Measurable Residual Disease Monitoring by Locked Nucleic Acid Quantitative Real-Time PCR Assay for IDH1/2 Mutation in Adult AML
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. Detection of Gene Mutations
2.3. Monitoring of IDH1/2 MRD by LNA-qPCR Assay
2.4. Detection of NPM1 RT-qPCR MRD
2.5. Statistical Analysis
3. Results
3.1. Patient Characteristics
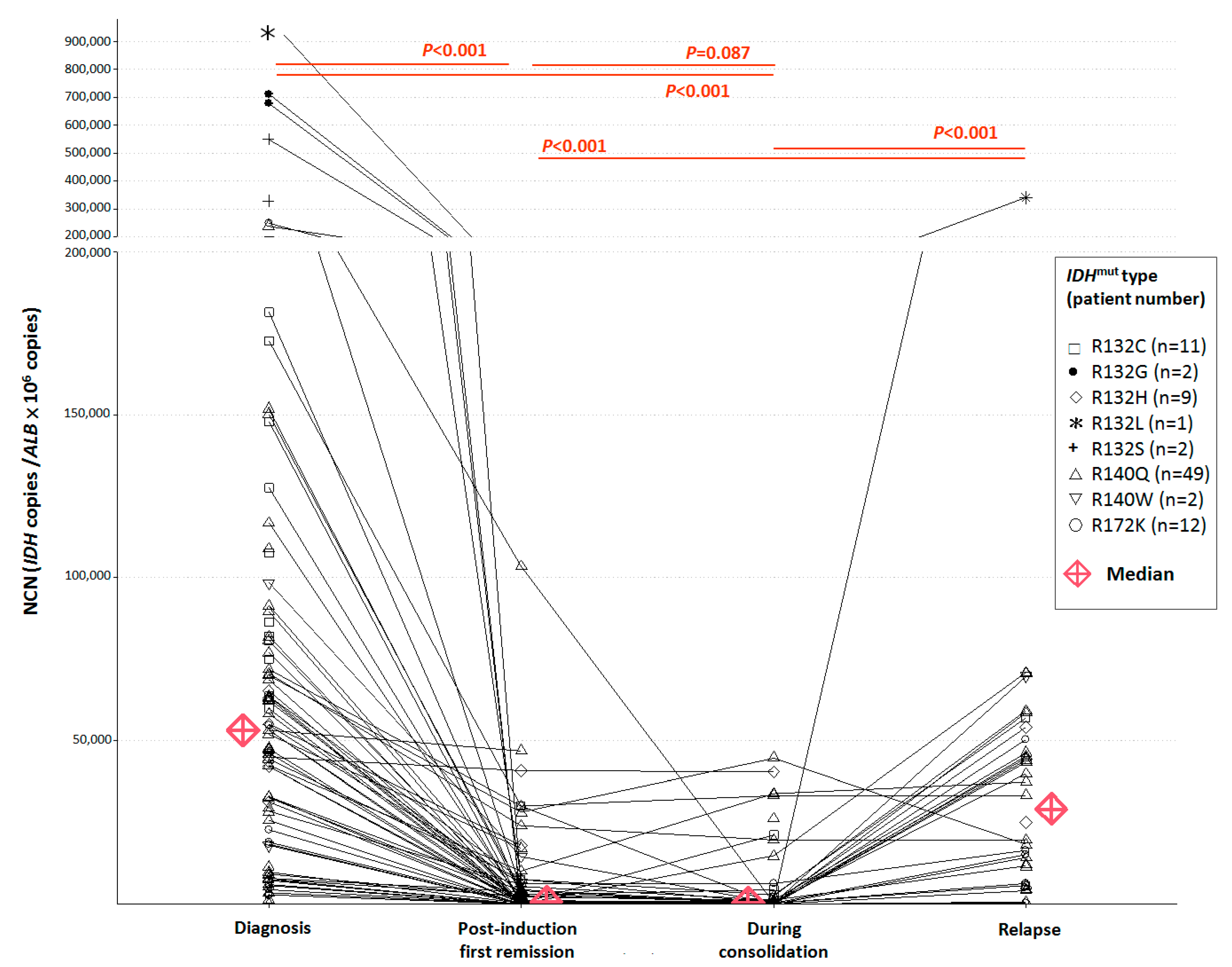
3.2. NCN of IDH1/2 Mutations in Pre-Treatment and Post-Treatment BM Samples
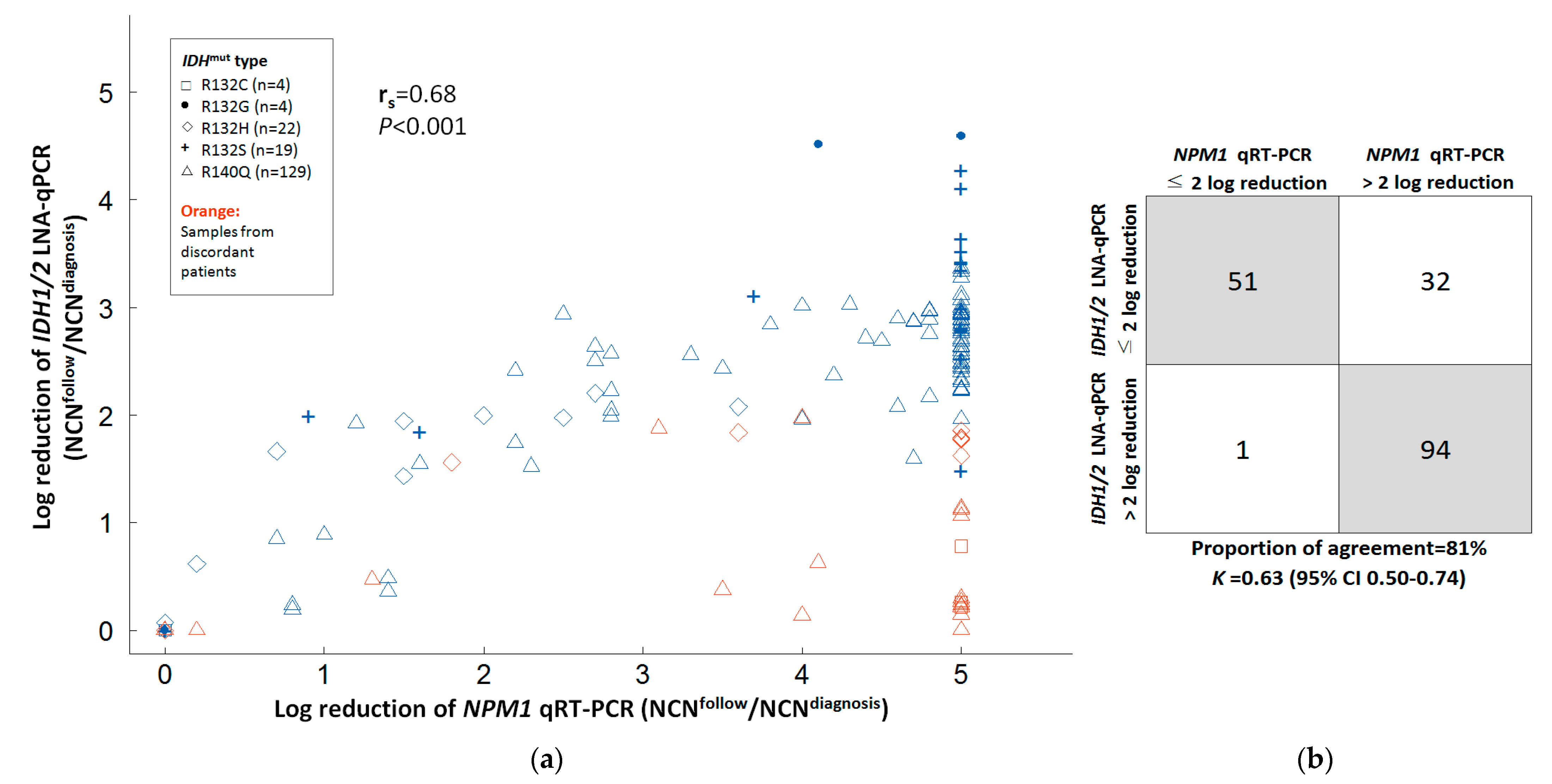
3.3. Correlation of IDH1/2 LNA-qPCR MRD with NPM1 RT-qPCR MRD and Hematologic Remission Status
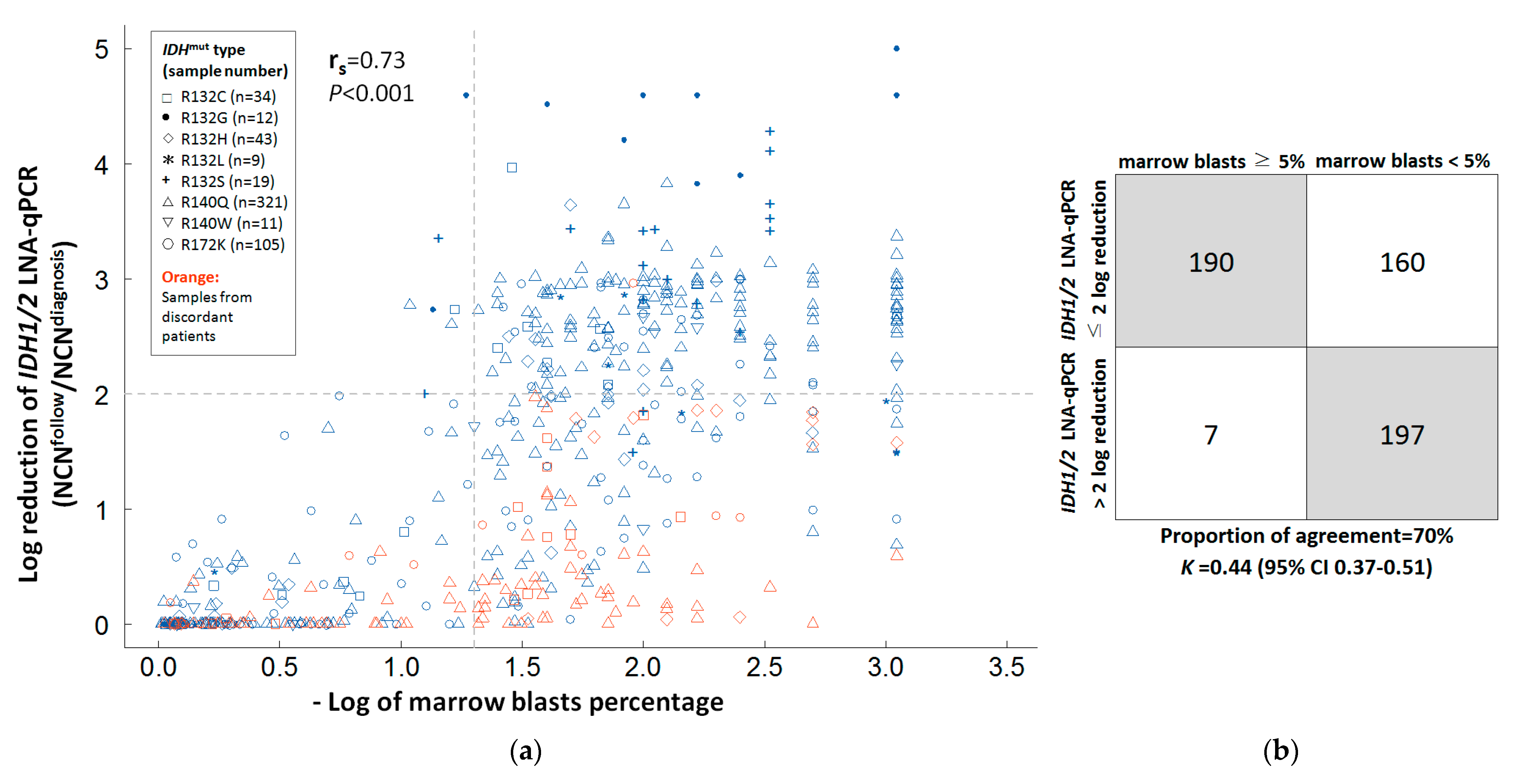
3.4. Correlation between IDH1/2 LNA-qPCR MRD, NPM1 RT-qPCR, and Marrow Blasts in Serial BM Samples
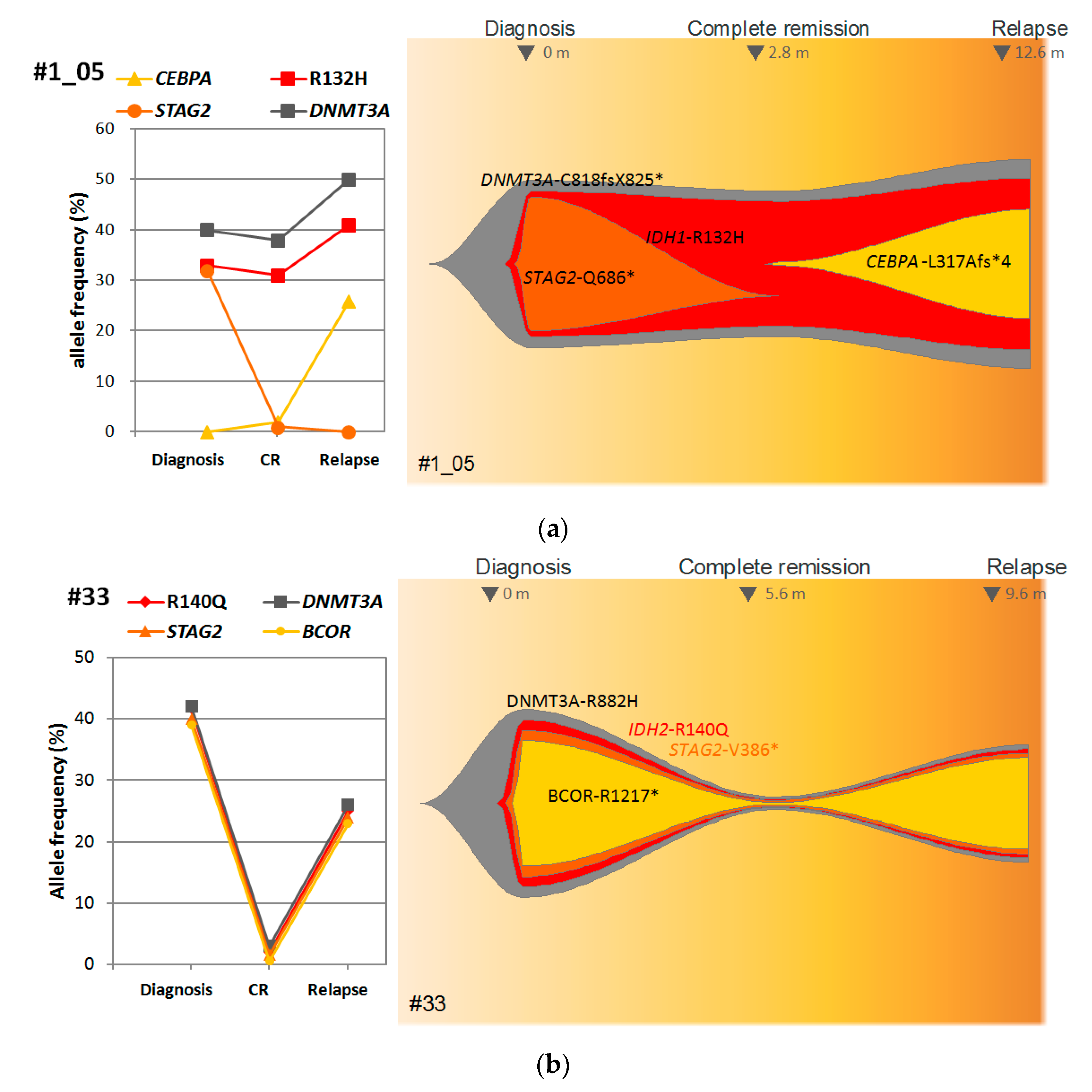
3.5. Correlation of IDH1/2 LNA-qPCR MRD with Clinical and Genetic Characteristics of AML Patients
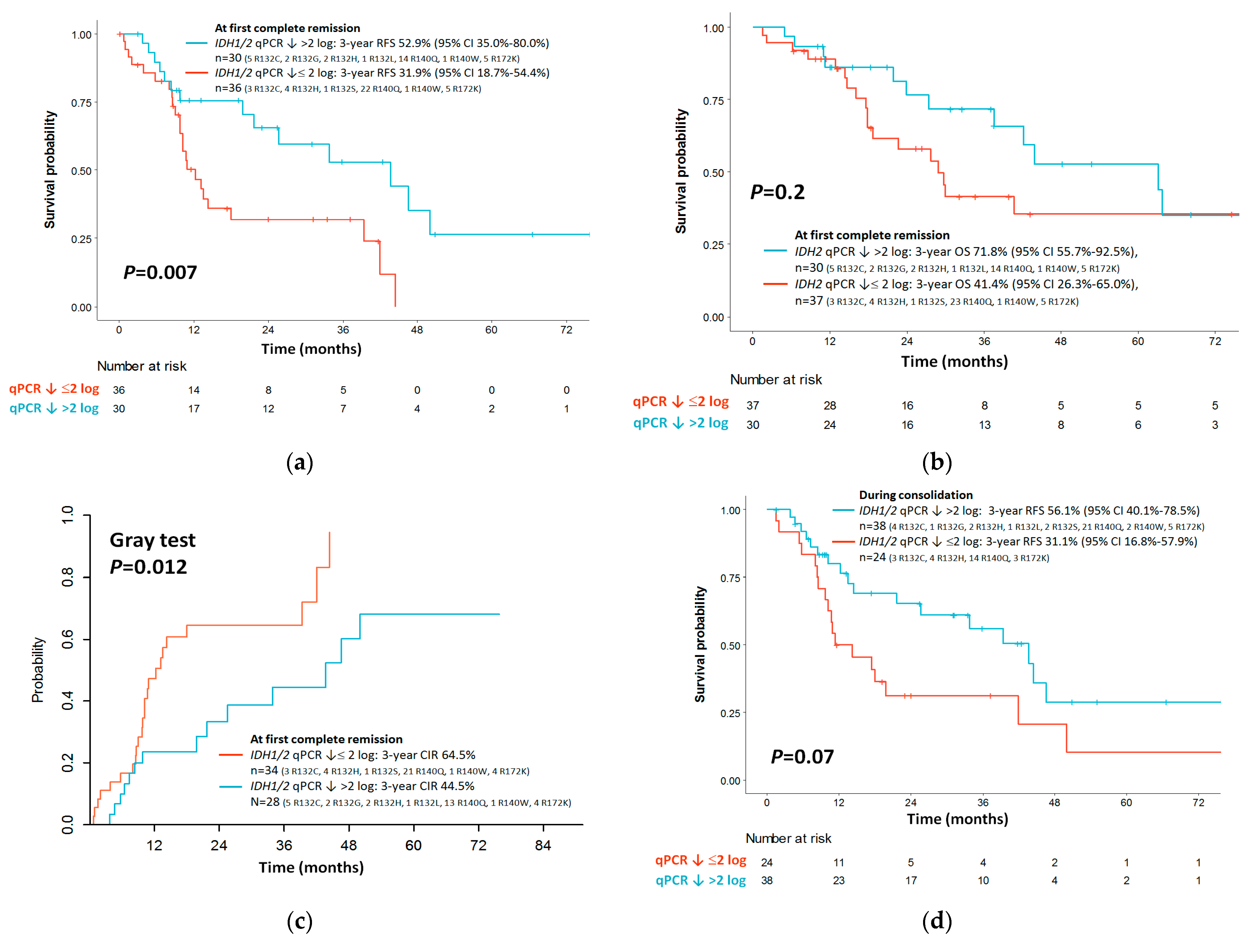
3.6. Outcome Impact of IDH1/2 LNA-qPCR MRD at the End of Induction
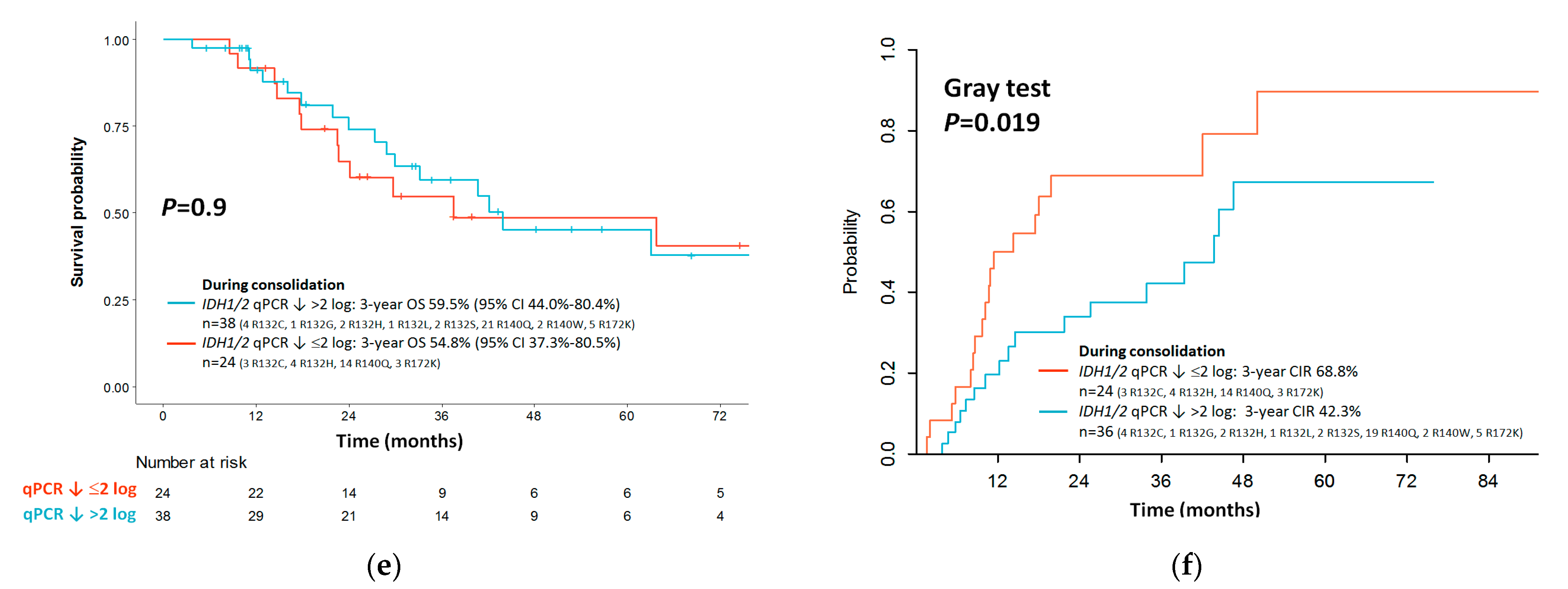
3.7. Outcome Impact of IDH1/2 LNA-qPCR MRD during Consolidation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Dohner, H.; Weisdorf, D.J.; Bloomfield, C.D. Acute Myeloid Leukemia. N. Engl. J. Med. 2015, 373, 1136–1152. [Google Scholar] [CrossRef] [PubMed]
- Ley, T.J.; Miller, C.; Ding, L.; Raphael, B.J.; Mungall, A.J.; Robertson, A.; Hoadley, K.; Triche, T.J., Jr.; Laird, P.W.; Baty, J.D.; et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Wei, A.H.; Appelbaum, F.R.; Craddock, C.; DiNardo, C.D.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Godley, L.A.; Hasserjian, R.P.; et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood 2022, 140, 1345–1377. [Google Scholar] [CrossRef] [PubMed]
- Kats, L.M.; Reschke, M.; Taulli, R.; Pozdnyakova, O.; Burgess, K.; Bhargava, P.; Straley, K.; Karnik, R.; Meissner, A.; Small, D.; et al. Proto-oncogenic role of mutant IDH2 in leukemia initiation and maintenance. Cell Stem Cell 2014, 14, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Ward, P.S.; Kapoor, G.S.; Rohle, D.; Turcan, S.; Abdel-Wahab, O.; Edwards, C.R.; Khanin, R.; Figueroa, M.E.; Melnick, A.; et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 2012, 483, 474–478. [Google Scholar] [CrossRef]
- Losman, J.A.; Looper, R.E.; Koivunen, P.; Lee, S.; Schneider, R.K.; McMahon, C.; Cowley, G.S.; Root, D.E.; Ebert, B.L.; Kaelin, W.G., Jr. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science 2013, 339, 1621–1625. [Google Scholar] [CrossRef]
- Gross, S.; Cairns, R.A.; Minden, M.D.; Driggers, E.M.; Bittinger, M.A.; Jang, H.G.; Sasaki, M.; Jin, S.; Schenkein, D.P.; Su, S.M.; et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J. Exp. Med. 2010, 207, 339–344. [Google Scholar] [CrossRef]
