Investigational Microbiological Therapy for Glioma
Abstract
Simple Summary
Abstract
1. Introduction
2. Microorganisms Influence Glioma Development
3. Exogenous Microorganisms and Their Derivatives for Glioma Treatment
3.1. Derivatives of Microorganisms Are Used to Treat Gliomas
3.2. Microbial-Targeted Therapy for Glioma
3.2.1. Potential of Bacteria
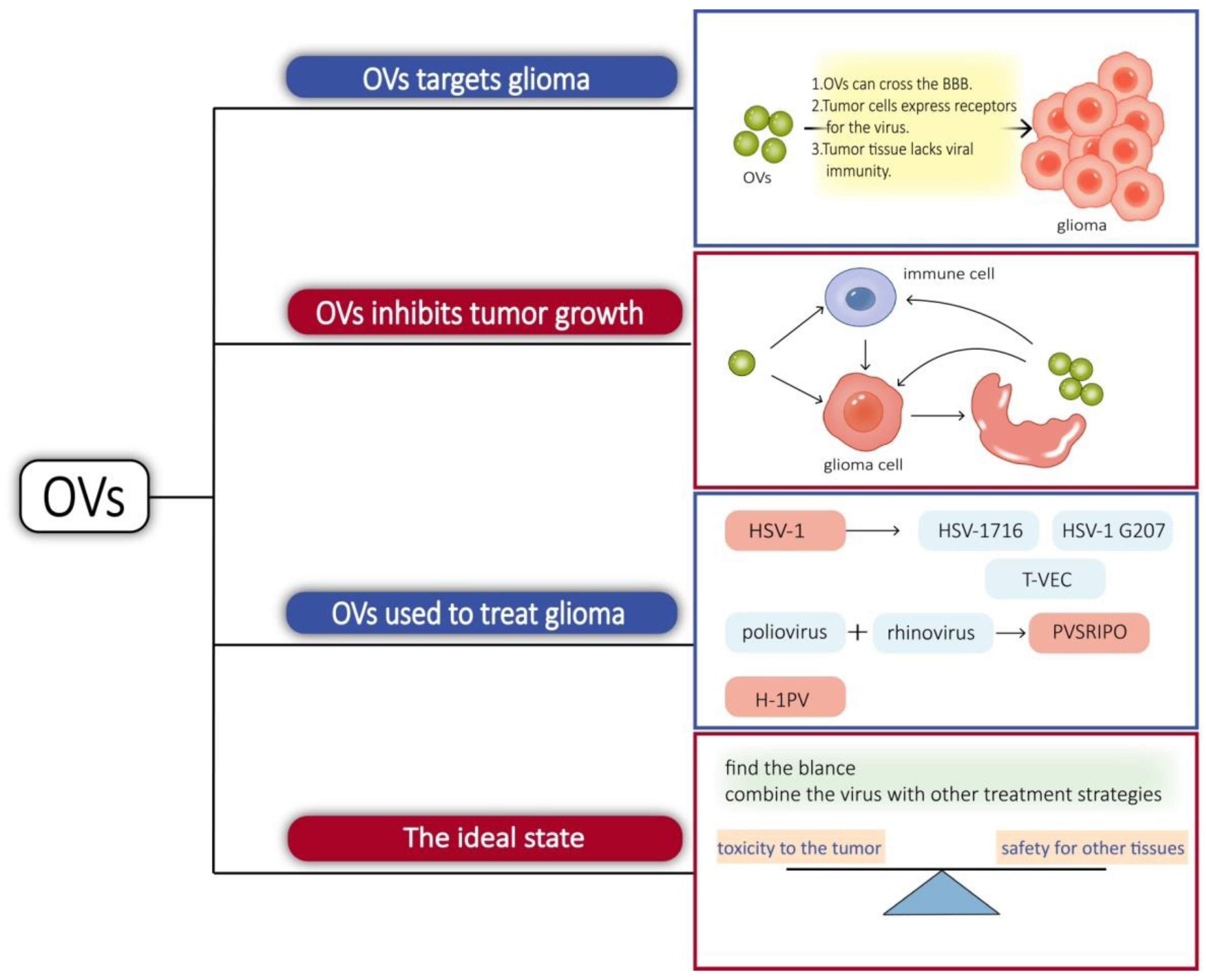
3.2.2. Oncolytic Virus
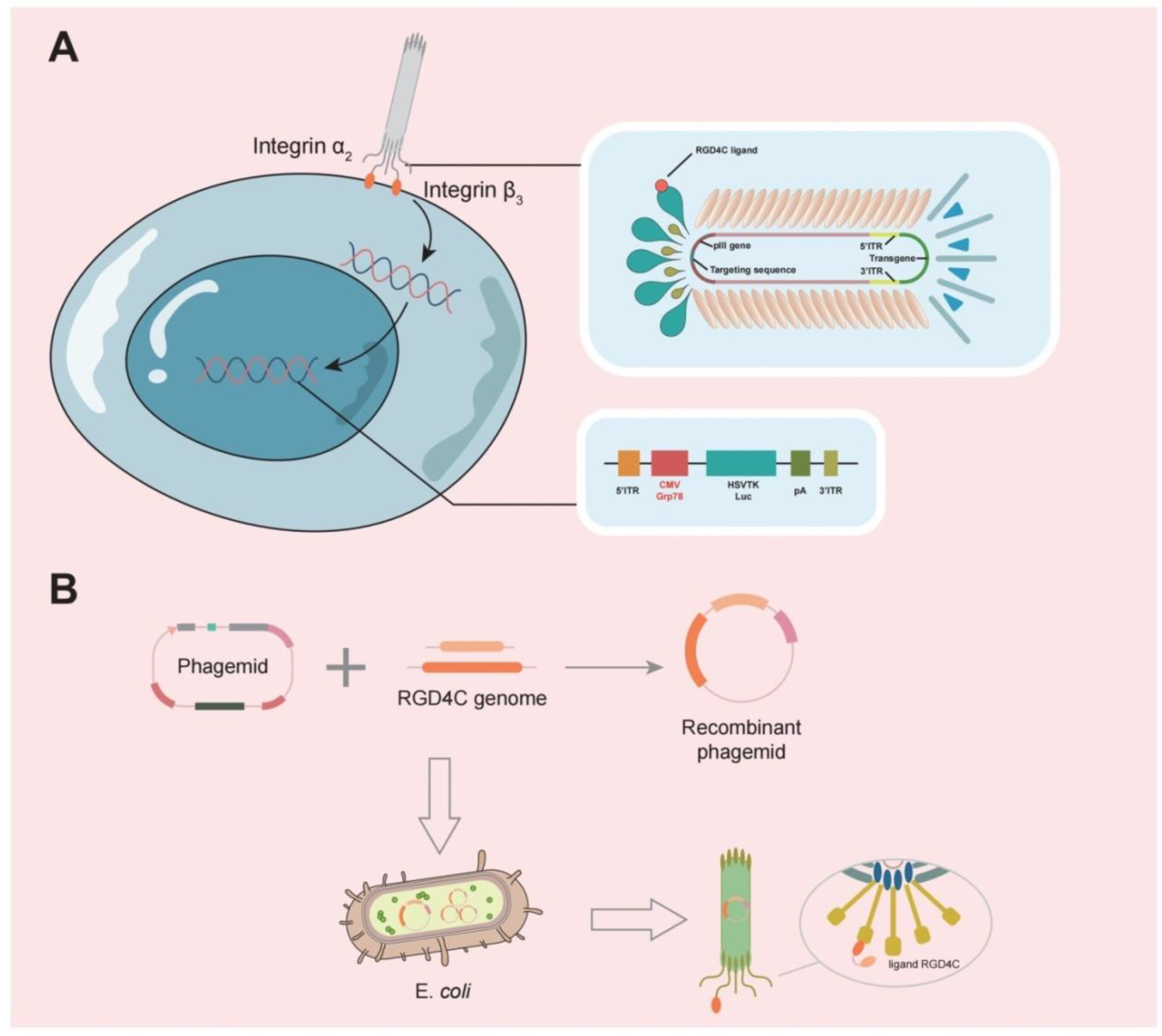
3.3. Phages Can Target Gliomas for Drug Delivery
4. Impact of the Local Environment
4.1. Glioma Microenvironment
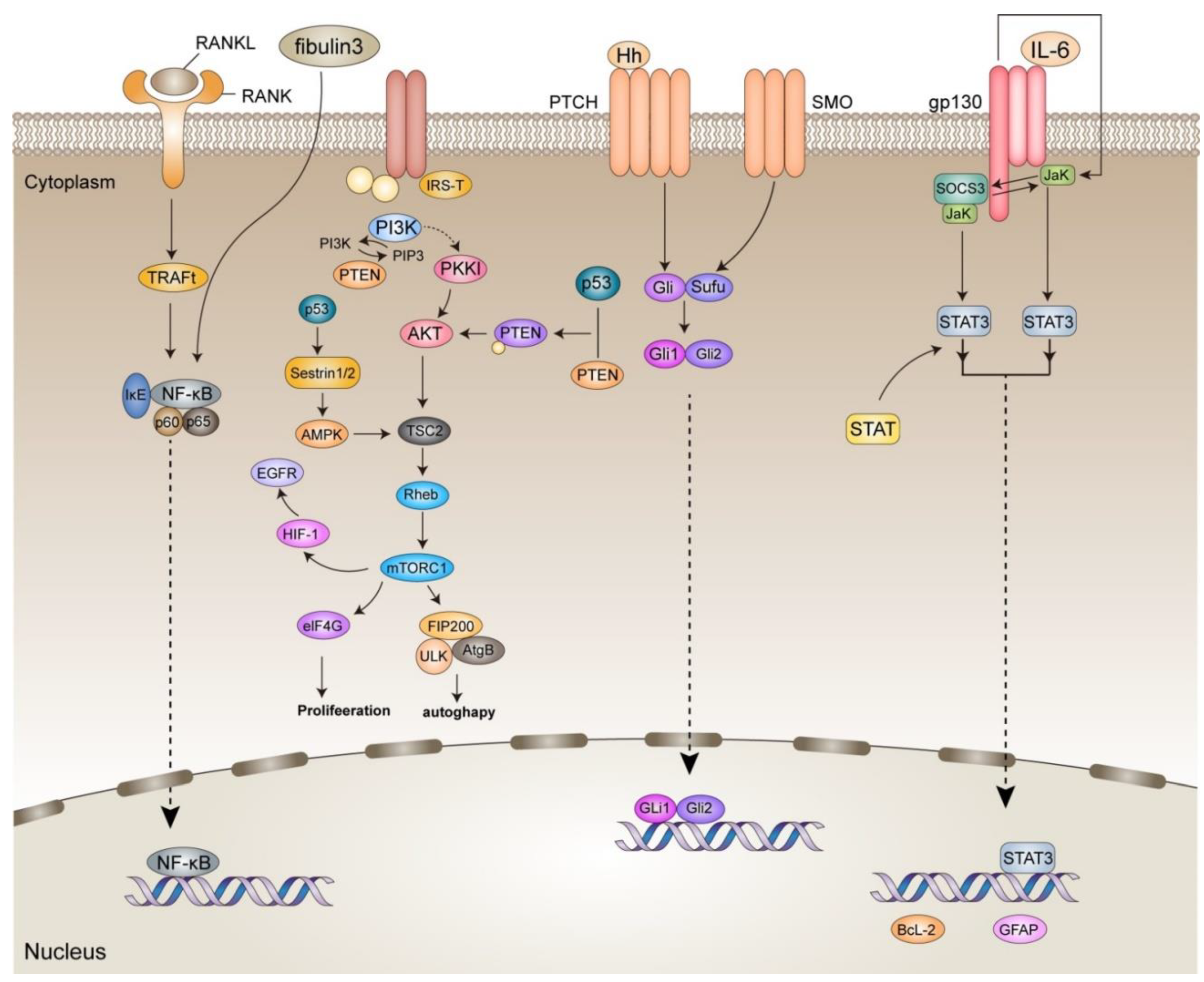
4.1.1. NF-κB
4.1.2. SHH
4.1.3. P53
4.1.4. JAK/STAT
4.1.5. PI3K/Akt
4.1.6. Microorganisms in Glioma
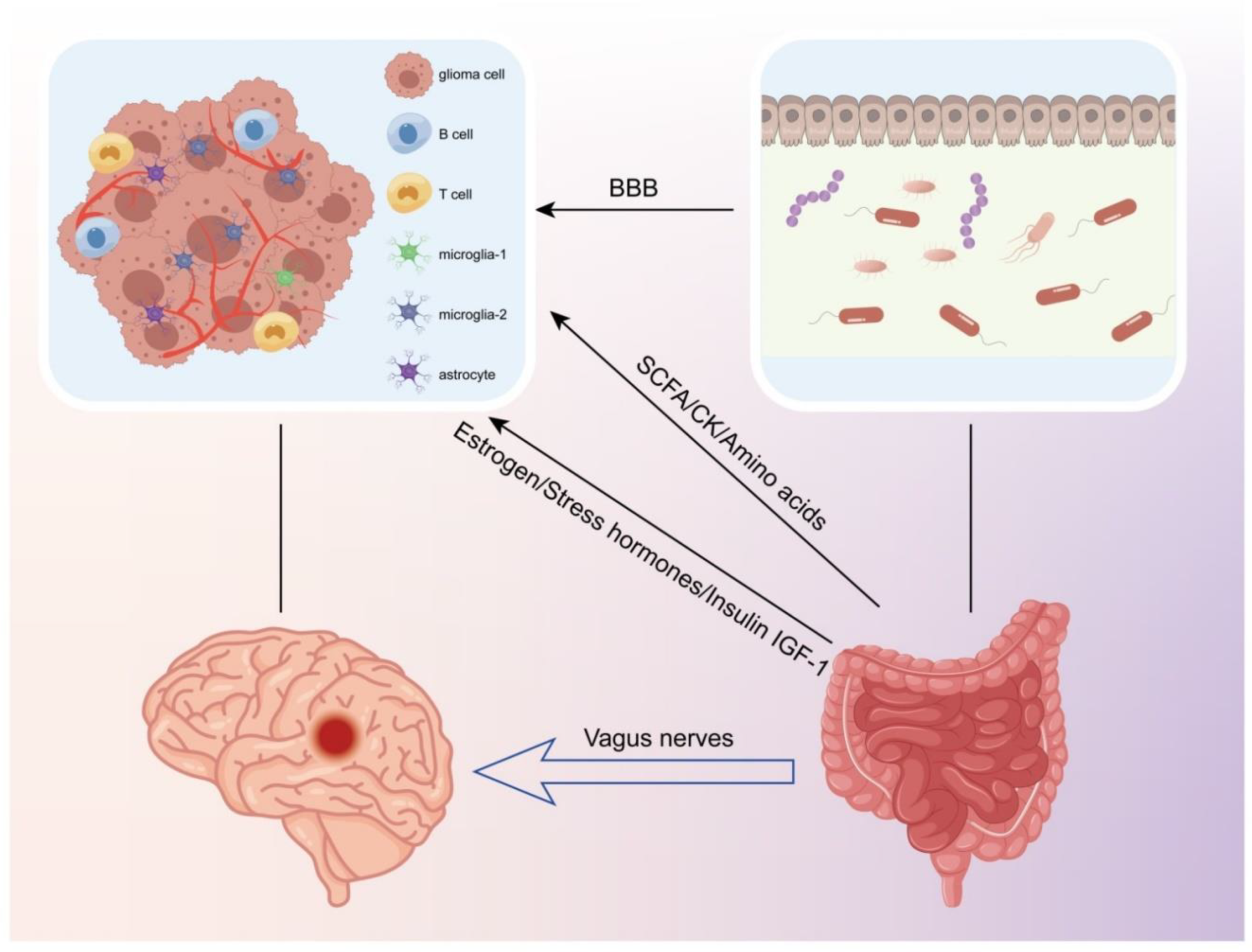
4.2. The Connection between the Gut Microbiota and Glioma
4.2.1. Metabolites Produced by Intestinal Flora
4.2.2. Intestinal Flora Regulate Hormone Release
4.2.3. Intestinal Flora Impact Neuronal Function
4.2.4. Gut Flora Regulate the Tumor Microenvironment
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PAMP | pathogen-associated molecular pattern |
| TSA | trichostatin A |
| MOX | Moxidectin |
| TLR | Toll-like receptor |
| NEAT1 | Nuclear-enriched abundant transcript 1 |
| STAT3 | Signal transducer and activator of transcription 3 |
| PE | pseudomonas exotoxin |
| CNF1 | Cytotoxic Necrotizing Factor 1 |
| TNF-α | tumor necrosis factor-α |
| HIF-1 | Hypoxia-inducible factor-1 |
| VEGF | vascular endothelial growth factor |
| IKK | IκB kinase |
| NLS | Nuclear localization sequence |
| RANKL | Receptor Activator of Nuclear Factor-κ B Ligand |
| CCL | C-C motif ligand |
| Gli | glioma-associated oncogene homolog |
| ECM | extracellular matrix |
| PTEN | Phosphatase and tensin homolog deleted on chromosome ten |
| IGF-1 | insulin-like growth factors -1 |
| SDF-1 | stromal cell-derived factor-1 |
| KPNA 2 | Karyopherin α2 |
| CXCR-4 | C-X-C-X-C chemokine receptor type 4 |
| GPCR | G protein-coupled receptors |
| MCT | H+-coupled monocarboxylate transporter |
| GF | germ-free |
| IDO | indoleamine 2,3-dioxygenase |
| AhR | aryl hydrocarbon receptor |
| IL4I1 | interleukin-4-induced-1 |
| GABA | γ-aminobutyric acid |
| 5-HT | 5-hydroxy tryptamine |
| α7 nAChR | nicotinic acetylcholine receptor |
| ACh | acetylcholine |
| GM-CSF | granulocyte-macrophage colony-stimulating factor |
| LCK | lymphocyte-specific protein tyrosine kinase |
| HIFU | high-intensity focused ultrasound |
| CTX | chlorotoxin |
| TIMP-2 | tissue inhibitor of metalloproteinase 2 |
| HSV-1 | Herpes simplex virus-1 |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Tang, L.; Li, X.; Fan, F.; Liu, Z. Immunotherapy for glioma: Current management and future application. Cancer Lett. 2020, 476, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Finch, A.; Solomou, G.; Wykes, V.; Pohl, U.; Bardella, C.; Watts, C. Advances in Research of Adult Gliomas. Int. J. Mol. Sci. 2021, 22, 924. [Google Scholar] [CrossRef]
- Chen, F.; Wendl, M.C.; Wyczalkowski, M.A.; Bailey, M.H.; Li, Y.; Ding, L. Moving pan-cancer studies from basic research toward the clinic. Nat. Cancer 2021, 2, 879–890. [Google Scholar] [CrossRef] [PubMed]
- Sepich-Poore, G.D.; Zitvogel, L.; Straussman, R.; Hasty, J.; Wargo, J.A.; Knight, R. The microbiome and human cancer. Science 2021, 371, eabc4552. [Google Scholar] [CrossRef]
- Rommasi, F. Bacterial-Based Methods for Cancer Treatment: What We Know and Where We Are. Oncol. Ther. 2022, 10, 23–54. [Google Scholar] [CrossRef] [PubMed]
- Mehrian-Shai, R.; Reichardt, J.; Harris, C.C.; Toren, A. The Gut–Brain Axis, Paving the Way to Brain Cancer. Trends Cancer 2019, 5, 200–207. [Google Scholar] [CrossRef]
- van Elsland, D.; Neefjes, J. Bacterial infections and cancer. EMBO Rep. 2018, 19, e46632. [Google Scholar] [CrossRef]
- Rosadi, F.; Fiorentini, C.; Fabbri, A. Bacterial protein toxins in human cancers. Pathog. Dis. 2016, 74, ftv105. [Google Scholar] [CrossRef]
- Emanuele Liardo, R.L.; Borzì, A.M.; Spatola, C.; Martino, B.; Privitera, G.; Basile, F.; Biondi, A.; Vacante, M. Effects of infections on the pathogenesis of cancer. Indian J. Med. Res. 2021, 153, 431–445. [Google Scholar]
- Singh, N.; Baby, D.; Rajguru, J.P.; Patil, P.B.; Thakkannavar, S.S.; Pujari, V.B. Inflammation and cancer. Ann. Afr. Med. 2019, 18, 121–126. [Google Scholar] [CrossRef]
- Pierce, E.S. Baseballs, tennis balls, livestock farm manure, the IDH1 mutation, endothelial cell proliferation and hypoxic pseudopalisading (granulomatous) necrosis: Mycobacterium avium subspecies paratuberculosis and the epidemiology, cellular metabolism and histology of diffuse gliomas, including glioblastoma. Open Veter.-J. 2019, 9, 5–12. [Google Scholar] [CrossRef]
- Farias, K.P.R.A.; Moreli, M.L.; Floriano, V.G.; da Costa, V.G. Evidence based on a meta-analysis of human cytomegalovirus infection in glioma. Arch. Virol. 2019, 164, 1249–1257. [Google Scholar] [CrossRef]
