A High MCM6 Proliferative Index in Atypical Meningioma Is Associated with Shorter Progression Free and Overall Survivals
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Population, Clinical Data, and WHO Grading
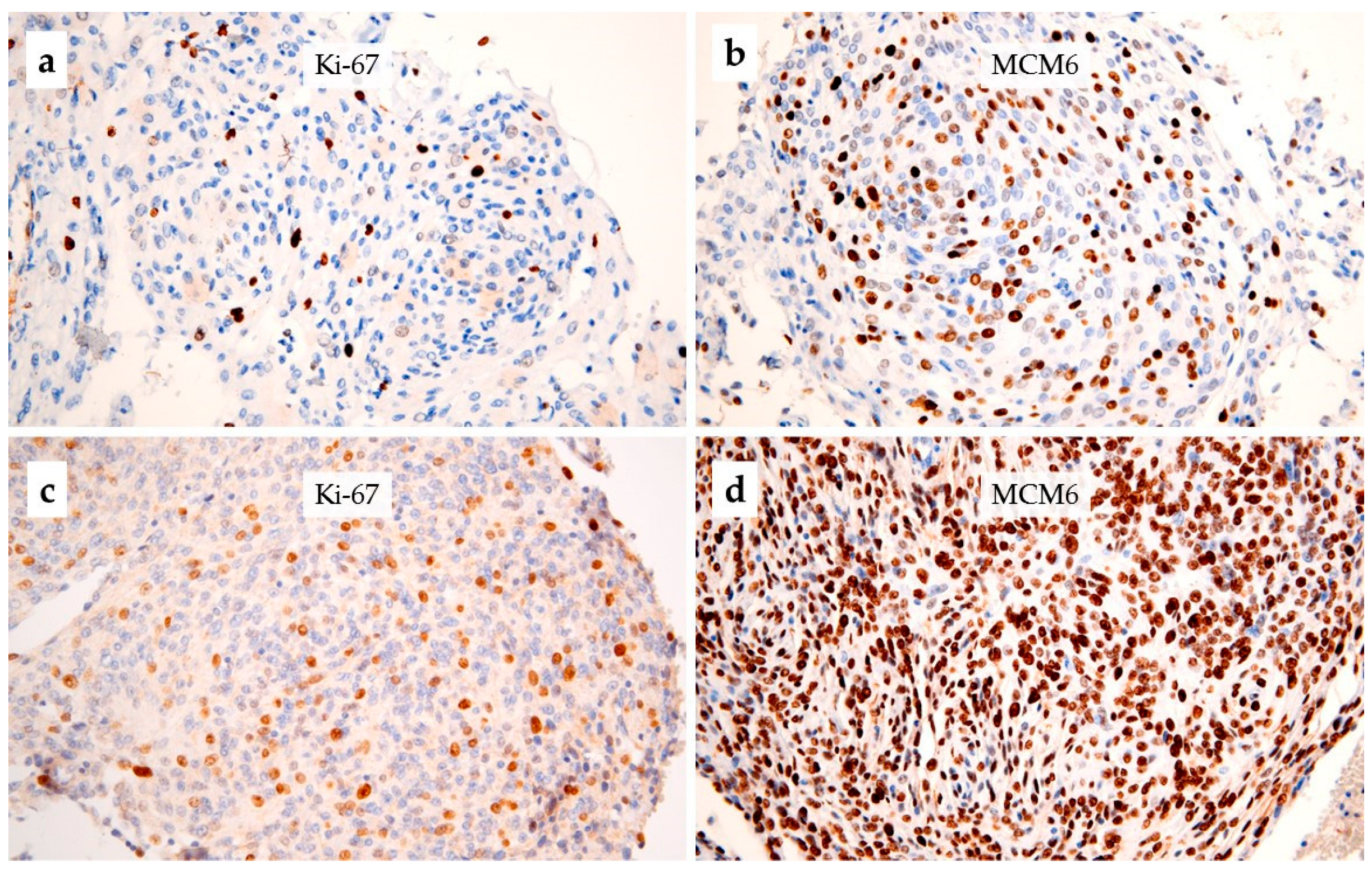
2.2. Immunohistochemistry
2.3. Methylome
2.4. Statistical Analysis
3. Results
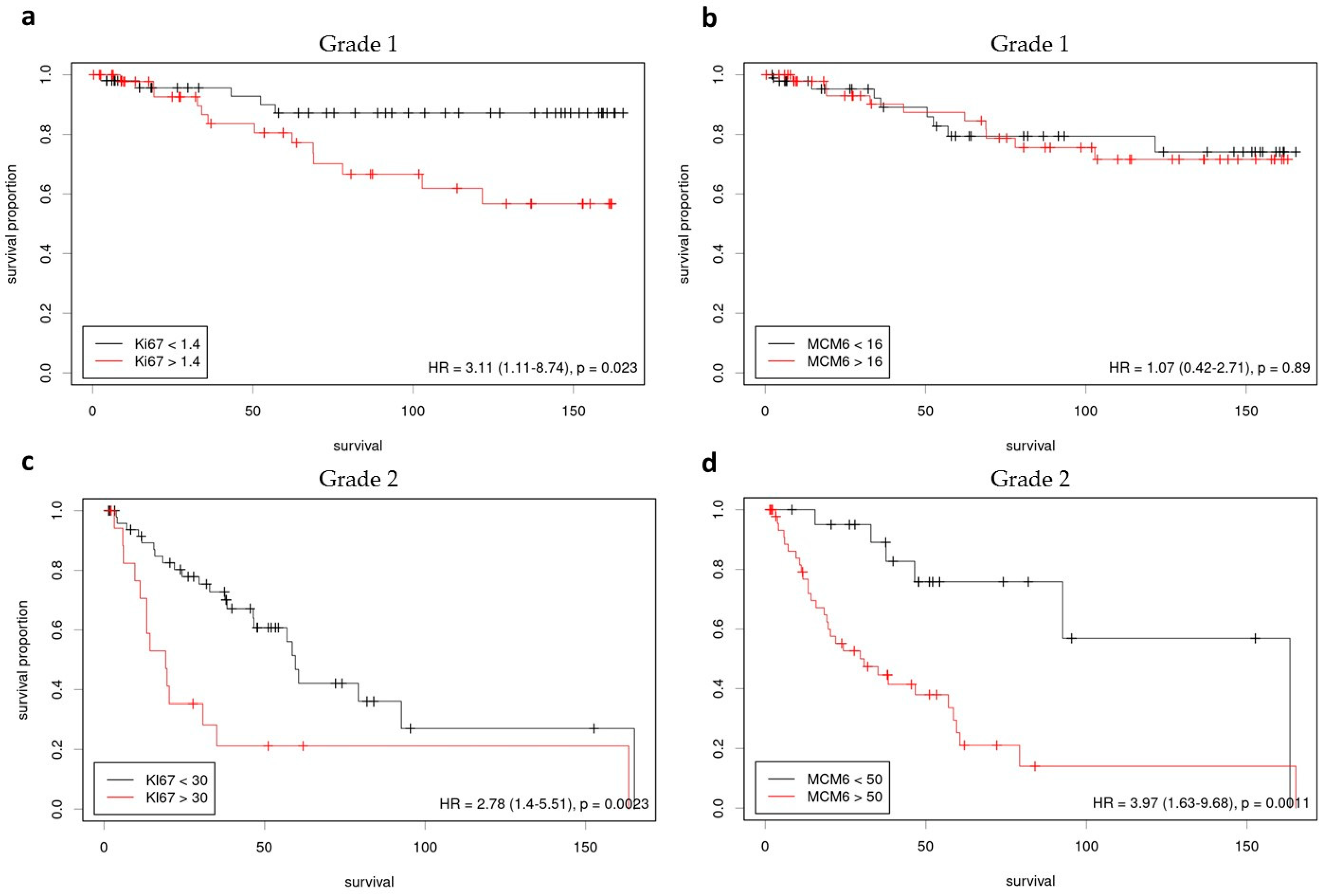
3.1. Grade 1 Meningiomas
3.2. Grade 2 Meningiomas
3.3. Correlation with Methylome and CNV
3.4. Reproducibility and Evaluation of the Potential Influence of Time-Related Tissue Degradation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baldi, I.; Engelhardt, J.; Bonnet, C.; Bauchet, L.; Berteaud, E.; Grüber, A.; Loiseau, H. Epidemiology of Meningiomas. Neurochirurgie 2018, 64, 5–14. [Google Scholar] [CrossRef] [PubMed]
- International Agency for Research on Cancer. WHO Classification of Tumours Editorial Board. Central Nervous System Tumours. In WHO Classification of Tumours Series, 5th ed.; International Agency for Research on Cancer: Lyon, France, 2021; Volume 6. [Google Scholar]
- Buerki, R.A.; Horbinski, C.M.; Kruser, T.; Horowitz, P.M.; James, C.D.; Lukas, R.V. An Overview of Meningiomas. Future Oncol. 2018, 14, 2161–2177. [Google Scholar] [CrossRef] [PubMed]
- Gauchotte, G.; Vigouroux, C.; Rech, F.; Battaglia-Hsu, S.-F.; Soudant, M.; Pinelli, C.; Civit, T.; Taillandier, L.; Vignaud, J.-M.; Bressenot, A. Expression of Minichromosome Maintenance MCM6 Protein in Meningiomas Is Strongly Correlated with Histologic Grade and Clinical Outcome. Am. J. Surg. Pathol. 2012, 36, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Prat-Acín, R.; Guarín-Corredor, M.J.; Galeano-Senabre, I.; Ayuso-Sacido, A.; Vera-Sempere, F. Value of KI-67/MIB-1 Labeling Index and Simpson Grading System to Predict the Recurrence of Who Grade I Intracranial Meningiomas Compared to Who Grade II. J. Clin. Neurosci. 2021, 86, 32–37. [Google Scholar] [CrossRef]