- Abbas, S.; Lugthart, S.; Kavelaars, F.G.; Schelen, A.; Koenders, J.E.; Zeilemaker, A.; van Putten, W.J.; Rijneveld, A.W.; Löwenberg, B.; Valk, P.J. Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia: Prevalence and prognostic value. Blood 2010, 116, 2122–2126. [Google Scholar] [CrossRef]
- Boissel, N.; Nibourel, O.; Renneville, A.; Gardin, C.; Reman, O.; Contentin, N.; Bordessoule, D.; Pautas, C.; de Revel, T.; Quesnel, B.; et al. Prognostic Impact of Isocitrate Dehydrogenase Enzyme Isoforms 1 and 2 Mutations in Acute Myeloid Leukemia: A Study by the Acute Leukemia French Association Group. J. Clin. Oncol. 2010, 28, 3717–3723. [Google Scholar] [CrossRef]
- Marcucci, G.; Maharry, K.; Wu, Y.Z.; Radmacher, M.D.; Mrózek, K.; Margeson, D.; Holland, K.B.; Whitman, S.P.; Becker, H.; Schwind, S.; et al. IDH1 and IDH2 Gene Mutations Identify Novel Molecular Subsets Within De Novo Cytogenetically Normal Acute Myeloid Leukemia: A Cancer and Leukemia Group B Study. J. Clin. Oncol. 2010, 28, 2348–2355. [Google Scholar] [CrossRef]
- Schnittger, S.; Haferlach, C.; Ulke, M.; Alpermann, T.; Kern, W.; Haferlach, T. IDH1 mutations are detected in 6.6% of 1414 AML patients and are associated with intermediate risk karyotype and unfavorable prognosis in adults younger than 60 years and unmutated NPM1 status. Blood 2010, 116, 5486–5496. [Google Scholar] [CrossRef] [PubMed]
- Paschka, P.; Schlenk, R.F.; Gaidzik, V.I.; Habdank, M.; Krönke, J.; Bullinger, L.; Späth, D.; Kayser, S.; Zucknick, M.; Götze, K.; et al. IDH1 and IDH2 Mutations Are Frequent Genetic Alterations in Acute Myeloid Leukemia and Confer Adverse Prognosis in Cytogenetically Normal Acute Myeloid Leukemia With NPM1 Mutation without FLT3 Internal Tandem Duplication. J. Clin. Oncol. 2010, 28, 3636–3643. [Google Scholar] [CrossRef] [PubMed]
- Green, C.L.; Evans, C.M.; Zhao, L.; Hills, R.K.; Burnett, A.K.; Linch, D.C.; Gale, R.E. The prognostic significance of IDH2 mutations in AML depends on the location of the mutation. Blood 2011, 118, 409–412. [Google Scholar] [CrossRef]
- Ivey, A.; Hills, R.K.; Simpson, M.A.; Jovanovic, J.V.; Gilkes, A.; Grech, A.; Patel, Y.; Bhudia, N.; Farah, H.; Mason, J.; et al. Assessment of Minimal Residual Disease in Standard-Risk AML. N. Engl. J. Med. 2016, 374, 422–433. [Google Scholar] [CrossRef]
- Terwijn, M.; van Putten, W.L.; Kelder, A.; van der Velden, V.H.; Brooimans, R.A.; Pabst, T.; Maertens, J.; Boeckx, N.; de Greef, G.E.; Valk, P.J.; et al. High prognostic impact of flow cytometric minimal residual disease detection in acute myeloid leukemia: Data from the HOVON/SAKK AML 42A study. J. Clin. Oncol. 2013, 31, 3889–3897. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.A.; O’Brien, M.A.; Hills, R.K.; Daly, S.B.; Wheatley, K.; Burnett, A.K. Minimal residual disease monitoring by quantitative RT-PCR in core binding factor AML allows risk stratification and predicts relapse: Results of the United Kingdom MRC AML-15 trial. Blood 2012, 120, 2826–2835. [Google Scholar] [CrossRef]
