Simple Summary
In adults, meningioma is the most common primary tumor of the brain. It is classified into three clinical grades of aggressiveness. Whereas disease recurrence after surgery and survival are associated with grade, it is worth investigating proliferation at a molecular level to identify markers capable of improving the clinical management of meningioma. In this study, we explore the DNA methylation profiles of 48 tumors of various grades and conduct statistical analyses on several proliferation indices and markers, such as mitotic index, grade, and Ki-67 or MCM6 expression levels. We identify differential methylation profiles between grades, loci highly correlated with cell growth and division, and a specific methylation signature of regulatory regions persistently associated with proliferation indices, grade, and survival. Finally, we report candidate genes under the control of these regions with potential prognostic and therapeutic value and deserving clinical evaluation.
Abstract
Meningiomas are the most common primary tumors of the central nervous system. Based on the 2021 WHO classification, they are classified into three grades reflecting recurrence risk and aggressiveness. However, the WHO’s histopathological criteria defining these grades are somewhat subjective. Together with reliable immunohistochemical proliferation indices, other molecular markers such as those studied with genome-wide epigenetics promise to revamp the current prognostic classification. In this study, 48 meningiomas of various grades were randomly included and explored for DNA methylation with the Infinium MethylationEPIC microarray over 850k CpG sites. We conducted differential and correlative analyses on grade and several proliferation indices and markers, such as mitotic index and Ki-67 or MCM6 immunohistochemistry. We also set up Cox proportional hazard models for extensive associations between CpG methylation and survival. We identified loci highly correlated with cell growth and a targeted methylation signature of regulatory regions persistently associated with proliferation, grade, and survival. Candidate genes under the control of these regions include SMC4, ESRRG, PAX6, DOK7, VAV2, OTX1, and PCDHA-PCDHB-PCDHG, i.e., the protocadherin gene clusters. This study highlights the crucial role played by epigenetic mechanisms in shaping dysregulated cellular proliferation and provides potential biomarkers bearing prognostic and therapeutic value for the clinical management of meningioma.
Keywords:
genome-wide DNA methylation; meningioma; methylome; proliferation signature; biomarkers; survival; Ki-67; MCM6 1. Introduction
Meningiomas are the most common primary tumors of the central nervous system in adults. The annual incidence rate ranges from 1.3/100,000 to 7.8/100,000 for cerebral meningiomas, with a tendency towards constant augmentation over the past few years [1]. The widely adopted WHO (World Health Organization) classification divides meningiomas into 15 subtypes and 3 grades of malignancy, mainly based upon histology [2]. Grade 1, 2, and 3 meningiomas represent about 70%, 20–30%, and 1–3% of reported cases, respectively. They correlate with recurrence risk (7–25% for grade 1, 29–59% for grade 2, and 50–94% for grade 3 [3]), as well as with 5-year and 10-year overall survival [4,5]. However, the histological criteria are rather subjective and are often associated with significant interobserver bias [6,7]; more reliable markers can thus improve the adequacy of treatments based on tumor grade. The last WHO classification (5th Edition, 2021) integrated a TERT promoter mutation or a homozygous deletion of CDKN2A and/or CDKN2B as new criteria for the recognition of grade 3 meningiomas. The standard of care is as follows: when deemed adequate, most patients undergo surgery, whereas adjuvant therapy is not systematic; for grade 2 and grade 3 meningioma, after surgery, conformational radiotherapy is recommended, particularly in cases of incomplete resection in grade 2 and in all grade 3 cases [8]. To date, no drug therapy has been validated for meningioma treatment.
The discovery of new molecular targets may present new therapeutic options in meningioma management. The study of the molecular landscape in meningioma is thus an important issue. However, until recently, few genetic variations have been described. These included the earliest finding of chromosome 22q deletion, which causes the loss of the tumor suppressor gene NF2 [9], the inactivation of which was observed in about half of the meningiomas studied [10]. More recently, several genes with recurrent mutations were identified in meningioma, including proapoptotic E3 ubiquitin ligase TNF receptor-associated factor 7 (TRAF7), pluripotency transcription factor Kruppel-like factor 4 (KLF4), proto-oncogene v-Akt murine thymoma viral oncogene homolog 1 (AKT1), Hedgehog pathway-signaling member “smoothened” (SMO), and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit A (PIK3CA). Approximately 40% of sporadic meningiomas harbor at least one of these variations [11,12]. Inhibitors of SMO, AKT1, and PIK3CA are therefore of therapeutic interest [8,10]. Other recently identified mutations are linked to a phosphatase tensin homolog on chromosome 10 (PTEN), as well as cyclin kinases CDKN2A/CDKN2B, main tumor suppressor genes predominantly implicated in meningioma progression [13,14,15]. However, approximately 20% of meningiomas present no known oncogenic mutation [10]. Other genetic rearrangements may be implicated; these include copy-number alteration and chromosomal abnormalities, both of which are associated with higher grade and poor tumor prognosis [16,17,18,19]. These unbalanced profiles can impact genes involved in cell cycle maintenance and progression, dysregulate major functional pathways, activate oncogenes, and inactivate tumor suppressor genes [13,20].
Recently, DNA methylation (DNAm) profiles have been studied to elaborate new prognostic classifications [21,22,23,24]. Two were landmark studies that greatly advanced our understanding of the role of DNAm in meningioma. One study investigated meningioma genome-wide DNAm patterns and classified them into three distinct and clinically relevant methylation classes (benign, intermediate, and malignant) and six methylation subclasses (benign-1, benign-2, benign-3, intermediate-A, intermediate-B, and malignant). This approach more efficiently predicted tumor recurrence than the WHO classification [23]. The other study identified four key molecular/phenotypic features associated with meningioma malignancy based on an integrative analysis of multi-omic data gathered from DNAm, somatic point mutations, copy-number aberrations, and mRNA abundance. These features presented immunogenic, benign NF2 wild-type, hypermetabolic, and proliferative characteristics of the tumor tissue and could be used to determine the most appropriate therapeutic strategy. This study further associated independent immunohistochemical markers with each molecular group. For instance, a high expression of MCM proteins—from the helicase complex involved in DNA replication, which can be used as a proliferation marker—was discovered to be characteristic of the proliferative group, with levels correlating with poor prognosis. This finding is consistent with our own observation that a high MCM6 index correlates with shorter progression-free survival [25], underscoring the crucial role of cell cycle progression and proliferation in meningioma progression.
Taken together, these insights prompted us to investigate the correlation between DNAm and meningioma cell proliferation and evaluate their levels of dependency. To achieve this, we studied tumor cell proliferation based on the immunohistochemical markers Ki-67 and MCM6, which are associated with histopathological factors such as WHO grade and mitotic index on one hand and genome-wide DNAm profiles of a meningioma cohort composed of 48 tumor tissues of various grades on the other hand. Our goal was to identify specific genes correlating with/presenting differentially methylated regions as a function of tumor proliferation. To conclude the potential clinical application of our results, we evaluated the association between DNAm and survival in an attempt to facilitate the discovery of potential new prognostic markers and/or therapeutic targets.
2. Materials and Methods
2.1. Population and Clinical Data
Forty-eight samples from surgical meningioma resections analyzed in the Department of Pathology of Nancy University Hospital (CHRU Nancy, France) between 2006 and 2018, with available frozen tissue, were randomly included, focusing on high-grade meningioma associated with a grade 1 control group. All samples were anonymized. The study was conducted in accordance with local ethical guidelines.
For each sample, we collected the WHO grade and other histopathological data from the pathology reports and clinical data from the medical records. Main clinical variables of interest were age, gender, localization of the tumor, quality of the excision (complete or not), treatment by chemotherapy or/and radiotherapy, recurrence of the meningioma and vital status. Because the date of first symptoms or diagnostic imaging was not available for all the patients, the diagnostic date was considered as the date of surgery. The progression date was considered as the day of the radiological exam during which progression was noted. The last consultation date and the last news date were also collected from the medical records. They were used to calculate overall survival (OS) and progression-free survival (PFS).
2.2. Histopathology
The WHO grade was established by a neuropathologist based on the WHO 2021 classification. The mitotic count per 10 high power fields (HPF; 1.6 mm2) was assessed independently by two pathologists, and the mean value was calculated. The level of agreement between the two raters was measured with the intraclass correlation coefficient. Both analyses were performed on hematoxylin, eosin, and saffron (HES) slides, blinded to the clinical and molecular data.
2.3. Immunohistochemistry
For each case, all immunohistochemical (IHC) staining was performed from the same block of formalin-fixed, paraffin-embedded (FFPE) tissue, which was selected after reviewing all the HES slides. Slides were manually prepared by paraffin sections of 4 µm, followed by deparaffinization, rehydration, and antigen retrieval. IHC staining was performed using the following antibodies: MCM6 (mouse monoclonal antibody, clone H-8, sc-3936-16, 1/2000 dilution, Santa Cruz) and Ki-67 (mouse monoclonal antibody, clone MIB-1, GA62661, prediluted, Dako, Agilent). Immunochemistry was performed with a Dako Omnis (Dako Agilent) automate using an Envision Flex revelation system (Dako Agilent). The labeling index (LI) for MCM6 and Ki-67 was calculated as the percentage of tumor cells with nuclear staining counted among a total of 1000 tumoral cells.
2.4. DNA Methylation Analysis
DNA was extracted with a Macherey Nagel DNA extraction kit (Macherey-466 Nagel, Düren, Germany). After qualitative control, 900 ng of the extracted DNA was used to perform analyses with an Illumina MethylationEPIC BeadChip following the manufacturer’s instructions, as previously described [24,25]. Raw data files (IDAT) were generated and used for downstream bioinformatics with the minfi package in R v4.1. The EPIC microarray interrogates 850K CpG sites, enabling a genome-wide methylome study including proximal promoters, distal regulatory regions, gene bodies, and intergenic features. Numerous preparation and filtering steps for quality control (as described in [26]), including technical checks and CpG removal from X and Y chromosomes, led to a 787,087 CpG working dataset. Normalization was carried out following the FunNorm procedure [27].