- Hu, M.; Yu, B.; Zhang, B.; Wang, B.; Qian, D.; Li, H.; Ma, J.; Liu, D.X. Human Cytomegalovirus Infection Activates Glioma Activating Transcription Factor 5 via microRNA in a Stress-Induced Manner. ACS Chem. Neurosci. 2021, 12, 3947–3956. [Google Scholar] [CrossRef] [PubMed]
- Egan, K.M.; Kim, Y.; Bender, N.; Hodge, J.M.; Coghill, A.E.; Smith-Warner, S.A.; Rollison, D.E.; Teras, L.R.; Grimsrud, T.K.; Waterboer, T. Prospective investigation of polyomavirus infection and the risk of adult glioma. Sci. Rep. 2021, 11, 9642. [Google Scholar] [CrossRef] [PubMed]
- Zang, J.; Zheng, M.H.; Cao, X.L.; Zhang, Y.Z.; Zhang, Y.F.; Gao, X.Y.; Cao, Y.; Shi, M.; Han, H.; Liang, L. Adenovirus infection promotes the formation of glioma stem cells from glioblastoma cells through the TLR9/NEAT1/STAT3 pathway. Cell Commun. Signal. 2020, 18, 135. [Google Scholar] [CrossRef]
- Schiller, J.T.; Lowy, D.R. An Introduction to Virus Infections and Human Cancer. Recent Results Cancer Res. 2021, 217, 1–11. [Google Scholar] [CrossRef]
- Cheng, S.-Y.; Chen, N.-F.; Kuo, H.-M.; Yang, S.-N.; Sung, C.-S.; Sung, P.-J.; Wen, Z.-H.; Chen, W.-F. Prodigiosin stimulates endoplasmic reticulum stress and induces autophagic cell death in glioblastoma cells. Apoptosis 2018, 23, 314–328. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.H.; Nguyen, V.H.; Jiang, S.-N.; Park, S.-H.; Tan, W.; Hong, S.H.; Shin, M.G.; Chung, I.-J.; Hong, Y.; Bom, H.-S.; et al. Two-step enhanced cancer immunotherapy with engineered Salmonella typhimurium secreting heterologous flagellin. Sci. Transl. Med. 2017, 9, eaak9537. [Google Scholar] [CrossRef]
- Mut, M.; Sherman, J.H.; Shaffrey, M.E.; Schiff, D. Cintredekin besudotox in treatment of malignant glioma. Expert Opin. Biol. Ther. 2008, 8, 805–812. [Google Scholar] [CrossRef]
- Hsu, Y.F.; Sheu, J.R.; Hsiao, G.; Lin, C.H.; Chang, T.H.; Chiu, P.T.; Wang, C.Y.; Hsu, M.J. p53 in trichostatin A induced C6 glioma cell death. Biochim. Biophys. Acta 2011, 1810, 504–513. [Google Scholar] [CrossRef]
- Song, D.; Liang, H.; Qu, B.; Li, Y.; Liu, J.; Chen, C.; Zhang, D.; Zhang, X.; Gao, A. Moxidectin inhibits glioma cell viability by inducing G0/G1 cell cycle arrest and apoptosis. Oncol. Rep. 2018, 40, 1348–1358. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Chai, W.; Song, T.; Ma, M.; Lian, X.-Y.; Zhang, Z. Anti-glioma Natural Products Downregulating Tumor Glycolytic Enzymes from Marine Actinomycete Streptomyces sp. ZZ406. Sci. Rep. 2018, 8, 72. [Google Scholar] [CrossRef]
- Guan, J.; Zhang, Z.; Hu, X.; Yang, Y.; Chai, Z.; Liu, X.; Liu, J.; Gao, B.; Lu, W.; Qian, J.; et al. Cholera Toxin Subunit B Enabled Multifunctional Glioma-Targeted Drug Delivery. Adv. Healthc. Mater. 2017, 6, 1700709. [Google Scholar] [CrossRef]
- Tantillo, E.; Colistra, A.; Vannini, E.; Cerri, C.; Pancrazi, L.; Baroncelli, L.; Costa, M.; Caleo, M. Bacterial Toxins and Targeted Brain Therapy: New Insights from Cytotoxic Necrotizing Factor 1 (CNF1). Int. J. Mol. Sci. 2018, 19, 1632. [Google Scholar] [CrossRef]
- Vannini, E.; Mori, E.; Tantillo, E.; Schmidt, G.; Caleo, M.; Costa, M. CTX-CNF1 Recombinant Protein Selectively Targets Glioma Cells In Vivo. Toxins 2021, 13, 194. [Google Scholar] [CrossRef]
- Fiedler, T.; Strauss, M.; Hering, S.; Redanz, U.; William, D.; Rosche, Y.; Classen, C.F.; Kreikemeyer, B.; Linnebacher, M.; Maletzki, C. Arginine deprivation by arginine deiminase of Streptococcus pyogenes controls primary glioblastoma growth in vitro and in vivo. Cancer Biol. Ther. 2015, 16, 1047–1055. [Google Scholar] [CrossRef]
- Foreman, P.M.; Friedman, G.K.; Cassady, K.A.; Markert, J.M. Oncolytic Virotherapy for the Treatment of Malignant Glioma. Neurotherapeutics 2017, 14, 333–344. [Google Scholar] [CrossRef]
- Wen, M.; Jung, S.; Moon, K.-S.; Jiang, S.N.; Li, S.-Y.; Min, J.-J. Targeting Orthotopic Glioma in Mice with Genetically Engineered Salmonella typhimurium. J. Korean Neurosurg. Soc. 2014, 55, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Wen, M.; Zheng, J.H.; Choi, J.M.; Pei, J.; Li, C.-H.; Li, S.-Y.; Kim, I.-Y.; Lim, S.-H.; Jung, T.-Y.; Moon, K.-S.; et al. Genetically-engineered Salmonella typhimurium expressing TIMP-2 as a therapeutic intervention in an orthotopic glioma mouse model. Cancer Lett. 2018, 433, 140–146. [Google Scholar] [CrossRef]
- Wang, Y.; Sheng, J.; Chai, J.; Zhu, C.; Li, X.; Yang, W.; Cui, R.; Ge, T. Filamentous Bacteriophage—A Powerful Carrier for Glioma Therapy. Front. Immunol. 2021, 12, 729336. [Google Scholar] [CrossRef]
- Pastorino, O.; Gentile, M.T.; Mancini, A.; Del Gaudio, N.; Di Costanzo, A.; Bajetto, A.; Franco, P.; Altucci, L.; Florio, T.; Stoppelli, M.P.; et al. Histone Deacetylase Inhibitors Impair Vasculogenic Mimicry from Glioblastoma Cells. Cancers 2019, 11, 747. [Google Scholar] [CrossRef]
- Pang, Z.; Gu, M.-D.; Tang, T. Pseudomonas aeruginosa in Cancer Therapy: Current Knowledge, Challenges and Future Perspectives. Front. Oncol. 2022, 12, 891187. [Google Scholar] [CrossRef]