- Krönke, J.; Schlenk, R.F.; Jensen, K.O.; Tschürtz, F.; Corbacioglu, A.; Gaidzik, V.I.; Paschka, P.; Onken, S.; Eiwen, K.; Habdank, M.; et al. Monitoring of minimal residual disease in NPM1-mutated acute myeloid leukemia: A study from the German-Austrian acute myeloid leukemia study group. J. Clin. Oncol. 2011, 29, 2709–2716. [Google Scholar] [CrossRef]
- Brambati, C.; Galbiati, S.; Xue, E.; Toffalori, C.; Crucitti, L.; Greco, R.; Sala, E.; Crippa, A.; Chiesa, L.; Soriani, N.; et al. Droplet digital polymerase chain reaction for DNMT3A and IDH1/2 mutations to improve early detection of acute myeloid leukemia relapse after allogeneic hematopoietic stem cell transplantation. Haematologica 2016, 101, e157–e161. [Google Scholar] [CrossRef]
- Ferret, Y.; Boissel, N.; Helevaut, N.; Madic, J.; Nibourel, O.; Marceau-Renaut, A.; Bucci, M.; Geffroy, S.; Celli-Lebras, K.; Castaigne, S.; et al. Clinical relevance of IDH1/2 mutant allele burden during follow-up in acute myeloid leukemia. A study by the French ALFA group. Haematologica 2018, 103, 822–829. [Google Scholar] [CrossRef]
- Ok, C.Y.; Loghavi, S.; Sui, D.; Wei, P.; Kanagal-Shamanna, R.; Yin, C.C.; Zuo, Z.; Routbort, M.J.; Tang, G.; Tang, Z.; et al. Persistent IDH1/2 mutations in remission can predict relapse in patients with acute myeloid leukemia. Haematologica 2019, 104, 305–311. [Google Scholar] [CrossRef]
- Shumilov, E.; Flach, J.; Joncourt, R.; Porret, N.; Wiedemann, G.; Novak, U.; Gfeller, E.; Jeker, B.; Amstutz, U.; Pabst, T.; et al. Clinical value of molecular MRD monitoring by next-generation sequencing in patients with IDH2 mutated AML. Leuk. Lymphoma 2019, 60, 2588–2590. [Google Scholar] [CrossRef] [PubMed]
- Debarri, H.; Lebon, D.; Roumier, C.; Cheok, M.; Marceau-Renaut, A.; Nibourel, O.; Geffroy, S.; Helevaut, N.; Rousselot, P.; Gruson, B.; et al. IDH1/2 but not DNMT3A mutations are suitable targets for minimal residual disease monitoring in acute myeloid leukemia patients: A study by the Acute Leukemia French Association. Oncotarget 2015, 6, 42345–42353. [Google Scholar] [CrossRef] [PubMed]
- Jeziskova, I.; Razga, F.; Toskova, M.; Dvorakova, D.; Timilsina, S.; Mayer, J.; Racil, Z. Quantitative detection of IDH2 mutation for minimal residual disease monitoring in patients with acute myeloid leukemia and its comparison with mutations in NPM1 gene. Leuk. Lymphoma 2013, 54, 867–870. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, E.; Besbes, S.; Renneville, A.; Nibourel, O.; Helevaut, N.; Preudhomme, C.; Soua, Z. Minimal Residual Disease assessment of IDH1/2 mutations in Acute Myeloid Leukemia by LNA-RQ-PCR. Tunis. Med. 2016, 94, 190–197. [Google Scholar]