Supervised analyses included statistical modeling with empirical Bayes for case/control studies (R limma package) and reported the differentially methylated CpGs, with p-values for each comparison adjusted for false discovery rate (FDR) following the Benjamini–Hochberg procedure. Differentially methylated regions (DMRs) between groups were determined using the same linear models with the dmrcate function of the R package DMRcate (parameters: lambda = 1000, C = 3, less than a 1000 bp gap, and at least 3 CpGs). Unbiased functional annotations on ontological terms (gene ontology, GO) were achieved at CpG and DMR levels with the R package missMethyl [28]. Spearman’s correlation tests were used for correlation analyses between CpG methylation levels and quantitative variables (mitotic index: number of mitoses, Ki-67 and MCM6 labeling indices: proliferative marker expressions). For each 787087 CpG, correlative metrics (Spearman’s Rho, p-value, and FDR by Benjamini–Hochberg) and beta-value statistics (average, median, standard deviation, methylation range max/min, and Q3/Q1) were computed and reported, along with CpG island (CGI) and gene contexts. By default, beta value is expressed from 0 (demethylated) to 1 (methylated), whereas methylation differentials, changes, and ranges are expressed in % and represent the real proportion of beta-value change: a 20% change signifies a 0.2 increase/decrease in beta value.
Cox proportional hazard models were used to evaluate the association between CpG methylation levels and survival, either for PFS or OS analyses (R survival and survminer packages). After selection of the top CpGs with univariate setups in PFS in OS, multivariate analyses were conducted using age and grade as covariates. For each 787,087 CpG, associative metrics (Wald test p-values and effect size) were computed and reported, along with CGI and gene annotations. The proportional hazard assumption was tested on each fitted univariate model. Two statistics were considered: the p-value of non-random distribution for the methylation variable (CpG) and the global p-value for the model. When both p-values > 0.05, the CpG was retained for further multivariate evaluation. Proportional hazard assumptions were not rechecked for age and sex, as we only considered the statistical significance of the methylation covariate.
In every statistical approach, an FDR < 0.05 was considered significant. Supervised analyses (statistical modeling and DMR search) were conducted on methylation M values. Unsupervised analyses (hierarchical clustering) and correlative studies were performed with methylation B values (beta values).
Samples also underwent brain tumor classification and copy-number variation (CNV) estimation according to Capper et al. [29] and Sahm et al. [23]. Raw methylation files (IDAT) were uploaded to the MolecularNeuropathology.org server using the v11b4 classifier for brain tumor classification and MNGv2.4 for meningioma subtype classification. The evaluations of copy-number aberrations (along with CNV plots), CDKN2A/B loss, and PTEN loss were extracted from the CNV profile section of the generated report.
3. Results
3.1. Clinicopathological Data
Our cohort was composed of 48 patients with tumor samples of diverse WHO 2021 grades, including grade 1 (21%; 10/48), grade 2 (atypical; 69%; 33/48), and grade 3 (anaplastic; 10%; 5/48) meningiomas. In comparison with the original pathology reports, the grade changed in one case due to the detection of a homozygous CDKN2A/B deletion, upgrading the case from grade 2 to 3. The median age was 57 years, with a male-to-female ratio of 0.5. Meningioma tissues were mainly localized in the convexity (69%) and skull base (25%). No neoadjuvant treatment was delivered. Twenty-two patients (46%) underwent radiotherapy (postoperative and/or after progression; Table S1), on patient (2%) received post-progression chemotherapy, and four patients received antiangiogenic (bevacizumab) therapy. Disease progression occurred in 19 cases (40%; median PFS time: 39 months (16;55)). Nine patients died during the follow-up, with a median OS time of 52 months (31;95). The mean mitotic index was 5.4 mitoses/mm2 (grade 1: 0.5/1.6 mm2; grade 2: 4.9/1.6 mm2; grade 3: 23/1.6 mm2), with good inter-observer agreement (intraclass correlation coefficient: 0.75). Average Ki-67 and MCM6 labeling indices were 21% and 51%, respectively (Table 1).

Table 1.
Clinical and pathological features.
3.2. Molecular Data Based on Molecular Neuropathology Classifiers and Copy-Number Variations
On MolecularNeuropathology.org, the brain tumor classification was successfully run on all 48 samples, reporting the methylation class “meningioma” with a valid score (calibrated score ˃ 0.9) for all samples (100%). The additional algorithm for meningioma subtype identification classified 31 meningiomas (65%) with a calibrated score ˃ 0.9: 13 in the benign class, 14 in the intermediate class, and 4 in the malignant class (Table 2). Grade 1 meningiomas were classified in benign and intermediate classes (80% and 20%, respectively). Among the 19 grade 2 cases with a calibrated score > 0.9, 5 were classified in benign (15%), 12 in intermediate (36%), and 2 in malignant (6%) classes.

Table 2.
Molecular features from Sahm et al.’s methylation classifier reports according to the WHO grade.
Copy-number variations (CNVs) identified by DNA methylation (DNAm) in the cohort of 48 meningiomas are represented in Figure 1. The total number of CNVs (complete or partial gain/deletion) averaged at 7.2 ± 5.7. The CNV profiles showed a homozygous loss of CDKN2A/B, loss of PTEN, and/or NF2 genes in 0%, 0%, and 50% of grade 1 meningiomas; 0%, 42%, and 91% of grade 2 meningiomas; and 40%,100%, and 100% of grade 3 meningiomas, respectively (Table 2).
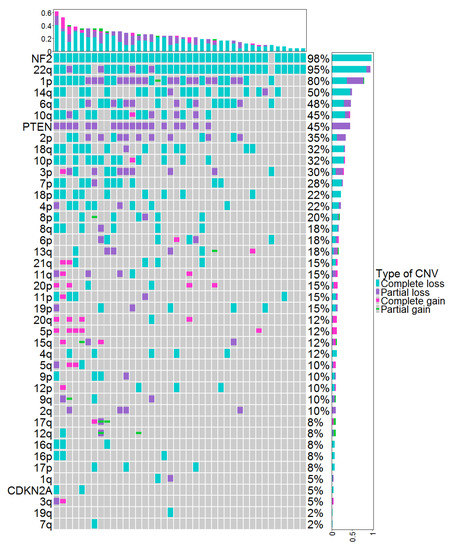
Figure 1.
CNVs identified by DNAm in the cohort of 48 meningiomas. Each column represents individual patients ordered from left to right by increasing frequency of genetic alterations. Genetic alterations are ordered on the y axis from top to bottom by decreasing frequency of genetic alterations. CNV: copy-number variation, DNAm: DNA methylation.
3.3. DNA Methylation and WHO Grade
At CpG resolution, differential analyses yielded 2099 significant hits between high-grade (grades 2 and 3; n = 38) and low-grade (grade 1; n = 10) meningiomas (Figure 2), with 1158 hypomethylated and 941 hypermethylated CpGs (moderated t-test; FDR < 0.05) (Figure S1). Of the total 2099 CpGs with FDR < 0.05, 2064 (98.3%) presented with a beta-value change > 1%, 1752 (83.5%) had a methylation differential > 5%, and 1641 (78.2%) showed > 10% change. Overall, hypomethylation occurred at already unmethylated and low methylation loci (beta value < 0.3 on a scale ranging from 0 to 1), with very low methylation dynamics (<10% change, a metric corresponding to 0.1 in beta-value change). Hypermethylation occurred to a greater extent (>50% change, i.e., >0.5 beta change) and mostly impacted medium-/high- methylation sites (beta value > 0.5) (Figure 3). Further analysis of areas spanning multiple CpGs areas consolidated these into 222 DMRs (with methylation differential > 10%), and in high grades, a majority of these were hypermethylated (200 DMRs) with only few hypomethylations (22 DMRs) (Table 3 and Table S2). Indeed, hypermethylated loci had rather restricted distribution, as 33.8% of these CpGs hit the same gene at least twice. Furthermore, they were distributed preferentially within CpG islands (CGI context) in promoter and intergenic regions (gene context), hinting at enhancer-linked functions. The hypomethylated loci were spread more evenly, with 88.5% hypomethylations located within the gene body of exclusive genes and predominantly in regions outside the CGI (open-sea probes) (Figure 3).
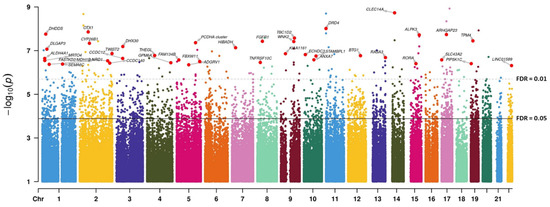
Figure 2.
Manhattan plot representing the association between DNAm at the CpG level and high-grade (>1) vs. low-grade meningiomas. The dashed and solid lines indicate FDR thresholds after p-value correction for genome-wide multitesting (Benjamini–Hochberg). FDR: false discovery rate.
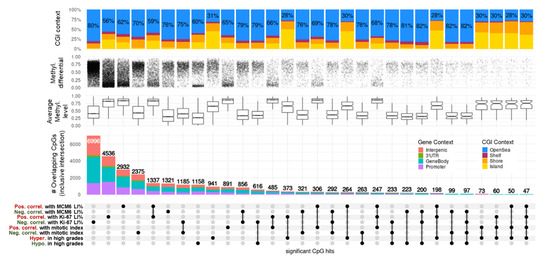
Figure 3.
UpSet plot summarizing the overlap between CpGs highly correlated with proliferation indices and grades in meningiomas. Every possible intersection is displayed (30 in total, including the 8 original hit lists shown on the bottom-left side). All intersections are inclusive. CpG regulatory contexts (gene and CGI annotations) are represented as proportions of barplot heights. Proportions of open-sea CpGs are displayed on CGI context bar plots. Average methylation levels are represented as boxplots of means (beta value) for each CpG of the intersected lists. Methylation differentials represent the range of each CpG beta value (max–min) and the densities of changes. DNAm: DNA methylation; methyl: methylation; CGI: CpG island; pos. correl: positive correlation; neg. correl: negative correlation; hypo: hypomethylation; hyper: hypermethylation; LI: labeling index.