- Hou, X.; Chen, S.; Zhang, P.; Guo, D.; Wang, B. Targeted Arginine Metabolism Therapy: A Dilemma in Glioma Treatment. Front. Oncol. 2022, 12, 938847. [Google Scholar] [CrossRef] [PubMed]
- Leschner, S.; Westphal, K.; Dietrich, N.; Viegas, N.; Jablonska, J.; Lyszkiewicz, M.; Lienenklaus, S.; Falk, W.; Gekara, N.O.; Loessner, H.; et al. Tumor Invasion of Salmonella enterica Serovar Typhimurium Is Accompanied by Strong Hemorrhage Promoted by TNF-α. PLoS ONE 2009, 4, e6692. [Google Scholar] [CrossRef]
- Zhou, S.; Gravekamp, C.; Bermudes, D.; Liu, K. Tumour-targeting bacteria engineered to fight cancer. Nat. Rev. Cancer 2018, 18, 727–743. [Google Scholar] [CrossRef]
- Duong, M.T.-Q.; Qin, Y.; You, S.-H.; Min, J.-J. Bacteria-cancer interactions: Bacteria-based cancer therapy. Exp. Mol. Med. 2019, 51, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Chen, C.; Shi, Y.; Wu, Q.; Gimple, R.C.; Fang, X.; Huang, Z.; Zhai, K.; Ke, S.Q.; Ping, Y.-F.; et al. Targeting Glioma Stem Cell-Derived Pericytes Disrupts the Blood-Tumor Barrier and Improves Chemotherapeutic Efficacy. Cell Stem Cell 2017, 21, 591–603.e4. [Google Scholar] [CrossRef]
- Al-Obaidi, M.M.J.; Desa, M.N.M. Mechanisms of Blood Brain Barrier Disruption by Different Types of Bacteria, and Bacterial-Host Interactions Facilitate the Bacterial Pathogen Invading the Brain. Cell Mol. Neurobiol. 2018, 38, 1349–1368. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Chen, Y.; Liu, X.; Min, J.-J.; Tan, W.; Zheng, J.H. Targeted cancer immunotherapy with genetically engineered oncolytic Salmonella typhimurium. Cancer Lett. 2020, 469, 102–110. [Google Scholar] [CrossRef]
- Finnie, J.W.; Uzal, F.A. Pathology and Pathogenesis of Brain Lesions Produced by Clostridium perfringens Type D Epsilon Toxin. Int. J. Mol. Sci. 2022, 23, 9050. [Google Scholar] [CrossRef]
- Cheng, X.; Zhang, X.; Zhou, Y.; Zhang, C.; Hua, Z.C. A Salmonella Typhimurium mutant strain capable of RNAi delivery: Higher tumor-targeting and lower toxicity. Cancer Biol. Ther. 2014, 15, 1068–1076. [Google Scholar] [CrossRef]
- Abedi, M.H.; Yao, M.S.; Mittelstein, D.R.; Bar-Zion, A.; Swift, M.B.; Lee-Gosselin, A.; Barturen-Larrea, P.; Buss, M.T.; Shapiro, M.G. Ultrasound-controllable engineered bacteria for cancer immunotherapy. Nat. Commun. 2022, 13, 1–11. [Google Scholar] [CrossRef]
- Flentie, K.; Kocher, B.; Gammon, S.T.; Novack, D.V.; McKinney, J.S.; Piwnica-Worms, D. A Bioluminescent Transposon Reporter-Trap Identifies Tumor-Specific Microenvironment-Induced Promoters in Salmonella for Conditional Bacterial-Based Tumor Therapy. Cancer Discov. 2012, 2, 624–637. [Google Scholar] [CrossRef]
- Rong, L.; Ni Li, N.; Zhang, Z. Emerging therapies for glioblastoma: Current state and future directions. J. Exp. Clin. Cancer Res. 2022, 41, 1–18. [Google Scholar] [CrossRef]
- Wollmann, G.; Ozduman, K.; van den Pol, A.N. Oncolytic virus therapy for glioblastoma multiforme: Concepts and candidates. Cancer J. 2012, 18, 69–81. [Google Scholar] [CrossRef]
- Suryawanshi, Y.; Schulze, A. Oncolytic Viruses for Malignant Glioma: On the Verge of Success? Viruses 2021, 13, 1294. [Google Scholar] [CrossRef]
- Merrill, M.K.; Bernhardt, G.; Sampson, J.; Wikstrand, C.J.; Bigner, D.D.; Gromeier, M. Poliovirus receptor CD155-targeted oncolysis of glioma. Neuro-Oncology 2004, 6, 208–217. [Google Scholar] [CrossRef]
- Stojdl, D.F.; Lichty, B.D.; Knowles, S.; Marius, R.; Atkins, H.; Sonenberg, N.; Bell, J.C. Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nat. Med. 2000, 6, 821–825. [Google Scholar] [CrossRef]
- Kamynina, M.; Tskhovrebova, S.; Fares, J.; Timashev, P.; Laevskaya, A.; Ulasov, I. Oncolytic Virus-Induced Autophagy in Glioblastoma. Cancers 2021, 13, 3482. [Google Scholar] [CrossRef]
- Achard, C.; Surendran, A.; Wedge, M.-E.; Ungerechts, G.; Bell, J.; Ilkow, C.S. Lighting a Fire in the Tumor Microenvironment Using Oncolytic Immunotherapy. EBioMedicine 2018, 31, 17–24. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Shin, D.H.; Sohoni, S.; Singh, S.K.; Rivera-Molina, Y.; Jiang, H.; Fan, X.; Gumin, J.; Lang, F.F.; Alvarez-Breckenridge, C.; et al. Reshaping the tumor microenvironment with oncolytic viruses, positive regulation of the immune synapse, and blockade of the immunosuppressive oncometabolic circuitry. J. Immunother. Cancer 2022, 10, e004935. [Google Scholar] [CrossRef]
- Rius-Rocabert, S.; García-Romero, N.; García, A.; Ayuso-Sacido, A.; Nistal-Villan, E. Oncolytic Virotherapy in Glioma Tumors. Int. J. Mol. Sci. 2020, 21, 7604. [Google Scholar] [CrossRef]
- Carpenter, A.; Aiken, R.; Hanft, S. Oncolytic virus in gliomas: A review of human clinical investigations. Ann. Oncol. 2021, 32, 968–982. [Google Scholar] [CrossRef]
- Gromeier, M.; Nair, S.K. Recombinant Poliovirus for Cancer Immunotherapy. Annu. Rev. Med. 2018, 69, 289–299. [Google Scholar] [CrossRef]