- Chou, W.C.; Peng, K.Y.; Lei, W.C.; Ko, B.S.; Tsay, W.; Kuo, C.H.; Tien, H.F. Persistence of mutant isocitrate dehydrogenase in patients with acute myeloid leukemia in remission. Leukemia 2012, 26, 527–529. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Heuser, M.; Freeman, S.D.; Ossenkoppele, G.J.; Buccisano, F.; Hourigan, C.S.; Ngai, L.L.; Tettero, J.M.; Bachas, C.; Baer, C.; Béné, M.C. 2021 Update on MRD in acute myeloid leukemia: A consensus document from the European LeukemiaNet MRD Working Party. Blood 2021, 138, 2753–2767. [Google Scholar] [CrossRef]
- Khoury, J.D.; Solary, E.; Abla, O.; Akkari, Y.; Alaggio, R.; Apperley, J.F.; Bejar, R.; Berti, E.; Busque, L.; Chan, J.K.C.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia 2022, 36, 1703–1719. [Google Scholar] [CrossRef]
- Kao, H.W.; Liang, D.C.; Wu, J.H.; Kuo, M.C.; Wang, P.N.; Yang, C.P.; Shih, Y.S.; Lin, T.H.; Huang, Y.H.; Shih, L.Y. Gene mutation patterns in patients with minimally differentiated acute myeloid leukemia. Neoplasia 2014, 16, 481–488. [Google Scholar] [CrossRef]
- Pongers-Willemse, M.J.; Verhagen, O.J.; Tibbe, G.J.; Wijkhuijs, A.J.; de Haas, V.; Roovers, E.; van der Schoot, C.E.; van Dongen, J.J. Real-time quantitative PCR for the detection of minimal residual disease in acute lymphoblastic leukemia using junctional region specific TaqMan probes. Leukemia 1998, 12, 2006–2014. [Google Scholar] [CrossRef]
- Gorello, P.; Cazzaniga, G.; Alberti, F.; Dell’Oro, M.G.; Gottardi, E.; Specchia, G.; Roti, G.; Rosati, R.; Martelli, M.F.; Diverio, D.; et al. Quantitative assessment of minimal residual disease in acute myeloid leukemia carrying nucleophosmin (NPM1) gene mutations. Leukemia 2006, 20, 1103–1108. [Google Scholar] [CrossRef]
- Di Giusto, D.A.; King, G.C. Strong positional preference in the interaction of LNA oligonucleotides with DNA polymerase and proofreading exonuclease activities: Implications for genotyping assays. Nucleic Acids Res. 2004, 32, e32. [Google Scholar] [CrossRef] [PubMed]
- Petrova, L.; Vrbacky, F.; Lanska, M.; Zavrelova, A.; Zak, P.; Hrochova, K. IDH1 and IDH2 mutations in patients with acute myeloid leukemia: Suitable targets for minimal residual disease monitoring? Clin. Biochem. 2018, 61, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Shao, Y.; Tian, L.; Flasch, D.A.; Mulder, H.L.; Edmonson, M.N.; Liu, Y.; Chen, X.; Newman, S.; Nakitandwe, J.; et al. Analysis of error profiles in deep next-generation sequencing data. Genome Biol. 2019, 20, 50. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Mosier, S.; Gocke, C.D.; Lin, M.T.; Eshleman, J.R. Cytosine deamination is a major cause of baseline noise in next-generation sequencing. Mol. Diagn. Ther. 2014, 18, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Shlush, L.I.; Zandi, S.; Mitchell, A.; Chen, W.C.; Brandwein, J.M.; Gupta, V.; Kennedy, J.A.; Schimmer, A.D.; Schuh, A.C.; Yee, K.W.; et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 2014, 506, 328–333. [Google Scholar] [CrossRef]
- Cappelli, L.V.; Meggendorfer, M.; Baer, C.; Nadarajah, N.; Hutter, S.; Jeromin, S.; Dicker, F.; Kern, W.; Haferlach, T.; Haferlach, C.; et al. Indeterminate and oncogenic potential: CHIP vs. CHOP mutations in AML with NPM1 alteration. Leukemia 2022, 36, 394–402. [Google Scholar] [CrossRef]
- Cloos, J.; Goemans, B.F.; Hess, C.J.; van Oostveen, J.W.; Waisfisz, Q.; Corthals, S.; de Lange, D.; Boeckx, N.; Hählen, K.; Reinhardt, D.; et al. Stability and prognostic influence of FLT3 mutations in paired initial and relapsed AML samples. Leukemia 2006, 20, 1217–1220. [Google Scholar] [CrossRef]
- Murphy, K.M.; Levis, M.; Hafez, M.J.; Geiger, T.; Cooper, L.C.; Smith, B.D.; Small, D.; Berg, K.D. Detection of FLT3 internal tandem duplication and D835 mutations by a multiplex polymerase chain reaction and capillary electrophoresis assay. J. Mol. Diagn. 2003, 5, 96–102. [Google Scholar] [CrossRef]
- Carbonell, D.; Chicano, M.; Cardero, A.J.; Gómez-Centurión, I.; Bailén, R.; Oarbeascoa, G.; Martínez-Señarís, D.; Franco, C.; Muñiz, P.; Anguita, J.; et al. FLT3-ITD Expression as a Potential Biomarker for the Assessment of Treatment Response in Patients with Acute Myeloid Leukemia. Cancers 2022, 14, 4006. [Google Scholar] [CrossRef]
- Levis, M.J.; Perl, A.E.; Altman, J.K.; Gocke, C.D.; Bahceci, E.; Hill, J.; Liu, C.; Xie, Z.; Carson, A.R.; McClain, V.; et al. A next-generation sequencing-based assay for minimal residual disease assessment in AML patients with FLT3-ITD mutations. Blood Adv. 2018, 2, 825–831. [Google Scholar] [CrossRef]
- Scholl, S.; Krause, C.; Loncarevic, I.F.; Müller, R.; Kunert, C.; Wedding, U.; Sayer, H.G.; Clement, J.H.; Höffken, K. Specific detection of Flt3 point mutations by highly sensitive real-time polymerase chain reaction in acute myeloid leukemia. J. Lab. Clin. Med. 2005, 145, 295–304. [Google Scholar] [CrossRef]
- Schnittger, S.; Kern, W.; Tschulik, C.; Weiss, T.; Dicker, F.; Falini, B.; Haferlach, C.; Haferlach, T. Minimal residual disease levels assessed by NPM1 mutation-specific RQ-PCR provide important prognostic information in AML. Blood 2009, 114, 2220–2231. [Google Scholar] [CrossRef] [PubMed]