- Nagahama, A.; Yashiro, M.; Kawashima, T.; Nakajo, K.; Morisako, H.; Uda, T.; Naito, K.; Ichinose, T.; Ohata, K.; Goto, T. Combination of P53 and Ki67 as a Promising Predictor of Postoperative Recurrence of Meningioma. Anticancer Res. 2021, 41, 203–210. [Google Scholar] [CrossRef]
- Møller, M.L.; Braendstrup, O. No Prediction of Recurrence of Meningiomas by PCNA and Ki-67 Immunohistochemistry. J. Neurooncol. 1997, 34, 241–246. [Google Scholar] [CrossRef]
- Mengel, M.; von Wasielewski, R.; Wiese, B.; Rüdiger, T.; Müller-Hermelink, H.K.; Kreipe, H. Inter-Laboratory and Inter-Observer Reproducibility of Immunohistochemical Assessment of the Ki-67 Labelling Index in a Large Multi-Centre Trial. J. Pathol. 2002, 198, 292–299. [Google Scholar] [CrossRef]
- Labib, K.; Kearsey, S.E.; Diffley, J.F. MCM2-7 Proteins Are Essential Components of Prereplicative Complexes That Accumulate Cooperatively in the Nucleus during G1-Phase and Are Required to Establish, but Not Maintain, the S-Phase Checkpoint. Mol. Biol. Cell 2001, 12, 3658–3667. [Google Scholar] [CrossRef]
- Gu, Y.; Hu, X.; Liu, X.; Cheng, C.; Chen, K.; Wu, Y.; Wu, Z. MCM6 Indicates Adverse Tumor Features and Poor Outcomes and Promotes G1/S Cell Cycle Progression in Neuroblastoma. BMC Cancer 2021, 21, 784. [Google Scholar] [CrossRef]
- Hendricks, A.; Gieseler, F.; Nazzal, S.; Bräsen, J.H.; Lucius, R.; Sipos, B.; Claasen, J.H.; Becker, T.; Hinz, S.; Burmeister, G.; et al. Prognostic Relevance of Topoisomerase II α and Minichromosome Maintenance Protein 6 Expression in Colorectal Cancer. BMC Cancer 2019, 19, 429. [Google Scholar] [CrossRef]
- Pouget, C.; Hergalant, S.; Lardenois, E.; Lacomme, S.; Houlgatte, R.; Carpentier, C.; Dehais, C.; Rech, F.; Taillandier, L.; Sanson, M.; et al. Ki-67 and MCM6 Labeling Indices Are Correlated with Overall Survival in Anaplastic Oligodendroglioma, IDH1-Mutant and 1p/19q-Codeleted: A Multicenter Study from the French POLA Network. Brain Pathol. 2020, 30, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Sadeghian, D.; Saffar, H.; Mahdavi Sharif, P.; Soleimani, V.; Jahanbin, B. MCM6 versus Ki-67 in Diagnosis of Luminal Molecular Subtypes of Breast Cancers. Diagn. Pathol. 2022, 17, 24. [Google Scholar] [CrossRef] [PubMed]
