cancer cells.
In addition, it was found that lncRNAs regulate gastric cancer cells via diverse contexts, such as regulating the EMT process and PI3K/AKT pathway [22,31,40]. The EMT is considered as a key role in human malignancies activated during tumor metastasis [41]. Given the abovementioned findings, the results suggested that the knockdown of LOC441461 increased the motility of gastric cancer cells. The downregulation of LOC441461 increased the proportion of migrating/invading cells and promoted wound closure. The upregulation of tumor-necrosis-factor-related apoptosis-inducing ligand (TRAIL) signaling also promotes the EMT in various cancers [42,43,44,45]. By contrast, the interaction of RUNX1 with cofactors was significantly decreased in the absence of LOC441461, and RUNX1 is a suppressor of the EMT [46,47]. Thus, the EMT was promoted in relation to the depletion of LOC441461, as indicated by TRAIL signaling and RUNX1 interactions.
Multiple studies have observed that lncRNAs are, overall, more numerous in the nucleus than their cytoplasm [16,31,34,35,48]. Interestingly, the instability of nuclear lncRNAs can act as an oncogene or tumor suppressor and modulate transcription factors, which are key players in transcriptional regulation [16,31,48]. We investigated the potential mechanism underlying the modulation of gene expression by transcription factors that interact with LOC441461 in gastric cancer cells. Transcription factor enrichment analysis was performed to identify transcription factors that interact with LOC441461. Hierarchical clustering based on the expression of the target genes of 11 transcription factors affected by LOC441461 resulted in the classification of the target genes of seven transcription factors, RELA, IRF1, ESR1, AR, POU5F1, TRIM28, and GATA1, into two groups (Figures S3 and S4). Hierarchical clustering of the target genes of seven transcription factors also clearly divided them into two groups, in correlation with the expression of LOC441461. RELA, also called p65, is associated with NF-κB heterodimer formation, nuclear translocation, and activation [49]. The dysregulation of NF-κB/RELA promotes distant metastasis in gastric cancer, suggesting its potential as a novel therapeutic target [50]. IRF1 not only acts as a viral infection response transcription factor [51], but also plays a role in cell cycle arrest, enhancing 5-FU sensitivity and suppressing the EMT in gastric cancer [52,53,54]. In the absence of LOC441461, IRF1 expression was upregulated (Figure S5). ESR1 and androgen receptor (AR) upregulation is associated with a poor prognosis in gastric cancer patients [55]. POU5F1, a POU class 5 homeobox 1 member, is associated with a poor prognosis, and its expression is significantly increased after chemotherapy in colorectal cancer [56,57]. Tripartite motif-containing 28 (TRIM28) is associated with a poor prognosis by promoting tumor progression and the activation of autophagy in glioma [58,59]. GATA1 or GATA-binding factor 1 promote the EMT in breast cancer [60].
According to the results of an expression analysis between TNM stages and a survival analysis, the expression of LOC441461 was associated with malignant characteristics of gastric cancer cells, including cell growth and motility. Specifically, the molecular function of LOC441461 was bioinformatically predicted by modulating the activity of seven transcription factors, including RELA, IRF1, ESR1, AR, POU5F1, TRIM28, and GATA1. Based on these facts, they can be applied to the novel treatment management of gastric cancer in various ways. First, upregulating the expression of LOC441461 in gastric cancer patients can suppress progression to a malignant status of gastric cancer. Second, modulating the activity of transcription factors interacting with LOC441461 could be a candidate therapeutic target to manage advanced gastric cancer. Recently, RNA biopharmaceuticals have been attracting attention as a breakthrough clinical application of the COVID-19 RNA vaccine [33,61]. Proving its application as a gastric cancer treatment by regulating the expression of LOC441461 will be a very interesting study in the future.