Table 3.
Top differentially methylated regions (DMRs; 15 hypermethylated, 10 hypomethylated) ordered by average methylation differential (Mean Diff.) between high-grade (2, 3) and low-grade (1) meningiomas.
We next conducted unbiased gene ontology analysis on the set of genes associated with the 2099 differentially methylated CpGs. The most enriched biological process ontologies in high-grade versus low-grade meningiomas concerned main functions such as morphogenesis, neurogenesis, and cell differentiation (all FDR < 1 × 10−13) (Table 4) and included growth (FDR = 2.80 × 10−3), cell leading edge (FDR = 2.89 × 10−3), cell cycle (FDR = 2.95 × 10−3), and apoptotic process (FDR = 4.97 × 10−6). Highly enriched GO terms comprised more specific processes, such as cell morphogenesis involved in neuron differentiation (FDR = 2.81 × 10−11, 464/555 genes), positive regulation of transcription by RNA polymerase II (FDR = 9.16 × 10−10, 893/1151 genes), regulation of nervous system development (FDR = 3.41 × 10−9, 712/894 genes), Rho protein signal transduction (FDR = 3.31 × 10−5, 159/191 genes), mitotic cell cycle checkpoint (FDR = 3.31 × 10−4, 132/164 genes), regulation of the extrinsic apoptotic pathway (FDR = 4.79 × 10−4, 121/149 genes), DNA damage checkpoint (FDR = 9.34 × 10−3, 117/142 genes), and cellular response to prostaglandin stimulus (FDR = 0.02, 21/21 genes) (Table S3).

Table 4.
Top 20 gene ontologies (biological processes) based on the highest significance (FDR) between high-grade (>1) and low-grade meningiomas.
Among the DMRs and multiple gene hits (Table 3 and Table S2, Figure 2), notable hypomethylated regions in high-grade meningiomas were linked to cell cycle, cell differentiation, and cell fate genes, such as SMC4/miR16 (four CpGs spanning 1036 bp, 41% average methylation differential (AMD)), PATJ (three CpGs spanning 155 bp, 31% AMD), and TP63 (nine CpGs spanning 650 bp, 23% AMD; coding for the tumor protein p63). DMR hypermethylation in high-grade meningiomas concerned important genes of neural development, such as CALCB (CpGs spanning 770 bp, 28% AMD), PAX6 (DMRs interspersed by 4.6 Kb and associated with alternative transcription start sites: (i) 15 CpGs, 2.76 kb, 27% AMD; and (ii) 9 CpGs, 1.21 kb, 17% AMD), the PCDH gene clusters (3 CpGs, 374 bp, 18% AMD), and WNK2 (8 CpGs, 1169 bp, 25% AMD). There was also a strong enrichment in SUZ12 (4.8-fold, q-value = 1.33 × 10−14) and EZH2 (5.9-fold, q-value = 2.87 × 10−4) target regions, both determinants of the polycomb repressor complex 2, which is involved in gene-silencing processes.
3.4. Correlation between DNAm and Mitotic Index
CpG methylation levels were significantly correlated with mitotic index (MI) in 891 occurrences for positive correlations (PC) and 2,375 (77%) for negative correlations (NC) (FDR < 0.05, Spearman test; Table S4, Figure 3). Moreover, with 94.2% very high correlations (|rho| > 2/3, 32/34 CpGs) being negative, DNAm was predominantly negatively correlated with MI. Most NC-linked CpGs were in the medium-methylation category (0.3 > beta value < 0.7) and displayed high methylation dynamics (60–90% variation). Conversely, PC with MI mostly covered methylated CpGs (beta-value > 0.7) with feeble methylation changes (<10%). Overall, these 3,266 correlated loci represented open-sea probes (70% in NC and 65% in PC) and were distributed evenly in proximal promoters, gene bodies, and intergenic contexts (Figure 3).
We obtained further molecular insights after cross-examination of significant loci reported between grade and MI. Intersection of grade-related CpGs with MI-related CpGs resulted in 73 hypermethylations/PC and 263 hypomethylations/NC. No other overlap existed between the four lists, implying the presence of a bidirectional DNAm regulatory mechanism for PC CpGs within CGI and regulatory regions and for NC CpGs outside CGIs in gene bodies (Figure 3). A list of the top correlated hits/genes with MI highlights the biologically relevant methylation candidate markers in meningiomas (Table 5, upper part). These hits were selected based not only on their very high methylation ranges (max–min > 50% and Q3–Q1 > 10%) but also on their gene-wise aggregation in the regulatory elements of the genes (annotations as islands, shores, and shelves within the CGI context, as well as in proximal or distal promoters in the gene context). These included CpGs linked to SMC4/miR16, p53 effector CD82, and probable methyltransferase METLL24 (seven, two, and two hits, respectively) for NC and CpGs linked to transcription factors ESRRG, PAX9, and OTX1 for PC (six, two, and two hits, respectively). Together with other dynamic and island-restricted CpGs, such as ARHGDIA (3 hits, NC), DOK7 (2 hits, NC), CAPN2 (2 hits, NC), the PCDH clusters (10 hits, PC) and PAX6 (10 hits, PC), these candidates constitute potential proliferative biomarkers associated with disease progression (Table S4).

Table 5.
Top correlated CpGs between DNAm levels (beta values) and mitotic indices, Ki-67 labeling indices, and MCM6 labeling indices in regulatory region contexts.
3.5. DNAm and Ki-67 Labeling Index
CpG methylation levels and Ki-67 labeling index (LI) correlated significantly in 11,532 occurrences, with 4536 PC and 6996 (61%) NC (FDR < 0.05; Table S5, Figure 3). DNAm was also largely negatively correlated with Ki-67 levels, with 88.4% NC at a very high cutoff (|rho| > 2/3, 38/43 CpGs). Similar to what was observed with MI, the majority of NC-linked CpGs were in the medium (0.3 > beta value < 0.7) methylation category and displayed high (50–90%) methylation dynamics, whereas DNAm PC with Ki-67 expression covered methylated CpGs (beta value > 0.7), mostly displaying moderate methylation variations (<25%). The proportion of CGI and known promoters was also higher in PC than in NC CpGs (Figure 3).
Ki-67 expression was highly correlated with MI (Spearman’s Rho = 0.71, p-value = 1.22 × 10−8). Surprisingly, less than half of the CpGs correlating with MI also correlated with Ki-67 levels (1185 for NC and 485 for PC). None of the loci in NC with Ki-67 overlapped with those in PC with MI and vice-versa. As with MI, we found no CpG overlap between NC with Ki-67 and hypermethylated sites in high grades, nor for CpGs in PC with Ki-67 and hypomethylated sites in high grades (Figure 3).
A list of the top correlated hits/genes relevant to Ki-67 LI is presented in Table 5 (middle part; same criteria as above with MI), including CpGs linked to SMC4/miR16 and the GTPases RAB33B and VAV2 (9, 2, and 4 hits, respectively) for NC and CpGs linked to long non-coding RNA CCDC140 and ESRRG and neuropeptide NPY for PC (10, 7, and 4 hits, respectively). Candidates such as DOK7 (3 hits, NC), the PCDH clusters (32 hits, PC), PAX6 (15 hits, PC), and TBR1 (6 hits, PC), among others with dynamic and island-restricted CpGs, constitute potential proliferative biomarkers in meningiomas (Table S5).
3.6. DNAm and MCM6 Labeling Index
CpG methylation levels and MCM6 LI correlated significantly in 4253 occurrences, with 2932 (69%) PC and 1321 NC (FDR < 0.05; Table S6, Figure 3). In contrast to what was observed with MI and Ki-67 LI, DNAm was generally positively correlated with MCM6 levels, with 70% PC at a very high cutoff (|rho| > 2/3, 7/10 CpGs). NC-linked CpGs were in the low/medium (beta-value < 0.4) methylation category and displayed high (50–90%) dynamics, whereas PC with MCM6 expression covered highly methylated CpGs (beta value > 0.8), most of which displayed moderate methylation variations (<25%). The proportion of island CpGs was also much higher in PC than in NC (Figure 3).
Nevertheless, MCM6 expression correlated positively with MI (Rho = 0.6, p-value = 5.31 × 10−6) and Ki-67 LI (Rho = 0.69, p-value = 7.77 × 10−8). Less than one-fifth of the CpGs correlating with MCM6 levels also correlated with MI (321 for NC and 306 for PC), with still no overlap between PC in MI and NC in MCM6 and vice-versa. Conversely, more than a half of the CpGs correlating with MCM6 levels also correlated with Ki-67 levels (856 for NC and 1,337 for PC). Again, no overlap was found between PC in MCM6 and NC in Ki-67 and vice-versa. Finally, as with MI and Ki-67, we found no CpG overlap between those in NC with MCM6 and hypermethylated sites in high grades, nor for CpGs in PC with MCM6 and hypomethylated sites in high grades (Figure 3). Thus, the controlled correlation structure observed between top loci associated with MI and WHO grades held and propagated with Ki-67 and MCM6 markers. The biologically relevant methylation markers based on the top correlated hits with MCM6 LI included CpGs linked with DNA replication inhibitor GMNN and ephrin EFNA1 (a single hit for both) for NC CpGs linked with brain transcription repressor TBR1 and PCDH gene clusters for PC (2 and 27 hits, respectively) (Table 5, lower part). Along with other candidates also associated with WHO grades, such as SMC4 (three hits, NC) and PAX6 (four hits, PC), these genes may constitute other interesting progression biomarkers in meningiomas (Table S6).