- Holl, E.K.; Brown, M.C.; Boczkowski, D.; McNamara, M.A.; George, D.J.; Bigner, D.D.; Gromeier, M.; Nair, S.K. Recombinant oncolytic poliovirus, PVSRIPO, has potent cytotoxic and innate inflammatory effects, mediating therapy in human breast and prostate cancer xenograft models. Oncotarget 2016, 7, 79828–79841. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, T.; Kulkarni, A.; Bretscher, C.; Nazarov, P.V.; Hossain, J.A.; Ystaas, L.A.R.; Miletic, H.; Röth, R.; Niesler, B.; Marchini, A. Oncolytic H-1 Parvovirus Hijacks Galectin-1 to Enter Cancer Cells. Viruses 2022, 14, 1018. [Google Scholar] [CrossRef] [PubMed]
- Immidisetti, A.; Nwagwu, C.; Adamson, D.; Patel, N.; Carbonell, A.-M. Clinically Explored Virus-Based Therapies for the Treatment of Recurrent High-Grade Glioma in Adults. Biomedicines 2021, 9, 138. [Google Scholar] [CrossRef] [PubMed]
- Geletneky, K.; Leoni, A.-L.; Pohlmeyer-Esch, G.; Loebhard, S.; Leuchs, B.; Hoefer, C.; Jochims, K.; Dahm, M.; Huber, B.; Rommelaere, J.; et al. Bioavailability, biodistribution, and CNS toxicity of clinical-grade parvovirus H1 after intravenous and intracerebral injection in rats. Comp. Med. 2015, 65, 36–45. [Google Scholar] [PubMed]
- Geletneky, K.; Hajda, J.; Angelova, A.L.; Leuchs, B.; Capper, D.; Bartsch, A.J.; Neumann, J.-O.; Schöning, T.; Hüsing, J.; Beelte, B.; et al. Oncolytic H-1 Parvovirus Shows Safety and Signs of Immunogenic Activity in a First Phase I/IIa Glioblastoma Trial. Mol. Ther. 2017, 25, 2620–2634. [Google Scholar] [CrossRef]
- Shoaf, M.L.; Desjardins, A. Oncolytic Viral Therapy for Malignant Glioma and Their Application in Clinical Practice. Neurotherapeutics 2022, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Desjardins, A.; Gromeier, M.; Herndon, J.E., 2nd; Beaubier, N.; Bolognesi, D.P.; Friedman, A.H.; Friedman, H.S.; McSherry, F.; Muscat, A.M.; Nair, S.; et al. Recurrent Glioblastoma Treated with Recombinant Poliovirus. N. Engl. J. Med. 2018, 379, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Long, X.; Liu, J.; Cheng, P. Glioblastoma microenvironment and its reprogramming by oncolytic virotherapy. Front. Cell. Neurosci. 2022, 16, 819363. [Google Scholar] [CrossRef] [PubMed]
- Kadhim, Z.A.; Sulaiman, G.M.; Al-Shammari, A.M.; Khan, R.A.; Al Rugaie, O.; Mohammed, H.A. Oncolytic Newcastle Disease Virus Co-Delivered with Modified PLGA Nanoparticles Encapsulating Temozolomide against Glioblastoma Cells: Developing an Effective Treatment Strategy. Molecules 2022, 27, 5757. [Google Scholar] [CrossRef]
- Mahmoud, A.B.; Ajina, R.; Aref, S.; Darwish, M.; Alsayb, M.; Taher, M.; AlSharif, S.A.; Hashem, A.M.; Alkayyal, A.A. Advances in immunotherapy for glioblastoma multiforme. Front. Immunol. 2022, 13, 944452. [Google Scholar] [CrossRef]
- Asija, S.; Chatterjee, A.; Yadav, S.; Chekuri, G.; Karulkar, A.; Jaiswal, A.K.; Goda, J.S.; Purwar, R. Combinatorial approaches to effective therapy in glioblastoma (GBM): Current status and what the future holds. Int. Rev. Immunol. 2022, 41, 582–605. [Google Scholar] [CrossRef]
- Wu, L.-P.; Ahmadvand, D.; Su, J.; Hall, A.; Tan, X.; Farhangrazi, Z.S.; Moghimi, S.M. Crossing the blood-brain-barrier with nanoligand drug carriers self-assembled from a phage display peptide. Nat. Commun. 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Saw, P.E.; Song, E.-W. Phage display screening of therapeutic peptide for cancer targeting and therapy. Protein Cell 2019, 10, 787–807. [Google Scholar] [CrossRef]
- Zhang, M.; Lu, W. Enhanced glioma-targeting and stability of L GICP peptide coupled with stabilized peptide D A7R. Acta Pharm. Sin. B 2018, 8, 106–115. [Google Scholar] [CrossRef]
- Przystal, J.M.; Waramit, S.; Pranjol, Z.I.; Yan, W.; Chu, G.; Chongchai, A.; Samarth, G.; Olaciregui, N.G.; Tabatabai, G.; Carcaboso, A.M.; et al. Efficacy of systemic temozolomide-activated phage-targeted gene therapy in human glioblastoma. EMBO Mol. Med. 2019, 11, e8492. [Google Scholar] [CrossRef]
- Yu, H.; Lin, L.; Zhang, Z.; Zhang, H.; Hu, H. Targeting NF-κB pathway for the therapy of diseases: Mechanism and clinical study. Signal. Transduct. Target Ther. 2020, 5, 209. [Google Scholar] [CrossRef]
- Kim, J.K.; Jin, X.; Sohn, Y.W.; Jin, X.; Jeon, H.Y.; Kim, E.J.; Ham, S.W.; Jeon, H.M.; Chang, S.Y.; Oh, S.Y.; et al. Tumoral RANKL activates astrocytes that promote glioma cell invasion through cytokine signaling. Cancer Lett. 2014, 353, 194–200. [Google Scholar] [CrossRef]
- Nandhu, M.S.; Kwiatkowska, A.; Bhaskaran, V.; Hayes, J.; Hu, B.; Viapiano, M.S. Tumor-derived fibulin-3 activates pro-invasive NF-κB signaling in glioblastoma cells and their microenvironment. Oncogene 2017, 36, 4875–4886. [Google Scholar] [CrossRef]
- Skoda, A.M.; Simovic, D.; Karin, V.; Kardum, V.; Vranic, S.; Serman, L. The role of the Hedgehog signaling pathway in cancer: A comprehensive review. Bosn. J. Basic Med. Sci. 2018, 18, 8–20. [Google Scholar] [CrossRef]
- Jeng, K.; Jeng, C.; Jeng, W.; Sheen, I.; Li, S.; Leu, C.; Tsay, Y.; Chang, C. Sonic Hedgehog signaling pathway as a potential target to inhibit the progression of hepatocellular carcinoma (Review). Oncol. Lett. 2019, 18, 4377–4384. [Google Scholar] [CrossRef]