- Vigouroux, C.; Casse, J.-M.; Battaglia-Hsu, S.-F.; Brochin, L.; Luc, A.; Paris, C.; Lacomme, S.; Gueant, J.-L.; Vignaud, J.-M.; Gauchotte, G. Methyl(R217)HuR and MCM6 Are Inversely Correlated and Are Prognostic Markers in Non Small Cell Lung Carcinoma. Lung. Cancer 2015, 89, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Saydam, O.; Senol, O.; Schaaij-Visser, T.B.M.; Pham, T.V.; Piersma, S.R.; Stemmer-Rachamimov, A.O.; Wurdinger, T.; Peerdeman, S.M.; Jimenez, C.R. Comparative Protein Profiling Reveals Minichromosome Maintenance (MCM) Proteins as Novel Potential Tumor Markers for Meningiomas. J. Proteome Res. 2010, 9, 485–494. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, H.; Wang, Y.; Wang, Q. MCM2-7 in Clear Cell Renal Cell Carcinoma: MCM7 Promotes Tumor Cell Proliferation. Front. Oncol. 2021, 11, 782755. [Google Scholar] [CrossRef]
- Zheng, Y.; Shi, Y.; Yu, S.; Han, Y.; Kang, K.; Xu, H.; Gu, H.; Sang, X.; Chen, Y.; Wang, J. GTSE1, CDC20, PCNA, and MCM6 Synergistically Affect Regulations in Cell Cycle and Indicate Poor Prognosis in Liver Cancer. Anal. Cell Pathol. 2019, 2019, 1038069. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, M.; Zhou, Z.; Wang, W.; Duan, J.; Wu, G. Expression and Prognostic Value of MCM Family Genes in Osteosarcoma. Front. Mol. Biosci. 2021, 8, 668402. [Google Scholar] [CrossRef]
- Pierre, C.; Agopiantz, M.; Brunaud, L.; Battaglia-Hsu, S.-F.; Max, A.; Pouget, C.; Nomine, C.; Lomazzi, S.; Vignaud, J.-M.; Weryha, G.; et al. COPPS, a Composite Score Integrating Pathological Features, PS100 and SDHB Losses, Predicts the Risk of Metastasis and Progression-Free Survival in Pheochromocytomas/Paragangliomas. Virchows. Arch. 2019, 474, 721–734. [Google Scholar] [CrossRef]