3.7. DNAm Proliferative Signature in Meningiomas
We did not observe a tight relationship between MI and Ki-67 DNAm markers and even less so with MCM6 levels, as only a small proportion of hits intersected (Figure 3). However, the overlapping structures were conservative between the three correlation experiments. Hence, we next evaluated the dependencies and the complementarities of these measurements (MI, Ki-67, and MCM6 LI) and derived a DNAm proliferation signature in meningiomas. This 310-CpG signature was obtained by cumulating top hits from the three positive (247 intersected CpGs) and three negative (292 intersected CpGs) correlation analyses (Figure 3) and by limiting the results to highly dynamic variables only (standard deviation > 10%; Table S7). Hierarchical clustering revealed a strongly correlated CpG structure, with an overall progressive sample demethylation according to both proliferation—with a very good adequation with MI—and disease progression, as the signature was also associated with WHO grade (Figure 4). Indeed, benign meningiomas (left side of the heat map) were methylated for a large open-sea CpG set and unmethylated at island CpGs, which constituted one-fifth of the signature. Samples with very low MCM6 LI (<30%) aggregated within this cluster. Grade 3 samples belonged to another cluster (right side of the heat map) of extreme methylation values for these CpGs. This cluster also aggregated samples with very high Ki-67 LI (>70%).
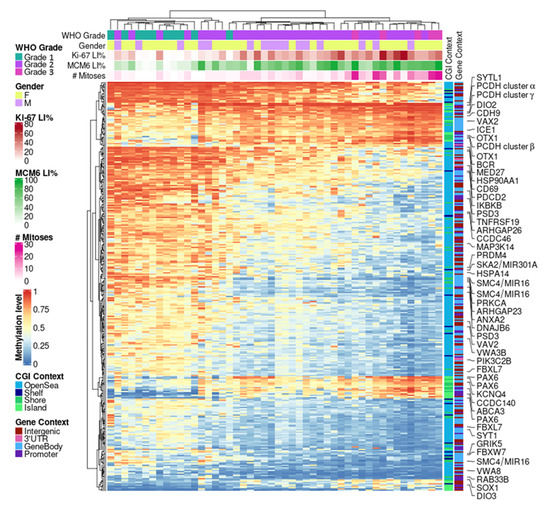
Figure 4.
DNAm proliferation signature in meningiomas. Hierarchical clustering heat map depicting 310 CpGs highly correlated between DNAm and three proliferative markers: mitotic index, Ki-67, and MCM6 expressions (metrics: Euclidean distance and complete linkage). CGI: CpG island; DNAm: DNA methylation; LI: labeling index; WHO: World Health Organization.
This DNAm signature was validated on an external meningioma dataset (GSE200321) of 60 QC-passed samples of various grades and histological subtypes, which also included invasiveness information (Figure S2). Hierarchical clustering again revealed the same patterns of progressive demethylation at open-sea CpGs across samples with increasing invasive and aggressive properties, accompanied by a strong methylation of island CpGs. Notably, chordoid meningioma is a morphological subtype designated as WHO grade 2 but mostly present with a low proliferation profile comparable to that of grade 1 meningiomas. These results are in line with recent findings suggesting that histology alone may not justify a grade 2 designation for chordoid meningiomas [30].
3.8. Associations between DNAm and Survival
We next conducted survival analyses with Cox proportional hazard models to identify specific loci associated with PFS and OS. We first examined the DNAm proliferative signature described above. As expected and independent of WHO grade and age, these 310 CpGs were all associated with survival, albeit better for PFS, with p-values in the range of [1.34 × 10−5–1.47 × 10−3], than for OS, with p-values in the range of [1.69 × 10−3–2.8 × 10−2] (Table S7; Wald test of multivariate settings).
We then examined genome-wide univariate associations with survival and observed a stronger DNAm association with PFS (69 hits, p < 1 × 10−5) than with OS (no hits, p < 1 × 10−5; 54 hits with p < 1 × 10−4) (Figure 5). Furthermore, PFS was associated massively with chr1q and chr20 loci, with 9.5- and 4.7-fold enrichments against background CpGs, respectively (both p < 2.2 × 10−16, chi-squared test; hits with p < 1 × 10−4). No such associations were found with OS. From these top univariate hits, we derived multivariate associations with PFS (Table S8) and OS (Table S9). One key conclusion is that for PFS, most loci (48/69) had a positive effect size, meaning that a methylation gain was linked to improved survival. This result echoes our findings based on grade/proliferation: the higher the proliferation (and the higher the grade), the lower the extent of DNAm (Figure 4). This trend was not observed for OS.
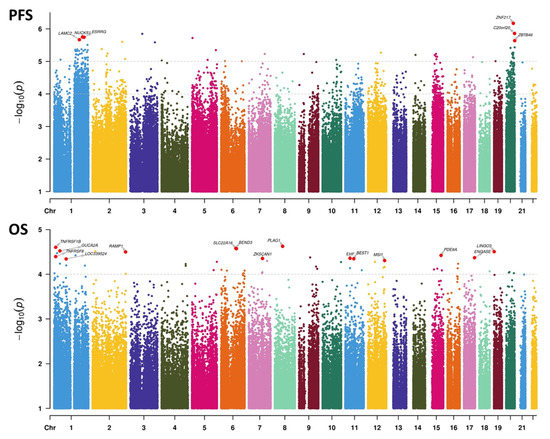
Figure 5.
Manhattan plots representing the association between DNAm at the CpG level and survival. Upper panel: associations with progression-free survival (PFS). Lower panel: associations with overall survival (OS). Univariate Cox statistics (Wald test). p-value cutoffs running multivariate analyses on selected CpGs are indicated by the topmost dashed lines in each panel. For the PFS panel, the second dashed line represents the threshold for computing hit enrichments over chromosomic regions.
4. Discussion
DNAm is an innovative tool that is increasingly used for the classification of brain tumors and useful to predict their clinical outcome [29]. In our setup of 48 samples with varied disease progression, all samples were recognized as meningiomas by the main brain tumor classification tool. However, regarding the specific meningioma algorithm, MNGv2.4, and despite our choice of using only freshly frozen tissues, we were still confronted with a high rate of non-classifiable meningiomas (35%). This rate was even higher for meningioma subclassification (46%), which limited its use. Our study unveiled the association between indices of proliferation with disease progression and DNAm and consequently helped to identify a targeted proliferation-relevant DNAm signature comprising only a few hundred CpGs. As this signature could easily be used to score or classify meningioma samples, we propose that such a methylation-based classifier could ameliorate their categorization.
In our attempt to link proliferation/grade/disease outcomes and DNAm, we observed two main dynamics associated with cell proliferation in meningiomas. First, there were few but strong methylations at CGIs located in proximal promoters and enhancers mostly affiliated with neural transcription factors and tumor suppressor genes. One such hit is the developmentally targeted epigenetic silencing by polycomb repressor complex 2; although it is difficult to extrapolate these previous results to meningiomas, this silencing is well-documented in brain cancers [31,32]. Second, there was an extended and progressive demethylation at open sea, mostly within gene bodies. This observation has also been reported in cancers [33,34] although in pediatric brain cancers, the enriched hits were found to be intergenic, with no gene affiliation [6]. This mechanism has been associated with chromosomal instability [35] and malignant progression of lower-grade glioma [36] and was predictive of response to standard chemotherapy in osteosarcoma [37].
Among the epigenetic changes between low- and high-grade meningiomas, we found expected pathways driving unchecked cell division, such as the hypomethylation of genes promoting cell cycle, growth, differentiation, and fate. We also reported dysregulation of regulatory pathways of DNA damage checkpoints and extrinsic apoptosis, along with genes with full driving potential, such as TP63 [38,39] and SMC4. SMC4 was found to be involved in tumor cell growth, migration, and invasion. It is also correlated with poor prognosis in some cancers. Indeed, its overexpression is suspected to play a role in numerous cancers, such as hepatocellular, colorectal, breast, and endometrial cancer [40,41,42,43,44]. In glioma, SMC4 overexpression promotes aggressive phenotypes by TGFβ/Smad signaling [45,46,47]. Hence, the hypomethylation of SMC4 in high-grade meningiomas could likely lead to overexpression, which is consistent with previous data indicating poor prognosis in other tumors. It would therefore be interesting to evaluate its predictive role in meningioma, as it was also highly correlated with all proliferation indices. Additionally, hypermethylated markers in high-grade meningiomas included proximal promoters of PAX6 and PCDH genes, in addition to correlation with proliferation markers. PAX6, a transcription factor playing an important role in the development of the central nervous system, constitutes a good candidate in our study, as in many regulatory loci, it was positively correlated with all proliferative markers. In glioblastoma, a few studies have suggested that a low PAX6 expression level should be considered prognostically pejorative [48,49,50]. The protocadherin (PCDH) family of proteins plays an important role in neural cell aggregation, cell recognition, and neural development. PCDHs are the “barcodes” of the cell, generating single-cell diversity in the brain [51]. They are encoded by combinations obtained by alternative splicing from the three mapped-in-tandem, multigene PCDH α, β, and γ clusters. This splicing is controlled through DNAm states of alternative promoters [52], PCDH cluster expression involving CTCF interaction, which can be affected by methylation alterations, leading to long-range epigenetic silencing in cancers [53]. Here, we highlight strong and spread-out PCDH promoter hypermethylations linked to disease progression and cell proliferation markers. We suggest that these aberrant methylations may be of diagnostic value in meningioma. Finally, our findings with respect to the hypermethylation of the tumor suppressor and familial meningioma WNK2 in grade 2 and 3 meningioma are consistent with previously published data, which suggested epigenetic alterations to be the dominant, grade-specific mechanism of gene inactivation [54,55].