- Okuda, H.; Tatsumi, K.; Morita-Takemura, S.; Nakahara, K.; Nochioka, K.; Shinjo, T.; Terada, Y.; Wanaka, A. Hedgehog Signaling Modulates the Release of Gliotransmitters from Cultured Cerebellar Astrocytes. Neurochem. Res. 2016, 41, 278–289. [Google Scholar] [CrossRef]
- Torrisi, F.; Alberghina, C.; Furno, D.L.; Zappalà, A.; Valable, S.; Volti, G.L.; Tibullo, D.; Vicario, N.; Parenti, R. Connexin 43 and Sonic Hedgehog Pathway Interplay in Glioblastoma Cell Proliferation and Migration. Biology 2021, 10, 767. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, J.; Xu, D.; Zhang, T.; Hu, W.; Feng, Z. Gain-of-function mutant p53 in cancer progression and therapy. J. Mol. Cell Biol. 2020, 12, 674–687. [Google Scholar] [CrossRef]
- Zhang, S.; Carlsen, L.; Borrero, L.H.; Seyhan, A.A.; Tian, X.; El-Deiry, W.S. Advanced Strategies for Therapeutic Targeting of Wild-Type and Mutant p53 in Cancer. Biomolecules 2022, 12, 548. [Google Scholar] [CrossRef]
- Trachootham, D.; Chen, G.; Zhang, W.; Lu, W.; Zhang, H.; Liu, J.; Huang, P. Loss of p53 in stromal fibroblasts promotes epithelial cell invasion through redox-mediated ICAM1 signal. Free Radic. Biol. Med. 2013, 58, 1–13. [Google Scholar] [CrossRef][Green Version]
- Biasoli, D.; Sobrinho, M.F.; Da Fonseca, A.C.C.; De Matos, D.G.; Romao, L.; de Moraes Maciel, R.; Rehen, S.K.; Moura-Neto, V.; Borges, H.L.; Lima, F.R.S. Glioblastoma cells inhibit astrocytic p53-expression favoring cancer malignancy. Oncogenesis 2014, 3, e123. [Google Scholar] [CrossRef] [PubMed]
- Swiatek-Machado, K.; Kaminska, B. STAT Signaling in Glioma Cells. Adv. Exp. Med. Biol. 2020, 1202, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Yamini, B. NF-κB, Mesenchymal Differentiation and Glioblastoma. Cells 2018, 7, 125. [Google Scholar] [CrossRef] [PubMed]
- Dumas, A.A.; Pomella, N.; Rosser, G.; Guglielmi, L.; Vinel, C.; Millner, T.O.; Rees, J.; Aley, N.; Sheer, D.; Wei, J.; et al. Microglia promote glioblastoma via mTOR-mediated immunosuppression of the tumour microenvironment. Embo J. 2020, 39, e103790. [Google Scholar] [CrossRef]
- Dzaye, O.D.A.; Hu, F.; Derkow, K.; Haage, V.; Euskirchen, P.; Harms, C.; Lehnardt, S.; Synowitz, M.; Wolf, S.A.; Kettenmann, H. Glioma Stem Cells but Not Bulk Glioma Cells Upregulate IL-6 Secretion in Microglia/Brain Macrophages via Toll-like Receptor 4 Signaling. J. Neuropathol. Exp. Neurol. 2016, 75, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef]
- de Groot, J.; Liang, J.; Kong, L.Y.; Wei, J.; Piao, Y.; Fuller, G.; Qiao, W.; Heimberger, A.B. Modulating antiangiogenic resistance by inhibiting the signal transducer and activator of transcription 3 pathway in glioblastoma. Oncotarget 2012, 3, 1036–1048. [Google Scholar] [CrossRef]
- Li, X.; Wu, C.; Chen, N.; Gu, H.; Yen, A.; Cao, L.; Wang, E.; Wang, L. PI3K/Akt/mTOR signaling pathway and targeted therapy for glioblastoma. Oncotarget 2016, 7, 33440–33450. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, X.; Zhao, P.; Zhao, H.; Gao, W.; Wang, L. Celastrol Suppresses Glioma Vasculogenic Mimicry Formation and Angiogenesis by Blocking the PI3K/Akt/mTOR Signaling Pathway. Front. Pharmacol. 2020, 11, 25. [Google Scholar] [CrossRef]
- Zhang, Z.-Q.; Wang, X.; Xue, B.-H.; Zhao, Y.; Xie, F.; Wang, S.-D.; Xue, C.; Wang, Y.; Zhang, Y.-S.; Qian, L.-J. Chronic stress promotes glioma cell proliferation via the PI3K/Akt signaling pathway. Oncol. Rep. 2021, 46, 1–12. [Google Scholar] [CrossRef]
- Ma, J.; Chen, C.; Li, M. Macrophages/Microglia in the Glioblastoma Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 5775. [Google Scholar] [CrossRef]
- Chanda, S.; Hale, W.D.; Zhang, B.; Wernig, M.; Südhof, T.C. Unique versus Redundant Functions of Neuroligin Genes in Shaping Excitatory and Inhibitory Synapse Properties. J. Neurosci. 2017, 37, 6816–6836. [Google Scholar] [CrossRef]
- Li, Z.; Gao, W.; Fei, Y.; Gao, P.; Xie, Q.; Xie, J.; Xu, Z. NLGN3 promotes neuroblastoma cell proliferation and growth through activating PI3K/AKT pathway. Eur. J. Pharmacol. 2019, 857, 172423. [Google Scholar] [CrossRef]
- Venkatesh, H.S.; Morishita, W.; Geraghty, A.C.; Silverbush, D.; Gillespie, S.M.; Arzt, M.; Tam, L.T.; Espenel, C.; Ponnuswami, A.; Ni, L.; et al. Electrical and synaptic integration of glioma into neural circuits. Nature 2019, 573, 539–545. [Google Scholar] [CrossRef]
- Venkatesh, H.S.; Tam, L.T.; Woo, P.J.; Lennon, J.; Nagaraja, S.; Gillespie, S.M.; Ni, J.; Duveau, D.Y.; Morris, P.J.; Zhao, J.J.; et al. Targeting neuronal activity-regulated neuroligin-3 dependency in high-grade glioma. Nature 2017, 549, 533–537. [Google Scholar] [CrossRef]
- Zhang, Z.; Huang, X.; Li, J.; Fan, H.; Yang, F.; Zhang, R.; Yang, Y.; Feng, S.; He, D.; Sun, W.; et al. Interleukin 10 promotes growth and invasion of glioma cells by up-regulating KPNA 2 in vitro. J. Cancer Res. Ther. 2019, 15, 927–932. [Google Scholar] [CrossRef]
- Radin, D.P.; Tsirka, S.E. Interactions between Tumor Cells, Neurons, and Microglia in the Glioma Microenvironment. Int. J. Mol. Sci. 2020, 21, 8476. [Google Scholar] [CrossRef]