5. Conclusions
In this study, we identified the lncRNA LOC441461 as a potential biomarker in patients with an advanced TNM stage, which has significant value in tumor development and progression. LOC441461 knockdown promoted growth, cell cycle progression, motility, and invasiveness in gastric cancer cells in vitro. Additionally, molecular links between the expression of LOC441461 and the nature of gastric cancer cells were investigated by measuring changes in gene expression modulated by transcription factors. Candidate transcription factors that may interact with LOC441461 were identified using bioinformatic methods and various databases. Although this study has the limitation that the exact mechanism of action with which LOC441461 interacted with seven transcription factors was only predicted by bioinformatical methods, and not validated by experimental methods, the findings in this study may lead to the development of novel therapeutic strategies for patients with gastric cancer.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers14051149/s1, Figure S1. Knockdown of LOC441461 by transfection with si-LOC441461 or a scrambled negative control (N.C) in SNU216 cells. The proliferation ability of N.C- and si-LOC441461-transfected cells was examined using the CCK-8 solution at every 24 h. All experiments were performed in triplicate, and data are expressed as the mean ± standard deviation. Figure S2. MKN74 or SNU216 gastric cancer cells were seeded in Transwell plates to evaluate migration and invasion. (A) The results of migration and invasion assays with MKN74 and SNU216 cells stained with 0.01% crystal violet solution. Scale bar = 200 μm. (B) Cell movement was quantified by elution with 33% acetic acid and measuring the absorbance at a wavelength of 590 nm using a plate reader. All experiments were performed in triplicate, and data were expressed as the mean ± standard deviation. Figure S3. Heatmap of the expression of the target genes of transcription factors dividing genes into LOC441461 knockdown and negative control groups. Figure S4. Heatmap of the expression of the target genes of transcription factors that did not divide genes into LOC441461 knockdown and negative control groups. Figure S5. IRF1 mRNA expression was upregulated in the LOC441461 knockdown group. qRT-PCR analysis of MKN74 cells transfected with a scrambled negative control siRNA (N.C) or si-LOC441461. The data were normalized to GAPDH. All experiments were performed in triplicate and expressed as the mean ± standard deviation (*** p < 0.001; Student’s t-test). Table S1. siRNA sequences and target RNA. Table S2. Primer set used in qRT-PCR targeting LOC441461, including TRAIL, cyclin D1, IRF1, U6, and GAPDH. Table S3. Results of IG analysis using TCGA STAD RNA-seq data and TNM stage information from clinical data obtained between stage I and IV. Table S4. Statistical analysis (t-tests) of the expression of genes with an IG >0 at TNM stages I and IV. Table S5. The results of a logrank test. Table S6. Differentially expressed genes between the LOC441461 knockdown and N.C groups. KD = knockdown of LOC441461. N.C = negative controls. Table S7. Results of pathway enrichment analysis using the Reactome database and differentially expressed genes. Blue indicates a downregulated pathway in the LOC441461 knockdown group. Red indicates an upregulated pathway in the LOC441461 knockdown group. Table S8. Molecules that interact with LOC441461. Table S9. Significantly-enriched transcription factors. Table S10. Target genes of each transcription factor.
Author Contributions
Conceptualization, S.-s.L., J.P., S.O. and K.K.; Data Curation, S.-s.L., J.P. and K.K.; Formal Analysis, S.-s.L., J.P. and S.O.; Funding Acquisition, K.K.; Investigation, S.O. and K.K.; Methodology, S.-s.L. and J.P.; Project Administration, K.K.; Software, J.P.; Supervision, K.K.; Visualization, S.-s.L. and J.P.; Writing—Original Draft, S.-s.L. and J.P.; Writing—Review and Editing, S.O. and K.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Ministry of Science, Technology, and Information, Republic of Korea (grant no. 2019R1F1A105492013).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
RNA-seq data have been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/ accessed on 18 November 2021) under the accession number GSE193700 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE193700 accessed on 18 November 2021).
Acknowledgments
Parts of the results are based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga (accessed on 18 November 2021). This study was supported by a Basic Research Project grant from the Ministry of Science, Technology, and Information, Republic of Korea (grant no. 2019R1F1A105492013).
Conflicts of Interest
The authors declare that they have no competing interests.