- Krönke, J.; Bullinger, L.; Teleanu, V.; Tschürtz, F.; Gaidzik, V.I.; Kühn, M.W.; Rücker, F.G.; Holzmann, K.; Paschka, P.; Kapp-Schwörer, S.; et al. Clonal evolution in relapsed NPM1-mutated acute myeloid leukemia. Blood 2013, 122, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Papadaki, C.; Dufour, A.; Seibl, M.; Schneider, S.; Bohlander, S.K.; Zellmeier, E.; Mellert, G.; Hiddemann, W.; Spiekermann, K. Monitoring minimal residual disease in acute myeloid leukaemia with NPM1 mutations by quantitative PCR: Clonal evolution is a limiting factor. Br. J. Haematol. 2009, 144, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Pettersson, L.; Levéen, P.; Axler, O.; Dvorakova, D.; Juliusson, G.; Ehinger, M. Improved minimal residual disease detection by targeted quantitative polymerase chain reaction in Nucleophosmin 1 type a mutated acute myeloid leukemia. Genes Chromosomes Cancer 2016, 55, 750–766. [Google Scholar] [CrossRef]
- Thol, F.; Kölking, B.; Damm, F.; Reinhardt, K.; Klusmann, J.H.; Reinhardt, D.; von Neuhoff, N.; Brugman, M.H.; Schlegelberger, B.; Suerbaum, S.; et al. Next-generation sequencing for minimal residual disease monitoring in acute myeloid leukemia patients with FLT3-ITD or NPM1 mutations. Genes Chromosomes Cancer 2012, 51, 689–695. [Google Scholar] [CrossRef] [PubMed]
- Hubmann, M.; Köhnke, T.; Hoster, E.; Schneider, S.; Dufour, A.; Zellmeier, E.; Fiegl, M.; Braess, J.; Bohlander, S.K.; Subklewe, M.; et al. Molecular response assessment by quantitative real-time polymerase chain reaction after induction therapy in NPM1-mutated patients identifies those at high risk of relapse. Haematologica 2014, 99, 1317–1325. [Google Scholar] [CrossRef] [PubMed]
- Bill, M.; Jentzsch, M.; Bischof, L.; Kohlschmidt, J.; Grimm, J.; Schmalbrock, L.K.; Backhaus, D.; Brauer, D.; Goldmann, K.; Franke, G.N.; et al. Impact of IDH1 and IDH2 mutation detection at diagnosis and in remission in patients with AML receiving allogeneic transplantation. Blood Adv. 2022. online ahead of print. [Google Scholar] [CrossRef]
- Bataller, A.; Oñate, G.; Diaz-Beyá, M.; Sampol, A.; Coll, R.; Vall-Llovera, F.; Oliver-Caldés, A. Acute myeloid leukemia with NPM1 mutation and favorable European LeukemiaNet category: Outcome after preemptive intervention based on measurable residual disease. Br. J. Haematol. 2020, 191, 52–61. [Google Scholar] [CrossRef]
- Platzbecker, U.; Middeke, J.M.; Sockel, K.; Herbst, R.; Wolf, D.; Baldus, C.D.; Oelschlägel, U.; Mütherig, A.; Fransecky, L.; Noppeney, R.; et al. Measurable residual disease-guided treatment with azacitidine to prevent haematological relapse in patients with myelodysplastic syndrome and acute myeloid leukaemia (RELAZA2): An open-label, multicentre, phase 2 trial. Lancet Oncol. 2018, 19, 1668–1679. [Google Scholar] [CrossRef]

| Target | Reverse Primer (5′-3′) | Forward Primer (5′-3′) | Probe (5′-3′) |