- Liang, J.; Li, T.; Zhao, J.; Wang, C.; Sun, H. Current understanding of the human microbiome in glioma. Front. Oncol. 2022, 12, 781741. [Google Scholar] [CrossRef]
- Lin, Z.; Zu, X.-P.; Xie, H.-S.; Jin, H.-Z.; Yang, N.; Liu, X.-R.; Zhang, W.-D. Research progress in mechanism of intestinal microorganisms in human diseases. Yao xue xue bao = Acta Pharm. Sin. 2016, 51, 843–852. [Google Scholar]
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef]
- Patrizz, A.; Dono, A.; Zorofchian, S.; Hines, G.; Takayasu, T.; Husein, N.; Otani, Y.; Arevalo, O.; Choi, H.A.; Savarraj, J.; et al. Glioma and temozolomide induced alterations in gut microbiome. Sci. Rep. 2020, 10, 21002. [Google Scholar] [CrossRef]
- Dono, A.; Patrizz, A.; McCormack, R.M.; Putluri, N.; Ganesh, B.P.; Kaur, B.; McCullough, L.D.; Ballester, L.Y.; Esquenazi, Y. Glioma induced alterations in fecal short-chain fatty acids and neurotransmitters. CNS Oncol. 2020, 9, CNS57. [Google Scholar] [CrossRef]
- D’Alessandro, G.; Antonangeli, F.; Marrocco, F.; Porzia, A.; Lauro, C.; Santoni, A.; Limatola, C. Author response for “Gut microbiota alterations affect glioma growth and innate immune cells involved in tumor immunosurveillance in mice”. Eur. J. Immunol. 2019, 50, 705–711. [Google Scholar] [CrossRef]
- Aglae, H.; Philippe, A.; Croyal, M.; Philippe, N.; Stéphanie, B.C.; Michel, N.; Yves, D.; Dominique, C.; Laetitia, A. Late-Stage Glioma Is Associated with Deleterious Alteration of Gut Bacterial Metabolites in Mice. Metabolites. Metabolites 2022, 12, 290. [Google Scholar]
- Lymperopoulos, A.; Suster, M.S.; Borges, J.I. Short-Chain Fatty Acid Receptors and Cardiovascular Function. Int. J. Mol. Sci. 2022, 23, 3303. [Google Scholar] [CrossRef]
- Puddu, A.; Sanguineti, R.; Montecucco, F.; Viviani, G.L. Evidence for the Gut Microbiota Short-Chain Fatty Acids as Key Pathophysiological Molecules Improving Diabetes. Mediat. Inflamm. 2014, 2014, 1–9. [Google Scholar] [CrossRef]
- Silva, Y.P.; Bernardi, A.; Frozza, R.L. The Role of Short-Chain Fatty Acids from Gut Microbiota in Gut-Brain Communication. Front. Endocrinol. (Lausanne) 2020, 11, 25. [Google Scholar] [CrossRef]
- López-Bañuelos, L.; Vega, L. Inhibition of acetylation, is it enough to fight cancer? Crit. Rev. Oncol. Hematol. 2022, 176, 103752. [Google Scholar] [CrossRef]
- Wolf, I.M.L.; Fan, Z.; Rauh, M.; Seufert, S.; Hore, N.; Buchfelder, M.; Savaskan, N.E.; Eyüpoglu, I.Y. Histone deacetylases inhibition by SAHA/Vorinostat normalizes the glioma microenvironment via xCT equilibration. Sci. Rep. 2014, 4, 6226. [Google Scholar] [CrossRef]
- Erny, D.; Hrabě de Angelis, A.L.; Jaitin, D.; Wieghofer, P.; Staszewski, O.; David, E.; Keren-Shaul, H.; Mahlakoiv, T.; Jakobshagen, K.; Buch, T.; et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 2015, 18, 965–977. [Google Scholar] [CrossRef]
- Anderson, G.; Maes, M. Gut Dysbiosis Dysregulates Central and Systemic Homeostasis via Suboptimal Mitochondrial Function: Assessment, Treatment and Classification Implications. Curr. Top. Med. Chem. 2020, 20, 524–539. [Google Scholar] [CrossRef] [PubMed]
- Wierzbicki, M.; Sawosz, E.; Strojny, B.; Jaworski, S.; Grodzik, M.; Chwalibog, A. NF-κB-related decrease of glioma angiogenic potential by graphite nanoparticles and graphene oxide nanoplatelets. Sci. Rep. 2018, 8, 14733. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Chae, C.W.; Lee, H.J.; Jung, Y.H.; Choi, G.E.; Kim, J.S.; Lim, J.R.; Lee, J.E.; Cho, J.H.; Park, H.; et al. Sodium butyrate inhibits high cholesterol-induced neuronal amyloidogenesis by modulating NRF2 stabilization-mediated ROS levels: Involvement of NOX2 and SOD1. Cell Death Dis. 2020, 11, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Roh, H.S.; Kim, J.E.; Park, S.D.; Park, W.H.; Moon, J.-Y. Compound K attenuates stromal cell-derived growth factor 1 (SDF-1)-induced migration of C6 glioma cells. Nutr. Res. Pr. 2016, 10, 259–264. [Google Scholar] [CrossRef]
- Gao, J.; Xu, K.; Liu, H.; Liu, G.; Bai, M.; Peng, C.; Li, T.; Yin, Y. Impact of the Gut Microbiota on Intestinal Immunity Mediated by Tryptophan Metabolism. Front. Cell. Infect. Microbiol. 2018, 8, 13. [Google Scholar] [CrossRef]
- Németh, H.; Toldi, J.; Vécsei, L. Role of kynurenines in the central and peripheral nervous systems. Curr. Neurovasc. Res. 2005, 2, 249–260. [Google Scholar] [CrossRef]
- Gao, K.; Mu, C.-L.; Farzi, A.; Zhu, W.-Y. Tryptophan Metabolism: A Link between the Gut Microbiota and Brain. Adv. Nutr. Int. Rev. J. 2020, 11, 709–723. [Google Scholar] [CrossRef]
- Sadik, A.; Patterson, L.F.S.; Öztürk, S.; Mohapatra, S.R.; Panitz, V.; Secker, P.F.; Pfänder, P.; Loth, S.; Salem, H.; Prentzell, M.T.; et al. IL4I1 Is a Metabolic Immune Checkpoint that Activates the AHR and Promotes Tumor Progression. Cell 2020, 182, 1252–1270.e34. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, H.; Sun, Q.; Geng, R.; Yuan, F.; Liu, B.; Chen, Q. Immunomodulatory Effects of Tryptophan Metabolism in the Glioma Tumor Microenvironment. Front. Immunol. 2021, 12, 730289. [Google Scholar] [CrossRef]