When considering mitotic activity in meningiomas, we found negative correlations with DNAm for ARHGDIA and DOK7 and positive correlations for ESRRG and OTX1 (both correlating with all proliferation indices). ARHGDIA codes for a Rho GDP dissociation inhibitor protein with antiapoptotic activity and is involved in cellular processes, such as cell proliferation, cell cycle progression, and cell migration [56]. Its expression is altered in various cancer, such as breast, prostate, and hepatocellular cancer [57,58,59]. In glioma, its level was reported to be correlated with positive prognosis and was considered both an independent predictive marker for OS [56] and an actionable therapeutic target [60]. DOK7 was also reported to be correlated with the Ki-67 index and may function as a tumor suppressor gene, which is consistent with our findings. However, in breast carcinoma, significant DOK7 promoter hypomethylation implicated its role in early tumorigenesis [61,62]. In lung cancer, lower DOK7 expression was associated with lower survival. In in vitro studies, DOK7 was reported to inhibit proliferation and migration by downregulating the PI3K/AKT/mTOR pathway [63,64]. Similarly, DOK7 was found to be downregulated in glioma, and its overexpression inhibited both in vitro and in vivo proliferation of glioma cells [65]. ESRRG is a transcriptional activator of DNMT1 (DNA methyltransferase 1). Its downregulation correlates with poor clinical outcome in gastric cancer, where it acts by suppressing cell growth and tumorigenesis. It also antagonizes Wnt signaling [66]. ESRRG promoter hypermethylation was used as a diagnostic and prognostic biomarker in laryngeal squamous cell carcinoma [67]. OTX1 is a transcription factor that is required for proper brain and sensory organ development in mice [68], where it regulates cell cycle progression, with its knockdown leading to diminished neurons but increased astrocytes [69]. Few studies have linked OTX1 overexpression with tumorigenesis and growth in cancers [70,71], and to the best of our knowledge, it has not been yet reported as an epigenetic marker in brain cancers.
Between DNAm and Ki-67 levels, we report negative correlations for VAV2, a guanine exchange factor playing an important role in angiogenesis. Its recruitment by phosphorylated EPHA2 is critical for EFNA1-induced RAC1 GTPase activation, as well as vascular endothelial cell migration and assembly. Interestingly, we also found EFNA1 DNAm to be correlated with MCM6 expression. VAV2 is overexpressed in numerous cancers [72,73,74], but its dysregulation has not yet been reported in brain cancers. It is implicated in both cutaneous and head and neck squamous cell carcinomas, where it promotes regenerative proliferation [75]. In esophageal squamous cell carcinoma, it is required for DNA repair and radiotherapy resistance [76]. Because it is a Rho guanine exchange factor, VAV2 is an attractive pharmacological target, with druggable catalytic sites and a more restricted tissue distribution pattern than other Rho proteins. TBR1 DNAm is positively correlated with the Ki-67 and MCM6 indices. TBR1 is a transcriptional repressor involved in multiple aspects of cortical development, including neuronal migration and axonal projection. Recurrent variations of TRB1 have been described in medulloblastoma, and a high frequency of its copy-number loss has been detected in glioblastoma, suggesting a possible involvement in tumorigenesis or progression [77,78]. To the best of our knowledge, no data are available in the literature with respect to any implication of these genes in meningioma.
With respect to MCM6 levels, we found negative correlations in GMNN- (geminin) and EFNA1- linked DNAm. GMNN was shown to inhibit DNA replication by preventing the incorporation of MCM complexes into prereplication complexes (pre-RC) [79,80,81,82,83]. Further evidence implies its participation in the inhibition of the transcriptional activity of a subset of Hox proteins, linking GMNN to proliferative cell cycle control [83]. EFNA1 codes for the receptor protein-tyrosine kinase Ephrin A1, which is known to mediate developmental events and is involved in migration, repulsion, and adhesion during neuronal, vascular, and epithelial development in the nervous system and in erythropoiesis. In the context of tumor biology, it was shown to be regulated by hypoxia, and its main function involves angiogenesis and tumor neovascularization [84]. The recruitment of VAV2 is critical for EFNA1-induced RAC1 GTPase activation and vascular endothelial cell migration and assembly.
When correlating proliferative indices (mitotic, Ki-67, and MCM6) with DNAm levels, we found that the positive and negative correlations exclusively occurred in the hyper- and hypomethylated DMRs differentiating WHO grades. This interrelation hints at a tight epigenetic control of cell proliferation. Thus, either on islands or in open-sea regions, non-concerted modification of methylation does not seem to be the key determinant driving cell division in meningioma. Moreover, fewer overlaps than expected were found between hits with our three chosen markers of proliferation, suggesting their complementing diagnostic value. As expected, because these markers are expressed during different stages of the cell cycle with different magnitudes of expression level, Ki-67 was found to perform well with high-grade and highly proliferative meningiomas, whereas MCM6 expression better delineated low-grade, low-proliferation tumors. Overall, the mitotic index was progressively correlated with demethylation in our proliferation signature.
Due to the limited number of samples in our dataset, the survival statistics were capped to a threshold of associations incompatible with genome-wide multitesting corrections. To overcome this, we first focused on the subset of 310 CpGs with proliferation-dependent DNAm and found that all CpGs were associated with PFS or OS. We next looked at genome-wide results and found that PFS associations with DNAm were the strongest, with unexpectedly numerous hits of high significance in broad genomic regions such a chromosomes 1q and 20. Results with PFS also confirmed the directionality of our DNAm proliferation signature, with progressive demethylation associated with worse progression. The lack of such an observation in OS indicates that no high adequation exists between these proliferation markers and occurrence of death; however, this could also be explained by a smaller number of events and the inclusion of deaths due to another pathology (i.e., not specific to OS). OS may be more strongly associated with independent or indirectly linked loci, such as the MSI1 regulatory region. This gene encodes an RNA binding protein that could be involved in the maintenance of stem cells in the central nervous system and in cell proliferation. In glioblastoma, it was found to promote the expression of stem cell marker CD44 by impairing miRNA function [85]. In these cells, it was shown to be regulated and stabilized by HuR [86], another RNA binding protein and biomarker of interest in meningiomas [87].
In the present study, given the importance of proliferation in the prognosis of meningiomas, we designed an original approach focusing on the correlation between DNAm and cell proliferation. Proliferation is one of the main mechanisms of tumorigenesis and involves a considerable number of pathways shared by multiple types of tumors. However, epigenetic regulation is only a part of the mechanisms by which cells alter their gene expression. Although we do not have evidence of how DNAm changes affect gene expression, DNAm could potentially regulate downstream genes expression, which may contribute to tumorigenesis or progression. To better dissect the underlying molecular pathways of aberrant cellular growth in meningiomas, integrative and multi-omics studies are needed. As a first step, in this pilot mono-omic study integrating molecular and histological indices, we identified a proliferation signature encompassing hundreds of regulated genes, with several candidates serving as potential predictive and prognostic biomarkers or new therapeutic targets. Most notably, SMC4, DOK7, PAX6, ARHGDIA, ESRRG, VAV2, and OTX1, as well as the three protocadherin gene clusters PCDHα, β, and γ, may be particularly relevant and deserve further preclinical and clinical investigations.
5. Conclusions
In conclusion, this study highlights the crucial role played by epigenetic mechanisms in shaping dysregulated cellular proliferation in meningioma. It provides molecular biomarkers with potential to revamp the current prognostic classification, as well as new druggable targets, adding therapeutic value to clinical management. The reported findings are novel and show the additional value of DNAm evaluation in diagnosis and prognostication for patients with meningioma.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers14246227/s1, Table S1. Clinical treatments: preoperative or post-progression radiotherapy, post-progression chemotherapy, post-progression chemotherapy (no neoadjuvant treatment). Table S2. Differentially methylated regions in high grade (>1) vs. low-grade meningiomas. Table S3. GO functional annotations of differential CpGs between high-grade (2 and 3) and low-grade (1) meningiomas. Table S4. Correlation statistics between DNA methylation and mitotic index. Table S5. Correlation statistics between DNA methylation and Ki-67 labeling index. Table S6. Correlation statistics between DNA methylation and MCM6 labeling index. Table S7. DNA methylation proliferation signature in meningiomas. Table S8. Top CpGs associated with progression-free survival. Table S9. Top CpGs associated with overall survival. Figure S1. Principal component analysis of the top 2099 CpGs from the differential analysis between grade 1 and grade 2 + 3 meningiomas. Figure S2. Validation of the proliferative signature on an external methylation dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE200321 (accessed on 23 November 2022); from Daoud et al. [30]). Hierarchical clustering against the 310 CpGs and 60 samples of various meningioma grades and histological types (6 grade 1: 5 meningothelial and 1 transitional; 50 grade 2: 10 chordoid, 27 atypical, and 13 atypical and invasive; 4 grade 3: anaplastic). Raw data from this dataset were processed with the same analytical pipeline as the main dataset presented in this work.
Author Contributions
Conceptualization, S.H. and G.G.; Data curation, C.G., P.R., C.S., S.L., C.G. and G.G.; Formal analysis, S.H. and G.G.; Data analysis, C.G., M.D., S.H., C.S., M.D. and S.L.; Investigation, C.S., M.D., S.L., F.R., C.G., C.P. and G.G.; Methodology, S.H., S.-F.B.-H. and G.G.; Project administration, G.G.; Supervision, G.G.; Writing—original draft, C.S. and S.H.; Writing—review and editing, S.-F.B.-H., S.H. and G.G. All authors have read and agreed to the published version of the manuscript.
Funding
La Région Lorraine, Grand Est—projet de recherche d’intérêt régional 2016; INSERM U1256 NGERE.
Institutional Review Board Statement
This study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board (or Ethics Committee) of the CHRU Nancy Hospital (protocol code 2021PI0154-127; date of approval: 3 November 2021).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE200321. The data generated and presented in this study are available upon request from the corresponding author. The data are not publicly available due to privacy restrictions.