- Hotton, J.; Agopiantz, M.; Leroux, A.; Charra-Brunaud, C.; Marie, B.; Busby-Venner, H.; Morel, O.; Guéant, J.-L.; Vignaud, J.-M.; Battaglia-Hsu, S.-F.; et al. Minichromosome Maintenance Complex Component 6 (MCM6) Expression Correlates with Histological Grade and Survival in Endometrioid Endometrial Adenocarcinoma. Virchows. Arch. 2018, 472, 623–633. [Google Scholar] [CrossRef]
- Fortin, J.-P.; Labbe, A.; Lemire, M.; Zanke, B.W.; Hudson, T.J.; Fertig, E.J.; Greenwood, C.M.; Hansen, K.D. Functional Normalization of 450k Methylation Array Data Improves Replication in Large Cancer Studies. Genome. Biol. 2014, 15, 503. [Google Scholar] [CrossRef]
- Capper, D.; Jones, D.T.W.; Sill, M.; Hovestadt, V.; Schrimpf, D.; Sturm, D.; Koelsche, C.; Sahm, F.; Chavez, L.; Reuss, D.E.; et al. DNA Methylation-Based Classification of Central Nervous System Tumours. Nature 2018, 555, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Sahm, F.; Schrimpf, D.; Stichel, D.; Jones, D.T.W.; Hielscher, T.; Schefzyk, S.; Okonechnikov, K.; Koelsche, C.; Reuss, D.E.; Capper, D.; et al. DNA Methylation-Based Classification and Grading System for Meningioma: A Multicentre, Retrospective Analysis. Lancet Oncol. 2017, 18, 682–694. [Google Scholar] [CrossRef] [PubMed]
- Hergalant, S.; Saurel, C.; Divoux, M.; Rech, F.; Pouget, C.; Godfraind, C.; Rouyer, P.; Lacomme, S.; Battaglia-Hsu, S.-F.; Gauchotte, G. Correlation between DNA Methylation and Cell Proliferation Identifies New Candidate Predictive Markers in Meningioma. Cancers 2022, 14, 6227. [Google Scholar] [CrossRef] [PubMed]
- Budczies, J.; Klauschen, F.; Sinn, B.V.; Győrffy, B.; Schmitt, W.D.; Darb-Esfahani, S.; Denkert, C. Cutoff Finder: A Comprehensive and Straightforward Web Application Enabling Rapid Biomarker Cutoff Optimization. PLoS ONE 2012, 7, e51862. [Google Scholar] [CrossRef]
- Yang, X.; Han, H.; De Carvalho, D.D.; Lay, F.D.; Jones, P.A.; Liang, G. Gene Body Methylation Can Alter Gene Expression and Is a Therapeutic Target in Cancer. Cancer Cell 2014, 26, 577–590. [Google Scholar] [CrossRef]