- Jin, U.-H.; Karki, K.; Cheng, Y.; Michelhaugh, S.K.; Mittal, S.; Safe, S. The aryl hydrocarbon receptor is a tumor suppressor–like gene in glioblastoma. J. Biol. Chem. 2019, 294, 11342–11353. [Google Scholar] [CrossRef]
- Pei, Z.; Lee, K.-C.; Khan, A.; Erisnor, G.; Wang, H.-Y. Pathway analysis of glutamate-mediated, calcium-related signaling in glioma progression. Biochem. Pharmacol. 2020, 176, 113814. [Google Scholar] [CrossRef] [PubMed]
- Venkataramani, V.; Tanev, D.I.; Strahle, C.; Studier-Fischer, A.; Fankhauser, L.; Kessler, T.; Körber, C.; Kardorff, M.; Ratliff, M.; Xie, R.; et al. Glutamatergic synaptic input to glioma cells drives brain tumour progression. Nature 2019, 573, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Maus, A.; Peters, G.J. Glutamate and α-ketoglutarate: Key players in glioma metabolism. Amino Acids 2017, 49, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Dehhaghi, M.; Panahi, H.K.S.; Heng, B.; Guillemin, G.J. The Gut Microbiota, Kynurenine Pathway, and Immune System Interaction in the Development of Brain Cancer. Front. Cell Dev. Biol. 2020, 8, 562812. [Google Scholar] [CrossRef]
- Dai, F.; Yu, W.; Song, J.; Li, Q.; Wang, C.; Xie, S. Extracellular polyamines-induced proliferation and migration of cancer cells by ODC, SSAT, and Akt1-mediated pathway. Anti-Cancer Drugs 2017, 28, 457–464. [Google Scholar] [CrossRef]
- Neuman, H.; Debelius, J.W.; Knight, R.; Koren, O. Microbial endocrinology: The interplay between the microbiota and the endocrine system. FEMS Microbiol. Rev. 2015, 39, 509–521. [Google Scholar] [CrossRef]
- Hampl, R.; Bičíková, M.; Sosvorová, L. Hormones and the blood-brain barrier. Horm. Mol. Biol. Clin. Investig. 2015, 21, 159–164. [Google Scholar] [CrossRef]
- Hirtz, A.; Rech, F.; Dubois-Pot-Schneider, H.; Dumond, H. Astrocytoma: A Hormone-Sensitive Tumor? Int. J. Mol. Sci. 2020, 21, 9114. [Google Scholar] [CrossRef]
- Altiok, N.; Ersoz, M.; Koyuturk, M. Estradiol induces JNK-dependent apoptosis in glioblastoma cells. Oncol. Lett. 2011, 2, 1281–1285. [Google Scholar] [CrossRef]
- Castracani, C.C.; Longhitano, L.; Distefano, A.; Anfuso, D.; Kalampoka, S.; La Spina, E.; Astuto, M.; Avola, R.; Caruso, M.; Nicolosi, D.; et al. Role of 17β-Estradiol on Cell Proliferation and Mitochondrial Fitness in Glioblastoma Cells. J. Oncol. 2020, 2020, 1–9. [Google Scholar] [CrossRef]
- Qi, X.; Yun, C.; Pang, Y.; Qiao, J. The impact of the gut microbiota on the reproductive and metabolic endocrine system. Gut Microbes 2021, 13, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Jarabo, P.; de Pablo, C.; Herranz, H.; Martín, F.A.; Casas-Tintó, S. Insulin signaling mediates neurodegeneration in glioma. Life Sci. Alliance 2021, 4, e202000693. [Google Scholar] [CrossRef] [PubMed]
- Verreault, M.; Vilchis, I.S.; Rosenberg, S.; Lemaire, N.; Schmitt, C.; Guehennec, J.; Royer-Perron, L.; Thomas, J.; Lam, T.T.; Dingli, F.; et al. Identification of growth hormone receptor as a relevant target for precision medicine in low-EGFR expressing glioblastoma. Clin. Transl. Med. 2022, 12, e939. [Google Scholar] [CrossRef] [PubMed]
- Lea, R.W.; Dawson, T.; Martinez-Moreno, C.G.; El-Abry, N.; Harvey, S. Growth hormone and cancer: GH production and action in glioma? Gen. Comp. Endocrinol. 2015, 220, 119–123. [Google Scholar] [CrossRef]
- Gao, C.; He, X.-F.; Xu, Q.-R.; Xu, Y.-J.; Shen, J. Sevoflurane downregulates insulin-like growth factor-1 to inhibit cell proliferation, invasion and trigger apoptosis in glioma through the PI3K/AKT signaling pathway. Anti-Cancer Drugs 2019, 30, 670–676. [Google Scholar] [CrossRef]
- Bonaz, B.; Bazin, T.; Pellissier, S. The Vagus Nerve at the Interface of the Microbiota-Gut-Brain Axis. Front. Neurosci. 2018, 12, 49. [Google Scholar] [CrossRef]
- Restaino, A.C.; Vermeer, P.D. Neural regulations of the tumor microenvironment. FASEB BioAdvances 2021, 4, 29–42. [Google Scholar] [CrossRef]
- Reijmen, E.; Vannucci, L.; De Couck, M.; De Grève, J.; Gidron, Y. Therapeutic potential of the vagus nerve in cancer. Immunol. Lett. 2018, 202, 38–43. [Google Scholar] [CrossRef]
- Dineley, K.T.; Pandya, A.A.; Yakel, J.L. Nicotinic ACh receptors as therapeutic targets in CNS disorders. Trends Pharmacol. Sci. 2015, 36, 96–108. [Google Scholar] [CrossRef]
- Zhang, Q.; Lu, Y.; Bian, H.; Guo, L.; Zhu, H. Activation of the α7 nicotinic receptor promotes lipopolysaccharide-induced conversion of M1 microglia to M2. Am. J. Transl. Res. 2017, 9, 971–985. [Google Scholar]
- Song, Y.; Gan, Y.; Wang, Q.; Meng, Z.; Li, G.; Shen, Y.; Wu, Y.; Li, P.; Yao, M.; Gu, J.; et al. Enriching the Housing Environment for Mice Enhances Their NK Cell Antitumor Immunity via Sympathetic Nerve–Dependent Regulation of NKG2D and CCR5. Cancer Res. 2017, 77, 1611–1622. [Google Scholar] [CrossRef] [PubMed]