References
- Smyth, E.C.; Nilsson, M.; Grabsch, H.I.; van Grieken, N.C.; Lordick, F. Gastric cancer. Lancet 2020, 396, 635–648. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Byrd, D.R.; Carducci, M.; Compton, C.; Fritz, A.; Greene, F. AJCC Cancer Staging Manual; Springer: New York, NY, USA, 2010. [Google Scholar]
- Yao, K.; Uedo, N.; Kamada, T.; Hirasawa, T.; Nagahama, T.; Yoshinaga, S.; Oka, M.; Inoue, K.; Mabe, K.; Yao, T.; et al. Guidelines for endoscopic diagnosis of early gastric cancer. Dig. Endosc. 2020, 32, 663–698. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Gong, Y.; Xu, H. Clinical and pathological staging of gastric cancer: Current perspectives and implications. Eur. J. Surg. Oncol. 2020, 46, e14–e19. [Google Scholar] [CrossRef] [PubMed]
- Roland, N.; Porter, G.; Fish, B.; Makura, Z. Tumour assessment and staging: United Kingdom national multidisciplinary guidelines. J. Laryngol. Otol. 2016, 130, S53–S58. [Google Scholar] [CrossRef] [PubMed]
- Sano, T.; Coit, D.G.; Kim, H.H.; Roviello, F.; Kassab, P.; Wittekind, C.; Yamamoto, Y.; Ohashi, Y. Proposal of a new stage grouping of gastric cancer for TNM classification: International Gastric Cancer Association staging project. Gastric Cancer 2017, 20, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Wittekind, C. The development of the TNM classification of gastric cancer. Pathol. Int. 2015, 65, 399–403. [Google Scholar] [CrossRef]
- Joshi, S.S.; Badgwell, B.D. Current treatment and recent progress in gastric cancer. CA Cancer J. Clin. 2021, 71, 264–279. [Google Scholar] [CrossRef]
- NCCN. Stomach Patient (NCCN Guidelines); NCCN: Bethesda, MA, USA, 2021. [Google Scholar]
- Yao, R.-W.; Wang, Y.; Chen, L.-L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef]
- Zhu, J.; Fu, H.; Wu, Y.; Zheng, X. Function of lncRNAs and approaches to lncRNA-protein interactions. Sci. China Life Sci. 2013, 56, 876–885. [Google Scholar] [CrossRef] [PubMed]
- Gao, N.; Li, Y.; Li, J.; Gao, Z.; Yang, Z.; Li, Y.; Liu, H.; Fan, T. Long non-coding RNAs: The regulatory mechanisms, research strategies, and future directions in cancers. Front. Oncol. 2020, 10, 2903. [Google Scholar] [CrossRef]
- Gil, N.; Ulitsky, I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat. Rev. Genet. 2020, 21, 102–117. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Romero-Barrios, N.; Legascue, M.F.; Benhamed, M.; Ariel, F.; Crespi, M. Splicing regulation by long noncoding RNAs. Nucleic Acids Res. 2018, 46, 2169–2184. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.-C.; Ni, J.-J.; Cui, W.-Y.; Wang, B.-Y.; Zhuo, W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am. J. Cancer Res. 2019, 9, 1354. [Google Scholar]
- Mathieu, E.-L.; Belhocine, M.; Dao, L.; Puthier, D.; Spicuglia, S. Functions of lncRNA in development and diseases. Med. Sci. 2014, 30, 790–796. [Google Scholar]
- Loewen, G.; Jayawickramarajah, J.; Zhuo, Y.; Shan, B. Functions of lncRNA HOTAIR in lung cancer. J. Hematol. Oncol. 2014, 7, 1–10. [Google Scholar] [CrossRef]
- Ren, S.; Liu, Y.; Xu, W.; Sun, Y.; Lu, J.; Wang, F.; Wei, M.; Shen, J.; Hou, J.; Gao, X. Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J. Urol. 2013, 190, 2278–2287. [Google Scholar] [CrossRef]
- Liu, Z.-Q.; He, W.-F.; Wu, Y.-J.; Zhao, S.-L.; Wang, L.; Ouyang, Y.-Y.; Tang, S.-Y. LncRNA SNHG1 promotes EMT process in gastric cancer cells through regulation of the miR-15b/DCLK1/Notch1 axis. BMC Gastroenterol. 2020, 20. [Google Scholar] [CrossRef]
- Cheng, T.; Wang, Y.; Bryant, S.H. FSelector: A Ruby gem for feature selection. Bioinformatics 2012, 28, 2851–2852. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Romanski, P.; Kotthoff, L.; Kotthoff, M.L. Package ‘FSelector’. 2013. Available online: http://cran.r-project.org/web/packages/FSelector/index.html (accessed on 18 November 2021).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Tang, Q.; He, J.; Li, L.; Yang, N.; Yu, S.; Wang, M.; Zhang, Y.; Lin, J.; Cui, T. RNAInter v4. 0: RNA interactome repository with redefined confidence scoring system and improved accessibility. Nucleic Acids Res. 2022, 50, D326–D332. [Google Scholar] [CrossRef]