- Slieker, R.C.; Bos, S.D.; Goeman, J.J.; Bovée, J.V.; Talens, R.P.; van der Breggen, R.; Suchiman, H.E.D.; Lameijer, E.-W.; Putter, H.; van den Akker, E.B.; et al. Identification and Systematic Annotation of Tissue-Specific Differentially Methylated Regions Using the Illumina 450k Array. Epigenetics Chromatin 2013, 6, 26. [Google Scholar] [CrossRef]
- Nassiri, F.; Liu, J.; Patil, V.; Mamatjan, Y.; Wang, J.Z.; Hugh-White, R.; Macklin, A.M.; Khan, S.; Singh, O.; Karimi, S.; et al. A Clinically Applicable Integrative Molecular Classification of Meningiomas. Nature 2021, 597, 119–125. [Google Scholar] [CrossRef]
- Wong, P.G.; Glozak, M.A.; Cao, T.V.; Vaziri, C.; Seto, E.; Alexandrow, M. Chromatin Unfolding by Cdt1 Regulates MCM Loading via Opposing Functions of HBO1 and HDAC11-Geminin. Cell Cycle 2010, 9, 4351–4363. [Google Scholar] [CrossRef]
- Liu, Y.; He, G.; Wang, Y.; Guan, X.; Pang, X.; Zhang, B. MCM-2 Is a Therapeutic Target of Trichostatin A in Colon Cancer Cells. Toxicol. Lett. 2013, 221, 23–30. [Google Scholar] [CrossRef]
- Liu, M.; Hu, Q.; Tu, M.; Wang, X.; Yang, Z.; Yang, G.; Luo, R. MCM6 Promotes Metastasis of Hepatocellular Carcinoma via MEK/ERK Pathway and Serves as a Novel Serum Biomarker for Early Recurrence. J. Exp. Clin. Cancer Res. 2018, 37, 10. [Google Scholar] [CrossRef]
- Hsu, E.-C.; Shen, M.; Aslan, M.; Liu, S.; Kumar, M.; Garcia-Marques, F.; Nguyen, H.M.; Nolley, R.; Pitteri, S.J.; Corey, E.; et al. MCM2-7 Complex Is a Novel Druggable Target for Neuroendocrine Prostate Cancer. Sci. Rep. 2021, 11, 13305. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, H.; Liu, W.; Yan, H.; Zhang, Y.; Cheung, A.H.K.; Zhang, J.; Chen, B.; Liang, L.; Zhou, Z.; et al. MCM6 Is a Critical Transcriptional Target of YAP to Promote Gastric Tumorigenesis and Serves as a Therapeutic Target. Theranostics 2022, 12, 6509–6526. [Google Scholar] [CrossRef] [PubMed]