Acknowledgments
The authors thank the team of the Pathology Department of Nancy (CHRU-ICL) for technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Baldi, I.; Engelhardt, J.; Bonnet, C.; Bauchet, L.; Berteaud, E.; Grüber, A.; Loiseau, H. Epidemiology of Meningiomas. Neurochirurgie 2018, 64, 5–14. [Google Scholar] [CrossRef] [PubMed]
- International Agency for Research on Cancer. WHO Classification of Tumours Editorial Board. Central Nervous System Tumours, 5th ed.; WHO Classification of Tumours Series; WHO: Geneva, Switzerland, 2021; Volume 6. [Google Scholar]
- Wang, N.; Osswald, M. Meningiomas: Overview and New Directions in Therapy. Semin. Neurol. 2018, 38, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Aizer, A.A.; Bi, W.L.; Kandola, M.S.; Lee, E.Q.; Nayak, L.; Rinne, M.L.; Norden, A.D.; Beroukhim, R.; Reardon, D.A.; Wen, P.Y.; et al. Extent of Resection and Overall Survival for Patients with Atypical and Malignant Meningioma: Extent of Resection and Recurrence in Meningioma. Cancer 2015, 121, 4376–4381. [Google Scholar] [CrossRef] [PubMed]
- van Alkemade, H.; de Leau, M.; Dieleman, E.M.T.; Kardaun, J.W.P.F.; van Os, R.; Vandertop, W.P.; van Furth, W.R.; Stalpers, L.J.A. Impaired Survival and Long-Term Neurological Problems in Benign Meningioma. Neuro-Oncology 2012, 14, 658–666. [Google Scholar] [CrossRef]
- Pathology Concordance Levels for Meningioma Classification and Grading in NRG Oncology RTOG Trial 0539. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4799683/ (accessed on 20 February 2022).
- Goldbrunner, R.; Minniti, G.; Preusser, M.; Jenkinson, M.D.; Sallabanda, K.; Houdart, E.; von Deimling, A.; Stavrinou, P.; Lefranc, F.; Lund-Johansen, M.; et al. EANO Guidelines for the Diagnosis and Treatment of Meningiomas. Lancet Oncol. 2016, 17, e383–e391. [Google Scholar] [CrossRef]
- Institut National du Cancer. Conduite a Tenir Devant des Patients Atteints de Méningiomes de Grade II et III/Synthèse, septembre 2020. e-cancer.fr. 2020. Available online: https://www.e-cancer.fr/Expertises-et-publications/Catalogue-des-publications/Conduites-a-tenir-devant-des-patients-atteints-d-un-meningiome-de-grade-II-et-III-Synthese (accessed on 1 November 2022).
- Rouleau, G.A.; Merel, P.; Lutchman, M.; Sanson, M.; Zucman, J.; Marineau, C.; Hoang-Xuan, K.; Demczuk, S.; Desmaze, C.; Plougastel, B. Alteration in a New Gene Encoding a Putative Membrane-Organizing Protein Causes Neuro-Fibromatosis Type 2. Nature 1993, 363, 515–521. [Google Scholar] [CrossRef]
- Bi, W.L.; Zhang, M.; Wu, W.W.; Mei, Y.; Dunn, I.F. Meningioma Genomics: Diagnostic, Prognostic, and Therapeutic Applications. Front. Surg. 2016, 3, 40. [Google Scholar] [CrossRef]
- Bi, W.L.; Mei, Y.; Agarwalla, P.K.; Beroukhim, R.; Dunn, I.F. Genomic and Epigenomic Landscape in Meningioma. Neurosurg. Clin. N. Am. 2016, 27, 167–179. [Google Scholar] [CrossRef]
- Proctor, D.T.; Ramachandran, S.; Lama, S.; Sutherland, G.R. Towards Molecular Classification of Meningioma: Evolving Treatment and Diagnostic Paradigms. World Neurosurg. 2018, 119, 366–373. [Google Scholar] [CrossRef]
- Galani, V.; Lampri, E.; Varouktsi, A.; Alexiou, G.; Mitselou, A.; Kyritsis, A.P. Genetic and Epigenetic Alterations in Meningiomas. Clin. Neurol. Neurosurg. 2017, 158, 119–125. [Google Scholar] [CrossRef]
- Yakubov, E.; Ghoochani, A.; Buslei, R.; Buchfelder, M.; Eyüpoglu, I.Y.; Savaskan, N. Hidden Association of Cowden Syndrome, PTEN Mutation and Meningioma Frequency. Oncoscience 2016, 3, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Alexiou, G.A.; Voulgaris, S. The Role of the PTEN Gene in Malignant Gliomas. Neurol. Neurochir. Pol. 2010, 44, 80–86. [Google Scholar] [CrossRef]
- Bi, W.L.; Greenwald, N.F.; Abedalthagafi, M.; Wala, J.; Gibson, W.J.; Agarwalla, P.K.; Horowitz, P.; Schumacher, S.E.; Esaulova, E.; Mei, Y.; et al. Genomic Landscape of High-Grade Meningiomas. NPJ Genomic Med. 2017, 2, 15. [Google Scholar] [CrossRef] [PubMed]
- Brastianos, P.K.; Horowitz, P.M.; Santagata, S.; Jones, R.T.; McKenna, A.; Getz, G.; Ligon, K.L.; Palescandolo, E.; Van Hummelen, P.; Ducar, M.D.; et al. Genomic Sequencing of Meningiomas Identifies Oncogenic SMO and AKT1 Mutations. Nat. Genet. 2013, 45, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Lekanne Deprez, R.H.; Riegman, P.H.; van Drunen, E.; Warringa, U.L.; Groen, N.A.; Stefanko, S.Z.; Koper, J.W.; Avezaat, C.J.; Mulder, P.G.; Zwarthoff, E.C. Cytogenetic, Molecular Genetic and Pathological Analyses in 126 Meningiomas. J. Neuropathol. Exp. Neurol. 1995, 54, 224–235. [Google Scholar] [CrossRef] [PubMed]
- Cordova, C.; Kurz, S.C. Advances in Molecular Classification and Therapeutic Opportunities in Meningiomas. Curr. Oncol. Rep. 2020, 22, 84. [Google Scholar] [CrossRef] [PubMed]
- Suvà, M.L.; Louis, D.N. Next-Generation Molecular Genetics of Brain Tumours. Curr. Opin. Neurol. 2013, 26, 681–687. [Google Scholar] [CrossRef]
- Katz, L.M.; Hielscher, T.; Liechty, B.; Silverman, J.; Zagzag, D.; Sen, R.; Wu, P.; Golfinos, J.G.; Reuss, D.; Neidert, M.C.; et al. Loss of Histone H3K27me3 Identifies a Subset of Meningiomas with Increased Risk of Recurrence. Acta Neuropathol. (Berl.) 2018, 135, 955–963. [Google Scholar] [CrossRef]
- Paramasivam, N.; Hübschmann, D.; Toprak, U.H.; Ishaque, N.; Neidert, M.; Schrimpf, D.; Stichel, D.; Reuss, D.; Sievers, P.; Reinhardt, A.; et al. Mutational Patterns and Regulatory Networks in Epigenetic Subgroups of Meningioma. Acta Neuropathol. (Berl.) 2019, 138, 295–308. [Google Scholar] [CrossRef]
- Sahm, F.; Schrimpf, D.; Stichel, D.; Jones, D.T.W.; Hielscher, T.; Schefzyk, S.; Okonechnikov, K.; Koelsche, C.; Reuss, D.E.; Capper, D.; et al. DNA Methylation-Based Classification and Grading System for Meningioma: A Multicentre, Retrospective Analysis. Lancet Oncol. 2017, 18, 682–694. [Google Scholar] [CrossRef]
- Nassiri, F.; Liu, J.; Patil, V.; Mamatjan, Y.; Wang, J.Z.; Hugh-White, R.; Macklin, A.M.; Khan, S.; Singh, O.; Karimi, S.; et al. A Clinically Applicable Integrative Molecular Classification of Meningiomas. Nature 2021, 597, 119–125. [Google Scholar] [CrossRef]
- Gauchotte, G.; Vigouroux, C.; Rech, F.; Battaglia-Hsu, S.-F.; Soudant, M.; Pinelli, C.; Civit, T.; Taillandier, L.; Vignaud, J.-M.; Bressenot, A. Expression of Minichromosome Maintenance MCM6 Protein in Meningiomas Is Strongly Correlated with Histologic Grade and Clinical Outcome. Am. J. Surg. Pathol. 2012, 36, 283–291. [Google Scholar] [CrossRef]
- Zgheib, R.; Battaglia-Hsu, S.-F.; Hergalant, S.; Quéré, M.; Alberto, J.-M.; Chéry, C.; Rouyer, P.; Gauchotte, G.; Guéant, J.-L.; Namour, F. Folate Can Promote the Methionine-Dependent Reprogramming of Glioblastoma Cells towards Pluripotency. Cell Death Dis. 2019, 10, 596. [Google Scholar] [CrossRef]
- Fortin, J.-P.; Labbe, A.; Lemire, M.; Zanke, B.W.; Hudson, T.J.; Fertig, E.J.; Greenwood, C.M.; Hansen, K.D. Functional Normalization of 450k Methylation Array Data Improves Replication in Large Cancer Studies. Genome Biol. 2014, 15, 503. [Google Scholar] [CrossRef]
- Phipson, B.; Maksimovic, J.; Oshlack, A. MissMethyl: An R Package for Analyzing Data from Illumina’s HumanMethylation450 Platform. Bioinforma. Oxf. Engl. 2016, 32, 286–288. [Google Scholar] [CrossRef]
- Capper, D.; Jones, D.T.W.; Sill, M.; Hovestadt, V.; Schrimpf, D.; Sturm, D.; Koelsche, C.; Sahm, F.; Chavez, L.; Reuss, D.E.; et al. DNA Methylation-Based Classification of Central Nervous System Tumours. Nature 2018, 555, 469–474. [Google Scholar] [CrossRef]