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57. [Google Scholar] [CrossRef]
- Lachmann, A.; Xu, H.; Krishnan, J.; Berger, S.I.; Mazloom, A.R.; Ma’ayan, A. ChEA: Transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics 2010, 26, 2438–2444. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Cui, H.; Jiang, Z.; Zeng, S.; Wu, H.; Zhang, Z.; Guo, X.; Dong, K.; Wang, J.; Shang, L.; Li, L. A new candidate oncogenic lncRNA derived from pseudogene WFDC21P promotes tumor progression in gastric cancer. Cell Death Dis. 2021, 12, 903. [Google Scholar] [CrossRef]
- Zhao, Z.; Liu, H.; Zhou, X.; Fang, D.; Ou, X.; Ye, J.; Peng, J.; Xu, J. Necroptosis-Related lncRNAs: Predicting Prognosis and the Distinction between the Cold and Hot Tumors in Gastric Cancer. J. Oncol. 2021, 2021, 1–16. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef]
- Bridges, M.C.; Daulagala, A.C.; Kourtidis, A. LNCcation: lncRNA localization and function. J. Cell Biol. 2021, 220, e202009045. [Google Scholar] [CrossRef]
- Ma, X.; Wang, G.; Fan, H.; Li, Z.; Chen, W.; Xiao, J.; Ni, P.; Liu, K.; Shen, K.; Wang, Y.; et al. Long noncoding RNA FAM225A promotes the malignant progression of gastric cancer through the miR-326/PADI2 axis. Cell Death Discov. 2022, 8, 20. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Shoorei, H.; Anamag, F.T.; Taheri, M. The role of non-coding RNAs in controlling cell cycle related proteins in cancer cells. Front. Oncol. 2020, 10, 2616. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, M.; Kitagawa, K.; Kotake, Y.; Niida, H.; Ohhata, T. Cell cycle regulation by long non-coding RNAs. Cell. Mol. Life Sci. 2013, 70, 4785–4794. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.-Y.; Tang, J.-N.; Xie, H.-X.; Du, Y.-A.; Huang, L.; Yu, P.-F.; Cheng, X.-D. 5-Fluorouracil chemotherapy of gastric cancer generates residual cells with properties of cancer stem cells. Int. J. Biol. Sci. 2015, 11, 284. [Google Scholar] [CrossRef]
- Sethy, C.; Kundu, C.N. 5-Fluorouracil (5-FU) resistance and the new strategy to enhance the sensitivity against cancer: Implication of DNA repair inhibition. Biomed. Pharmacother. 2021, 137, 111285. [Google Scholar] [CrossRef]
- Ba, M.-C.; Ba, Z.; Long, H.; Cui, S.-Z.; Gong, Y.-F.; Yan, Z.-F.; Lin, K.-P.; Wu, Y.-B.; Tu, Y.-N. LncRNA AC093818.1 accelerates gastric cancer metastasis by epigenetically promoting PDK1 expression. Cell Death Dis. 2020, 11, 64. [Google Scholar] [CrossRef]
- Bakir, B.; Chiarella, A.M.; Pitarresi, J.R.; Rustgi, A.K. EMT, MET, Plasticity, and Tumor Metastasis. Trends Cell Biol. 2020, 30, 764–776. [Google Scholar] [CrossRef]
- Peyre, L.; Meyer, M.; Hofman, P.; Roux, J. TRAIL receptor-induced features of epithelial-to-mesenchymal transition increase tumour phenotypic heterogeneity: Potential cell survival mechanisms. Br. J. Cancer 2021, 124, 91–101. [Google Scholar] [CrossRef]
- Zhang, H.; Qin, G.; Zhang, C.; Yang, H.; Liu, J.; Hu, H.; Wu, P.; Liu, S.; Yang, L.; Chen, X. TRAIL promotes epithelial-to-mesenchymal transition by inducing PD-L1 expression in esophageal squamous cell carcinomas. J. Exp. Clin. Cancer Res. 2021, 40, 1–16. [Google Scholar] [CrossRef]
- Trivedi, R.; Mishra, D.P. Trailing TRAIL resistance: Novel targets for TRAIL sensitization in cancer cells. Front. Oncol. 2015, 5, 69. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xu, C.; Kong, X.; Li, X.; Kong, X.; Wang, Y.; Ding, X.; Yang, Q. Trail resistance induces epithelial-mesenchymal transition and enhances invasiveness by suppressing PTEN via miR-221 in breast cancer. PLoS ONE 2014, 9, e99067. [Google Scholar] [CrossRef]