|---|---|---|---|
| IDH1-R132C | R-LNA-0: GATCCCCATAAGCATGACA | CGGTCTTCAGAGAAGCCATT | FAM-ATGGGTAAAACCTATCATC-MGB |
| IDH1-R132H | R-LNA-0: CTTGATCCCCATAAGCATGAT | ||
| IDH1-R132S | R-LNA-0: GATCCCCATAAGCATGACT | ||
| IDH1-R132G | R-LNA-0: GATCCCCATAAGCATGACC | ||
| IDH1-R132L | R-LNA-0: CTTGATCCCCATAAGCATGAA | ||
| IDH2-R140Q | R-LNA-0: GTCCCCCCCAGGATGTTCT | AGTTCAAGCTGAAGAAGATGTGG | FAM-AGTCCCAATGGAACTA-MGB |
| IDH2-R140W | R-LNA-0: GTCCCCCCCAGGATGTTCCA | ||
| IDH2-R172K | GTCGCCATGGGCGTGCT | TCCCACGCCTAGTCCCTGGCTG | FAM-AGCCCATCACCAT-MGB |
| ALB | CTCTCCTTCTCAGAAAGTGTGCATAT | TGAAACATACGTTCCCAAAGAGTTT | FAM-TGCTGAAACATTCACCTTCCATGCAGA -TAMRA |
| Variables | All (N = 88) | IDH1 (N = 25) | IDH2 (N = 63) | p |
|---|---|---|---|---|
| Median age, range (years) * | 54 (18–86) | 54 (36–68) | 55 (18–86) | 0.582 |
| Gender | 0.587 | |||
| Female, n (%)† | 47 (53) | 15 (60) | 32 (51) | |
| Male, n (%)† | 41 (47) | 10 (40) | 31 (49) | |
| Hemoglobin, range (g/dL) * | 7.9 (3.7–14.3) | 8.1 (3.7–12.6) | 7.8 (3.8–14.3) | 0.627 |
| White blood cell count, range (× 109/L) * | 12.8 (0.7–354.6) | 11.2 (0.9–162.0) | 13.2 (0.7–355.0) | 0.701 |
| Platelet count, range (× 109/L) * | 66 (0–382) | 47 (7–194) | 70 (0–382) | 0.134 |
| Circulating blasts, range (%) * | 47 (0–99) | 80 (6–99) | 37 (0–99) | 0.005 |
| Marrow blasts, range (%) * | 67 (19–96) | 86 (20–96) | 61 (19–94) | 0.627 |
| 2022 WHO AML classification † | ||||
| AML with NPM1 mutation | 42 (48) | 11 (44) | 31 (49) | |
| AML with CEBPA mutation | 2 (2) | 0 (0) | 2 (3) | |
| AML with TP53 mutation | 1 (1) | 1 (4) | 0 (0) | |
| AML, myelodysplasia-related | 18 (20) | 5 (20) | 13 (21) | |
| AML with minimal differentiation | 1 (1) | 0 (0) | 1 (2) | |
| AML without maturation | 6 (7) | 4 (16) | 2 (3) | |
| AML with maturation | 16 (18) | 3 (12) | 13 (21) | |
| Acute myelomonocytic leukemia | 2 (2) | 1 (4) | 1 (2) | |
| Cytogenetic risk † | 0.755 | |||
| Normal, n (%) | 56 (64) | 14 (56) | 42 (67) | |
| Intermediate risk, n (%) | 15 (17)) | 4 (16) | 11 (17)) | |
| Adverse risk, n (%) | 8 (9) | 3 (12) | 5 (8) | |
| Unknown, n (%) | 9 (10) | 4 (16) | 5 (8) | |
| 2022 ELN risk group † | 0.384 | |||
| Favorable, n (%) | 31 (35) | 6 (24) | 25 (40) | |
| Intermediate, n (%) | 32 (36) | 9 (36) | 23 (37) | |
| Unfavorable, n (%) | 22 (25) | 8 (32) | 14 (22) | |
| Unknown | 3 (4) | 2 (8) | 1 (2) | |
| Treatment † | 0.534 | |||
| Standard chemotherapy, n (%) | 84 (95) | 24 (96) | 60 (95) | |
| Azacitidine, n (%) | 2 (2) | 1 (4) | 1 (2) | |
| Low-dose cytarabine, n (%) | 2 (2) | 0 (0) | 2 (3) | |
| IDH1/2 mutation † | ||||
| IDH1-R132C/H/G/S/L, n (%) | 25 (28) | 11/9/2/2/1 | - | |
| IDH2-R140Q/W, n (%) | 51 (58) | - | 49/2 (81) | |
| IDH1-R172K, n (%) | 12 (14) | - | 12 (19) | |