- Daoud, E.V.; Zhu, K.; Mickey, B.; Mohamed, H.; Wen, M.; Delorenzo, M.; Tran, I.; Serrano, J.; Hatanpaa, K.J.; Raisanen, J.M.; et al. Epigenetic and Genomic Profiling of Chordoid Meningioma: Implications for Clinical Management. Acta Neuropathol. Commun. 2022, 10, 56. [Google Scholar] [CrossRef]
- Esteller, M. Epigenetic Gene Silencing in Cancer: The DNA Hypermethylome. Hum. Mol. Genet. 2007, 16, R50–R59. [Google Scholar] [CrossRef]
- Mack, S.C.; Hubert, C.G.; Miller, T.E.; Taylor, M.D.; Rich, J.N. An Epigenetic Gateway to Brain Tumor Cell Identity. Nat. Neurosci. 2016, 19, 10–19. [Google Scholar] [CrossRef]
- Rechache, N.S.; Wang, Y.; Stevenson, H.S.; Killian, J.K.; Edelman, D.C.; Merino, M.; Zhang, L.; Nilubol, N.; Stratakis, C.A.; Meltzer, P.S.; et al. DNA Methylation Profiling Identifies Global Methylation Differences and Markers of Adrenocortical Tumors. J. Clin. Endocrinol. Metab. 2012, 97, E1004–E1013. [Google Scholar] [CrossRef]
- Song, M.-A.; Tiirikainen, M.; Kwee, S.; Okimoto, G.; Yu, H.; Wong, L.L. Elucidating the Landscape of Aberrant DNA Methylation in Hepatocellular Carcinoma. PLoS ONE 2013, 8, e55761. [Google Scholar] [CrossRef]
- Rodriguez, J.; Frigola, J.; Vendrell, E.; Risques, R.-A.; Fraga, M.F.; Morales, C.; Moreno, V.; Esteller, M.; Capellà, G.; Ribas, M.; et al. Chromosomal Instability Correlates with Genome-Wide DNA Demethylation in Human Primary Colorectal Cancers. Cancer Res. 2006, 66, 8462–9468. [Google Scholar] [CrossRef]
- Nomura, M.; Saito, K.; Aihara, K.; Nagae, G.; Yamamoto, S.; Tatsuno, K.; Ueda, H.; Fukuda, S.; Umeda, T.; Tanaka, S.; et al. DNA Demethylation Is Associated with Malignant Progression of Lower-Grade Gliomas. Sci. Rep. 2019, 9, 1903. [Google Scholar] [CrossRef]
- Lietz, C.E.; Newman, E.T.; Kelly, A.D.; Xiang, D.H.; Zhang, Z.; Luscko, C.A.; Lozano-Calderon, S.A.; Ebb, D.H.; Raskin, K.A.; Cote, G.M.; et al. Genome-Wide DNA Methylation Patterns Reveal Clinically Relevant Predictive and Prognostic Subtypes in Human Osteosarcoma. Commun. Biol. 2022, 5, 213. [Google Scholar] [CrossRef]
- Cancino, G.I.; Yiu, A.P.; Fatt, M.P.; Dugani, C.B.; Flores, E.R.; Frankland, P.W.; Josselyn, S.A.; Miller, F.D.; Kaplan, D.R. P63 Regulates Adult Neural Precursor and Newly Born Neuron Survival to Control Hippocampal-Dependent Behavior. J. Neurosci. Off. J. Soc. Neurosci. 2013, 33, 12569–12585. [Google Scholar] [CrossRef]
- Dugani, C.B.; Paquin, A.; Fujitani, M.; Kaplan, D.R.; Miller, F.D. P63 Antagonizes P53 to Promote the Survival of Embryonic Neural Precursor Cells. J. Neurosci. Off. J. Soc. Neurosci. 2009, 29, 6710–6721. [Google Scholar] [CrossRef]
- Feng, X.-D.; Song, Q.; Li, C.-W.; Chen, J.; Tang, H.-M.; Peng, Z.-H.; Wang, X.-C. Structural Maintenance of Chromosomes 4 Is a Predictor of Survival and a Novel Therapeutic Target in Colorectal Cancer. Asian Pac. J. Cancer Prev. APJCP 2014, 15, 9459–9465. [Google Scholar] [CrossRef]
- Chen, Y.; Huang, F.; Deng, L.; Yuan, X.; Tao, Q.; Wang, T.; Li, D.; Fan, Y.; Peng, Q.; Tang, D. HIF-1-MiR-219-SMC4 Regulatory Pathway Promoting Proliferation and Migration of HCC under Hypoxic Condition. BioMed Res. Int. 2019, 2019, 8983704. [Google Scholar] [CrossRef]
- Yan, Y.; Liu, C.; Zhang, J.; Li, W.; Yin, X.; Dong, L.; Pang, S.; Li, X. SMC4 Knockdown Inhibits Malignant Biological Behaviors of Endometrial Cancer Cells by Regulation of FoxO1 Activity. Arch. Biochem. Biophys. 2021, 712, 109026. [Google Scholar] [CrossRef]
- Ma, R.-M.; Yang, F.; Huang, D.-P.; Zheng, M.; Wang, Y.-L. The Prognostic Value of the Expression of SMC4 MRNA in Breast Cancer. Dis. Markers 2019, 2019, 2183057. [Google Scholar] [CrossRef]
- Zhou, B.; Chen, H.; Wei, D.; Kuang, Y.; Zhao, X.; Li, G.; Xie, J.; Chen, P. A Novel MiR-219-SMC4-JAK2/Stat3 Regulatory Pathway in Human Hepatocellular Carcinoma. J. Exp. Clin. Cancer Res. CR 2014, 33, 55. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Zhou, J.; Zhong, D.; Zhou, Y.; Zhang, W.; Wu, W.; Zhao, Z.; Wang, W.; Xu, W.; He, L.; et al. Overexpression of SMC4 Activates TGFβ/Smad Signaling and Promotes Aggressive Phenotype in Glioma Cells. Oncogenesis 2017, 6, e301. [Google Scholar] [CrossRef] [PubMed]
- You, A.; Rao, G.; Wang, J.; Li, J.; Zhang, Y.; Gu, J.; Ge, X.; Zhang, K.; Gao, X.; Wu, X.; et al. MiR-433-3p Restrains the Proliferation, Migration and Invasion of Glioma Cells via Targeting SMC4. Brain Res. 2021, 1767, 147563. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, Z. The Clinical Significance and Transcription Regulation of a DNA Damage Repair Gene, SMC4, in Low-Grade Glioma via Integrated Bioinformatic Analysis. Front. Oncol. 2021, 11, 761693. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.Y.; Hu, Y.; Siegel, E.; Stanley, L.; Zhou, Y.-H. PAX6 Increases Glioma Cell Susceptibility to Detachment and Oxidative Stress. J. Neurooncol. 2007, 84, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Pavlakis, E.; Tonchev, A.B.; Kaprelyan, A.; Enchev, Y.; Stoykova, A. Interaction between Transcription Factors PAX6/PAX6-5a and Specific Members of MiR-183-96-182 Cluster, May Contribute to Glioma Progression in Glioblastoma Cell Lines. Oncol. Rep. 2017, 37, 1579–1592. [Google Scholar] [CrossRef]
- Zhou, Y.-H.; Wu, X.; Tan, F.; Shi, Y.-X.; Glass, T.; Liu, T.J.; Wathen, K.; Hess, K.R.; Gumin, J.; Lang, F.; et al. PAX6 Suppresses Growth of Human Glioblastoma Cells. J. Neurooncol. 2005, 71, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.V.; Maniatis, T. Clustered Protocadherins. Dev. Camb. Engl. 2013, 140, 3297–3302. [Google Scholar] [CrossRef]
- Tasic, B.; Nabholz, C.E.; Baldwin, K.K.; Kim, Y.; Rueckert, E.H.; Ribich, S.A.; Cramer, P.; Wu, Q.; Axel, R.; Maniatis, T. Promoter Choice Determines Splice Site Selection in Protocadherin Alpha and Gamma Pre-MRNA Splicing. Mol. Cell 2002, 10, 21–33. [Google Scholar] [CrossRef]
- Vega-Benedetti, A.F.; Loi, E.; Moi, L.; Blois, S.; Fadda, A.; Antonelli, M.; Arcella, A.; Badiali, M.; Giangaspero, F.; Morra, I.; et al. Clustered Protocadherins Methylation Alterations in Cancer. Clin. Epigenet. 2019, 11, 100. [Google Scholar] [CrossRef]
- Jun, P.; Hong, C.; Lal, A.; Wong, J.M.; McDermott, M.W.; Bollen, A.W.; Plass, C.; Held, W.A.; Smiraglia, D.J.; Costello, J.F. Epigenetic Silencing of the Kinase Tumor Suppressor WNK2 Is Tumor-Type and Tumor-Grade Specific. Neuro-Oncology 2009, 11, 414–422. [Google Scholar] [CrossRef]
- He, S.; Pham, M.H.; Pease, M.; Zada, G.; Giannotta, S.L.; Wang, K.; Mack, W.J. A Review of Epigenetic and Gene Expression Alterations Associated with Intracranial Meningiomas. Neurosurg. Focus 2013, 35, E5. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Wang, X.; Liu, J.; He, Y.; Liang, Z.; Xia, Z.; Cai, Y.; Zhou, L.; Zhu, H.; Liang, S. Downregulation of ARHGDIA Contributes to Human Glioma Progression through Activation of Rho GTPase Signaling Pathway. Tumour Biol. 2016, 37, 15783–15793. [Google Scholar] [CrossRef]
- Wang, H.; Wang, B.; Liao, Q.; An, H.; Li, W.; Jin, X.; Cui, S.; Zhao, L. Overexpression of RhoGDI, a Novel Predictor of Distant Metastasis, Promotes Cell Proliferation and Migration in Hepatocellular Carcinoma. FEBS Lett. 2014, 588, 503–508. [Google Scholar] [CrossRef]
- Yamashita, T.; Okamura, T.; Nagano, K.; Imai, S.; Abe, Y.; Nabeshi, H.; Yoshikawa, T.; Yoshioka, Y.; Kamada, H.; Tsutsumi, Y.; et al. Rho GDP-Dissociation Inhibitor Alpha Is Associated with Cancer Metastasis in Colon and Prostate Cancer. Pharm.-Int. J. Pharm. Sci. 2012, 67, 253–255. [Google Scholar]