- Hong, D.; Messier, T.L.; Tye, C.E.; Dobson, J.R.; Fritz, A.J.; Sikora, K.R.; Browne, G.; Stein, J.L.; Lian, J.B.; Stein, G.S. Runx1 stabilizes the mammary epithelial cell phenotype and prevents epithelial to mesenchymal transition. Oncotarget 2017, 8, 17610. [Google Scholar] [CrossRef] [PubMed]
- Hong, D. RUNX1 Control of Mammary Epithelial and Breast Cancer Cell Phenotypes; University of Massachusetts Medical School: Worcester, MA, USA, 2017. [Google Scholar]
- Marchese, F.P.; Raimondi, I.; Huarte, M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017, 18, 206. [Google Scholar] [CrossRef] [PubMed]
- Chanut, A.; Duguet, F.; Marfak, A.; David, A.; Petit, B.; Parrens, M.; Durand-Panteix, S.; Boulin-Deveza, M.; Gachard, N.; Youlyouz-Marfak, I. RelA and RelB cross-talk and function in Epstein–Barr virus transformed B cells. Leukemia 2014, 28, 871–879. [Google Scholar] [CrossRef]
- Ye, T.; Yang, M.; Huang, D.; Wang, X.; Xue, B.; Tian, N.; Xu, X.; Bao, L.; Hu, H.; Lv, T. MicroRNA-7 as a potential therapeutic target for aberrant NF-κB-driven distant metastasis of gastric cancer. J. Exp. Clin. Cancer Res. 2019, 38, 1–18. [Google Scholar] [CrossRef]
- Kimura, T.; Nakayama, K.; Penninger, J.; Kitagawa, M.; Harada, H.; Matsuyama, T.; Tanaka, N.; Kamijo, R.; Vilček, J.; Mak, T.W. Involvement of the IRF-1 transcription factor in antiviral responses to interferons. Science 1994, 264, 1921–1924. [Google Scholar] [CrossRef]
- Nozawa, H.; Oda, E.; Ueda, S.; Tamura, G.; Maesawa, C.; Muto, T.; Taniguchi, T.; Tanaka, N. Functionally inactivating point mutation in the tumor-suppressor IRF-1 gene identified in human gastric cancer. Int. J. Cancer 1998, 77, 522–527. [Google Scholar] [CrossRef]
- Gao, J.; Tian, Y.; Zhang, J. Overexpression of interferon regulatory factor 1 enhances chemosensitivity to 5-fluorouracil in gastric cancer cells. J. Cancer Res. Ther. 2012, 8, 57. [Google Scholar] [CrossRef]
- Jiang, L.; Liu, J.-y.; Shi, Y.; Tang, B.; He, T.; Liu, J.-j.; Fan, J.-y.; Wu, B.; Xu, X.-h.; Zhao, Y.-l. MTMR2 promotes invasion and metastasis of gastric cancer via inactivating IFNγ/STAT1 signaling. J. Exp. Clin. Cancer Res. 2019, 38, 1–16. [Google Scholar] [CrossRef]
- Chang, W.-C.; Huang, S.-F.; Lee, Y.-M.; Lai, H.-C.; Cheng, B.-H.; Cheng, W.-C.; Ho, J.Y.-P.; Jeng, L.-B.; Ma, W.-L. Cholesterol import and steroidogenesis are biosignatures for gastric cancer patient survival. Oncotarget 2017, 8, 692. [Google Scholar] [CrossRef] [PubMed]
- Saigusa, S.; Tanaka, K.; Toiyama, Y.; Yokoe, T.; Okugawa, Y.; Ioue, Y.; Miki, C.; Kusunoki, M. Correlation of CD133, OCT4, and SOX2 in rectal cancer and their association with distant recurrence after chemoradiotherapy. Ann. Surg. Oncol. 2009, 16, 3488–3498. [Google Scholar] [CrossRef] [PubMed]
- Bauer, L.; Langer, R.; Becker, K.; Hapfelmeier, A.; Ott, K.; Novotny, A.; Höfler, H.; Keller, G. Expression profiling of stem cell-related genes in neoadjuvant-treated gastric cancer: A NOTCH2, GSK3B and β-catenin gene signature predicts survival. PLoS ONE 2012, 7, e44566. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Zhang, M.; Jiang, Z.; Jiang, Y. TRIM28 activates autophagy and promotes cell proliferation in glioblastoma. OncoTargets Ther. 2019, 12, 397. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Li, H.; Gao, W. TRIM28 is overexpressed in glioma and associated with tumor progression. OncoTargets Ther. 2018, 11, 6447. [Google Scholar] [CrossRef]
- Li, Y.; Ke, Q.; Shao, Y.; Zhu, G.; Li, Y.; Geng, N.; Jin, F.; Li, F. GATA1 induces epithelial-mesenchymal transition in breast cancer cells through PAK5 oncogenic signaling. Oncotarget 2015, 6, 4345. [Google Scholar] [CrossRef]
- Ouranidis, A.; Vavilis, T.; Mandala, E.; Davidopoulou, C.; Stamoula, E.; Markopoulou, C.K.; Karagianni, A.; Kachrimanis, K. mRNA Therapeutic Modalities Design, Formulation and Manufacturing under Pharma 4.0 Principles. Biomedicines 2021, 10, 50. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).