| Median IDH1/2 NCN at diagnosis, range (/ALB × 106) | 53,228 (87–980,686) | 80,920 (31,343–980,686) | 46,156 (87–247,796) | << 0.001 |
| Concurrent gene mutations † | ||||
| FLT3-ITD, n (%) | 20 (23) | 8 (32) | 12 (19) | |
| FLT3-TKD, n (%) | 6 (7) | 2 (8) # | 4 (6) | |
| EZH2, n (%) | 1 (1) | 0 (0) | 1 (2) | |
| RUNX1, n (%) | 7 (8) | 4 (16) | 3 (5) | |
| ASXL1, n (%) | 4 (5) | 1 (4) | 3 (5) | |
| STAG2, n (%) | 4 (5) | 1 (4) | 3 (5) | |
| SF3B1, n (%) | 1 (1) | 0 (0) | 1 (2) | |
| BCOR, n (%) | 3 (4) | 1 (4) | 2 (3) | |
| SRSF2, n (%) | 1 (1) | 0 (0) | 1 (2) |
| Factors | With Correlation (n = 69) | Without Correlation (n = 19) | p |
|---|---|---|---|
| Age, range (years) * | 53 (18–86) | 61 (23–74) | 0.025 |
| NCN at diagnosis (/ALB × 106) * | 53,281 (1252–980,686) | 44,929 (87–181,539) | 0.903 |
| Marrow blasts, range (%) * | 71 (19–96) | 60 (23–94) | 0.231 |
| WBC, range (× 109/L) * | 20.4 (0.7–355.0) | 10.3 (1.4–162.0) | 0.271 |
| Hemoglobin, range (g/dL) * | 7.8 (3.7–13.6) | 8.1 (4.3–14.3) | 0.520 |
| Platelet, range (× 109/L) * | 65 (7–382) | 69 (0–201) | 0.594 |
| Circulating blasts, range (%) * | 52 (0–98) | 44 (0–99) | 0.294 |
| FLT3-ITD/TKD † | 0.012 | ||
| Positive | 24 | 1 | |
| Negative | 45 | 18 | |
| NPM1 mutation † | 0.458 | ||
| Positive | 31 | 11 | |
| Negative | 38 | 8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kao, H.-W.; Kuo, M.-C.; Huang, Y.-J.; Chang, H.; Hu, S.-F.; Huang, C.-F.; Hung, Y.-S.; Lin, T.-L.; Ou, C.-W.; Lien, M.-Y.; et al. Measurable Residual Disease Monitoring by Locked Nucleic Acid Quantitative Real-Time PCR Assay for IDH1/2 Mutation in Adult AML. Cancers 2022, 14, 6205. https://doi.org/10.3390/cancers14246205
Kao H-W, Kuo M-C, Huang Y-J, Chang H, Hu S-F, Huang C-F, Hung Y-S, Lin T-L, Ou C-W, Lien M-Y, et al. Measurable Residual Disease Monitoring by Locked Nucleic Acid Quantitative Real-Time PCR Assay for IDH1/2 Mutation in Adult AML. Cancers. 2022; 14(24):6205. https://doi.org/10.3390/cancers14246205
Chicago/Turabian StyleKao, Hsiao-Wen, Ming-Chung Kuo, Ying-Jung Huang, Hung Chang, Shu-Fen Hu, Chein-Fuang Huang, Yu-Shin Hung, Tung-Liang Lin, Che-Wei Ou, Ming-Yu Lien, and et al. 2022. "Measurable Residual Disease Monitoring by Locked Nucleic Acid Quantitative Real-Time PCR Assay for IDH1/2 Mutation in Adult AML" Cancers 14, no. 24: 6205. https://doi.org/10.3390/cancers14246205
APA StyleKao, H.-W., Kuo, M.-C., Huang, Y.-J., Chang, H., Hu, S.-F., Huang, C.-F., Hung, Y.-S., Lin, T.-L., Ou, C.-W., Lien, M.-Y., Wu, J.-H., Chen, C.-C., & Shih, L.-Y. (2022). Measurable Residual Disease Monitoring by Locked Nucleic Acid Quantitative Real-Time PCR Assay for IDH1/2 Mutation in Adult AML. Cancers, 14(24), 6205. https://doi.org/10.3390/cancers14246205