- Bozza, W.P.; Zhang, Y.; Hallett, K.; Rosado, L.A.R.; Zhang, B. RhoGDI Deficiency Induces Constitutive Activation of Rho GTPases and COX-2 Pathways in Association with Breast Cancer Progression. Oncotarget 2015, 6, 32723–32736. [Google Scholar] [CrossRef][Green Version]
- Harding, M.A.; Theodorescu, D. RhoGDI Signaling Provides Targets for Cancer Therapy. Eur. J. Cancer Oxf. Engl. 1990 2010, 46, 1252–1259. [Google Scholar] [CrossRef]
- Shirkavand, A.; Boroujeni, Z.N.; Aleyasin, S.A. Examination of Methylation Changes of VIM, CXCR4, DOK7, and SPDEF Genes in Peripheral Blood DNA in Breast Cancer Patients. Indian J. Cancer 2018, 55, 366–371. [Google Scholar] [CrossRef]
- Heyn, H.; Carmona, F.J.; Gomez, A.; Ferreira, H.J.; Bell, J.T.; Sayols, S.; Ward, K.; Stefansson, O.A.; Moran, S.; Sandoval, J.; et al. DNA Methylation Profiling in Breast Cancer Discordant Identical Twins Identifies DOK7 as Novel Epigenetic Biomarker. Carcinogenesis 2013, 34, 102–108. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, G.; Ye, L.; Yu, H.; Li, S.; Jiang, W.G. DOK7V1 Influences the Malignant Phenotype of Lung Cancer Cells through PI3K/AKT/MTOR and FAK/Paxillin Signaling Pathways. Int. J. Oncol. 2019, 54, 381–389. [Google Scholar] [CrossRef]
- Chen, G.; Yu, H.; Satherley, L.; Zabkiewicz, C.; Resaul, J.; Zhao, H.; Mu, H.; Zhi, X.; He, J.; Ye, L.; et al. The Downstream of Tyrosine Kinase 7 Is Reduced in Lung Cancer and Is Associated with Poor Survival of Patients with Lung Cancer. Oncol. Rep. 2017, 37, 2695–2701. [Google Scholar] [CrossRef] [PubMed]
- Hua, C.-D.; Bian, E.-B.; Chen, E.-F.; Yang, Z.-H.; Tang, F.; Wang, H.-L.; Zhao, B. Repression of Dok7 Expression Mediated by DNMT1 Promotes Glioma Cells Proliferation. Biomed. Pharmacother. Biomedecine Pharmacother. 2018, 106, 678–685. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.-H.; Choi, H.; Oshima, M.; Cheong, J.-H.; Kim, S.; Lee, J.H.; Park, Y.S.; Choi, H.-S.; Kweon, M.-N.; Pack, C.-G.; et al. Estrogen-Related Receptor Gamma Functions as a Tumor Suppressor in Gastric Cancer. Nat. Commun. 2018, 9, 1920. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Hu, Y.; Zhou, C.; Yuan, J.; Xu, J.; Hao, W.; Deng, H.; Ye, D. ESRRG Promoter Hypermethylation as a Diagnostic and Prognostic Biomarker in Laryngeal Squamous Cell Carcinoma. J. Clin. Lab. Anal. 2019, 33, e22899. [Google Scholar] [CrossRef]
- Acampora, D.; Mazan, S.; Avantaggiato, V.; Barone, P.; Tuorto, F.; Lallemand, Y.; Brûlet, P.; Simeone, A. Epilepsy and Brain Abnormalities in Mice Lacking the Otx1 Gene. Nat. Genet. 1996, 14, 218–222. [Google Scholar] [CrossRef]
- Huang, B.; Li, X.; Tu, X.; Zhao, W.; Zhu, D.; Feng, Y.; Si, X.; Chen, J.-G. OTX1 Regulates Cell Cycle Progression of Neural Progenitors in the Developing Cerebral Cortex. J. Biol. Chem. 2018, 293, 2137–2148. [Google Scholar] [CrossRef]
- Jiang, L.; Zuo, Z.; Lin, J.; Yang, C. Orthodenticle Homeobox OTX1 Is a Potential Prognostic Biomarker for Bladder Cancer. Bioengineered 2021, 12, 6559–6571. [Google Scholar] [CrossRef]
- Tu, X.-P.; Li, H.; Chen, L.-S.; Luo, X.-N.; Lu, Z.-M.; Zhang, S.-Y.; Chen, S.-H. OTX1 Exerts an Oncogenic Role and Is Negatively Regulated by MiR129-5p in Laryngeal Squamous Cell Carcinoma. BMC Cancer 2020, 20, 794. [Google Scholar] [CrossRef]
- Jiang, Y.; Prabakaran, I.; Wan, F.; Mitra, N.; Furstenau, D.K.; Hung, R.K.; Cao, S.; Zhang, P.J.; Fraker, D.L.; Guvakova, M.A. Vav2 Protein Overexpression Marks and May Predict the Aggressive Subtype of Ductal Carcinoma in Situ. Biomark. Res. 2014, 2, 22. [Google Scholar] [CrossRef]
- Citterio, C.; Menacho-Márquez, M.; García-Escudero, R.; Larive, R.M.; Barreiro, O.; Sánchez-Madrid, F.; Paramio, J.M.; Bustelo, X.R. The Rho Exchange Factors Vav2 and Vav3 Control a Lung Metastasis-Specific Transcriptional Program in Breast Cancer Cells. Sci. Signal. 2012, 5, ra71. [Google Scholar] [CrossRef]
- Tan, B.-B.; Li, Y.; Fan, L.-Q.; Zhao, Q.; Liu, Q.-W.; Liu, Y.; Wang, D.; Jia, N. Upregulated Vav2 in Gastric Cancer Tissues Promotes Tumor Invasion and Metastasis. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2017, 39, 1010428317698392. [Google Scholar] [CrossRef]
- Lorenzo-Martín, L.F.; Fernández-Parejo, N.; Menacho-Márquez, M.; Rodríguez-Fdez, S.; Robles-Valero, J.; Zumalave, S.; Fabbiano, S.; Pascual, G.; García-Pedrero, J.M.; Abad, A.; et al. VAV2 Signaling Promotes Regenerative Proliferation in Both Cutaneous and Head and Neck Squamous Cell Carcinoma. Nat. Commun. 2020, 11, 4788. [Google Scholar] [CrossRef]
- Liu, W.; Miao, C.; Zhang, S.; Liu, Y.; Niu, X.; Xi, Y.; Guo, W.; Chu, J.; Lin, A.; Liu, H.; et al. VAV2 Is Required for DNA Repair and Implicated in Cancer Radiotherapy Resistance. Signal Transduct. Target. Ther. 2021, 6, 322. [Google Scholar] [CrossRef]
- Jones, D.T.W.; Jäger, N.; Kool, M.; Zichner, T.; Hutter, B.; Sultan, M.; Cho, Y.-J.; Pugh, T.J.; Hovestadt, V.; Stütz, A.M.; et al. Dissecting the Genomic Complexity Underlying Medulloblastoma. Nature 2012, 488, 100–105. [Google Scholar] [CrossRef]
- Nakahara, Y.; Shiraishi, T.; Okamoto, H.; Mineta, T.; Oishi, T.; Sasaki, K.; Tabuchi, K. Detrended Fluctuation Analysis of Genome-Wide Copy Number Profiles of Glioblastomas Using Array-Based Comparative Genomic Hybridization. Neuro-Oncol. 2004, 6, 281–289. [Google Scholar] [CrossRef]
- McGarry, T.J.; Kirschner, M.W. Geminin, an Inhibitor of DNA Replication, Is Degraded during Mitosis. Cell 1998, 93, 1043–1053. [Google Scholar] [CrossRef]
- Miotto, B.; Struhl, K. HBO1 Histone Acetylase Activity Is Essential for DNA Replication Licensing and Inhibited by Geminin. Mol. Cell 2010, 37, 57–66. [Google Scholar] [CrossRef]
- Sugimoto, N.; Tatsumi, Y.; Tsurumi, T.; Matsukage, A.; Kiyono, T.; Nishitani, H.; Fujita, M. Cdt1 Phosphorylation by Cyclin A-Dependent Kinases Negatively Regulates Its Function without Affecting Geminin Binding. J. Biol. Chem. 2004, 279, 19691–19697. [Google Scholar] [CrossRef]
- Caillat, C.; Pefani, D.-E.; Gillespie, P.J.; Taraviras, S.; Blow, J.J.; Lygerou, Z.; Perrakis, A. The Geminin and Idas Coiled Coils Preferentially Form a Heterodimer That Inhibits Geminin Function in DNA Replication Licensing. J. Biol. Chem. 2013, 288, 31624–31634. [Google Scholar] [CrossRef]
- Zhou, B.; Liu, C.; Xu, Z.; Zhu, G. Structural Basis for Homeodomain Recognition by the Cell-Cycle Regulator Geminin. Proc. Natl. Acad. Sci. USA 2012, 109, 8931–8936. [Google Scholar] [CrossRef]
- Hao, Y.; Li, G. Role of EFNA1 in Tumorigenesis and Prospects for Cancer Therapy. Biomed. Pharmacother. 2020, 130, 110567. [Google Scholar] [CrossRef]
- Pötschke, R.; Haase, J.; Glaß, M.; Simmermacher, S.; Misiak, C.; Penalva, L.O.F.; Kühnöl, C.D.; Hüttelmaier, S. MSI1 Promotes the Expression of the GBM Stem Cell Marker CD44 by Impairing MiRNA-Dependent Degradation. Cancers 2020, 12, 3654. [Google Scholar] [CrossRef]
- Vo, D.T.; Abdelmohsen, K.; Martindale, J.L.; Qiao, M.; Tominaga, K.; Burton, T.L.; Gelfond, J.A.L.; Brenner, A.J.; Patel, V.; Trageser, D.; et al. The Oncogenic RNA-Binding Protein Musashi1 Is Regulated by HuR via MRNA Translation and Stability in Glioblastoma Cells. Mol. Cancer Res. MCR 2012, 10, 143–155. [Google Scholar] [CrossRef]
- Gauchotte, G.; Hergalant, S.; Vigouroux, C.; Casse, J.-M.; Houlgatte, R.; Kaoma, T.; Helle, D.; Brochin, L.; Rech, F.; Peyre, M.; et al. Cytoplasmic Overexpression of RNA-Binding Protein HuR Is a Marker of Poor Prognosis in Meningioma, and HuR Knockdown Decreases Meningioma Cell Growth and Resistance to Hypoxia. J. Pathol. 2017, 242, 421–434. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).