| Variable | Grade 1 n = 100 | Grade 2 n = 69 |
|---|---|---|
| Age (mean; min.-max.) | 54 (29–79) y.o. | 62 (33–91) y.o. |
| Sex | F: 83 M: 17 | F: 39 M: 30 |
| Gross total resection | 86% | 62% |
| Localization | Convexity: 32 Falcorial/parafalcorial: 15 Skullbase: 48 Spinal: 5 | Convexity: 46 Falcorial/parafalcorial: 8 Skullbase: 12 Ventricular: 3 |
| Adjuvant radiotherapy | 8% | 56% |
| Progression | 18% | 51% |
| Death | 11% | 21% |
| Marker | Inter-Observer | Inter-Laboratory |
|---|---|---|
| Ki-67 | 0.85 | 0.81 |
| MCM6 | 0.87 | 0.87 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gauchotte, G.; Bédel, C.; Lardenois, E.; Hergalant, S.; Cuglietta, L.; Pflaum, R.; Lacomme, S.; Pina, H.; Treffel, M.; Rech, F.; et al. A High MCM6 Proliferative Index in Atypical Meningioma Is Associated with Shorter Progression Free and Overall Survivals. Cancers 2023, 15, 535. https://doi.org/10.3390/cancers15020535
Gauchotte G, Bédel C, Lardenois E, Hergalant S, Cuglietta L, Pflaum R, Lacomme S, Pina H, Treffel M, Rech F, et al. A High MCM6 Proliferative Index in Atypical Meningioma Is Associated with Shorter Progression Free and Overall Survivals. Cancers. 2023; 15(2):535. https://doi.org/10.3390/cancers15020535
Chicago/Turabian StyleGauchotte, Guillaume, Charles Bédel, Emilie Lardenois, Sébastien Hergalant, Laura Cuglietta, Robin Pflaum, Stéphanie Lacomme, Héloïse Pina, Mathilde Treffel, Fabien Rech, and et al. 2023. "A High MCM6 Proliferative Index in Atypical Meningioma Is Associated with Shorter Progression Free and Overall Survivals" Cancers 15, no. 2: 535. https://doi.org/10.3390/cancers15020535
APA StyleGauchotte, G., Bédel, C., Lardenois, E., Hergalant, S., Cuglietta, L., Pflaum, R., Lacomme, S., Pina, H., Treffel, M., Rech, F., & Battaglia-Hsu, S.-F. (2023). A High MCM6 Proliferative Index in Atypical Meningioma Is Associated with Shorter Progression Free and Overall Survivals. Cancers, 15(2), 535. https://doi.org/10.3390/cancers15020535