- Luck, B.; Engevik, M.A.; Ganesh, B.; Lackey, E.P.; Lin, T.; Balderas, M.; Major, A.; Runge, J.; Luna, R.A.; Sillitoe, R.V.; et al. Bifidobacteria shape host neural circuits during postnatal development by promoting synapse formation and microglial function. Sci. Rep. 2020, 10, 7737. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Xu, J.; Yang, B.; Chen, K.; Kong, Y.; Fang, N.; Gong, T.; Wang, F.; Ling, Z.; Liu, J. Effect of Clostridium butyricum against Microglia-Mediated Neuroinflammation in Alzheimer’s Disease via Regulating Gut Microbiota and Metabolites Butyrate. Mol. Nutr. Food Res. 2020, 64, e1900636. [Google Scholar] [CrossRef] [PubMed]
- Lebovitz, Y.; Kowalski, E.A.; Wang, X.; Kelly, C.; Lee, M.; McDonald, V.; Ward, R.; Creasey, M.; Mills, W.; Basso, E.K.G.; et al. Lactobacillus rescues postnatal neurobehavioral and microglial dysfunction in a model of maternal microbiome dysbiosis. Brain Behav. Immun. 2019, 81, 617–629. [Google Scholar] [CrossRef]
- Grabowski, M.M.; Sankey, E.W.; Ryan, K.J.; Chongsathidkiet, P.; Lorrey, S.J.; Wilkinson, D.; Fecci, P.E. Immune suppression in gliomas. J. Neuro-Oncol. 2020, 151, 3–12. [Google Scholar] [CrossRef]
- Ding, X.; Zhou, J.; Zhao, L.; Chen, M.; Wang, S.; Zhang, M.; Zhang, X.; Jiang, G. Intestinal Flora Composition Determines Microglia Activation and Improves Epileptic Episode Progress. Front. Cell. Infect. Microbiol. 2022, 12, 835217. [Google Scholar] [CrossRef]
- Jia, W.; Jackson-Cook, C.; Graf, M.R. Tumor-infiltrating, myeloid-derived suppressor cells inhibit T cell activity by nitric oxide production in an intracranial rat glioma+vaccination model. J. Neuroimmunol. 2010, 223, 20–30. [Google Scholar] [CrossRef]
- Srivastava, M.K.; Sinha, P.; Clements, V.K.; Rodriguez, P.; Ostrand-Rosenberg, S. Myeloid-Derived Suppressor Cells Inhibit T-Cell Activation by Depleting Cystine and Cysteine. Cancer Res. 2010, 70, 68–77. [Google Scholar] [CrossRef]
- Feng, S.; Cheng, X.; Zhang, L.; Lu, X.; Chaudhary, S.; Teng, R.; Frederickson, C.; Champion, M.M.; Zhao, R.; Cheng, L.; et al. Myeloid-derived suppressor cells inhibit T cell activation through nitrating LCK in mouse cancers. Proc. Natl. Acad. Sci. USA 2018, 115, 10094–10099. [Google Scholar] [CrossRef]
- Law, A.M.K.; Valdes-Mora, F.; Gallego-Ortega, D. Myeloid-Derived Suppressor Cells as a Therapeutic Target for Cancer. Cells 2020, 9, 561. [Google Scholar] [CrossRef]
- Brown, R.L.; Sequeira, R.P.; Clarke, T.B. The microbiota protects against respiratory infection via GM-CSF signaling. Nat. Commun. 2017, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Jayakumar, A.; Bothwell, A.L. Stat6 Promotes Intestinal Tumorigenesis in a Mouse Model of Adenomatous Polyposis by Expansion of MDSCs and Inhibition of Cytotoxic CD8 Response. Neoplasia 2017, 19, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Carmeliet, P.; Jain, R.K. Molecular mechanisms and clinical applications of angiogenesis. Nature 2011, 473, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Nguyen, M.; Khetrapal, V.; Sonnert, N.D.; Martin, A.L.; Chen, H.; Kriegel, M.A.; Palm, N.W. Within-host evolution of a gut pathobiont facilitates liver translocation. Nature 2022, 607, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.; Li, D.; Zhao, M.; Liu, C.; Liu, J.; Zeng, A.; Shi, X.; Cheng, S.; Pan, B.; Zheng, L.; et al. Gut flora-dependent metabolite Trimethylamine-N-oxide accelerates endothelial cell senescence and vascular aging through oxidative stress. Free Radic. Biol. Med. 2018, 116, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Tumur, Z.; Niwa, T. Indoxyl Sulfate Inhibits Nitric Oxide Production and Cell Viability by Inducing Oxidative Stress in Vascular Endothelial Cells. Am. J. Nephrol. 2009, 29, 551–557. [Google Scholar] [CrossRef]
- Vicentini, F.A.; Keenan, C.M.; Wallace, L.E.; Woods, C.; Cavin, J.-B.; Flockton, A.R.; Macklin, W.B.; Belkind-Gerson, J.; Hirota, S.A.; Sharkey, K.A. Intestinal microbiota shapes gut physiology and regulates enteric neurons and glia. Microbiome 2021, 9, 1–24. [Google Scholar] [CrossRef]
- Zhao, Z.; Ning, J.; Bao, X.-Q.; Shang, M.; Ma, J.; Li, G.; Zhang, D. Fecal microbiota transplantation protects rotenone-induced Parkinson’s disease mice via suppressing inflammation mediated by the lipopolysaccharide-TLR4 signaling pathway through the microbiota-gut-brain axis. Microbiome 2021, 9, 1–27. [Google Scholar] [CrossRef]
- Naomi, R.; Embong, H.; Othman, F.; Ghazi, H.F.; Maruthey, N.; Bahari, H. Probiotics for Alzheimer’s Disease: A Systematic Review. Nutrients 2021, 14, 20. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Helmink, B.A.; Spencer, C.N.; Reuben, A.; Wargo, J.A. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell 2018, 33, 570–580. [Google Scholar] [CrossRef]
- Jiang, H.; Zeng, W.; Zhang, X.; Pei, Y.; Zhang, H.; Li, Y. The role of gut microbiota in patients with benign and malignant brain tumors: A pilot study. Bioengineered 2022, 13, 7846–7858. [Google Scholar] [CrossRef] [PubMed]
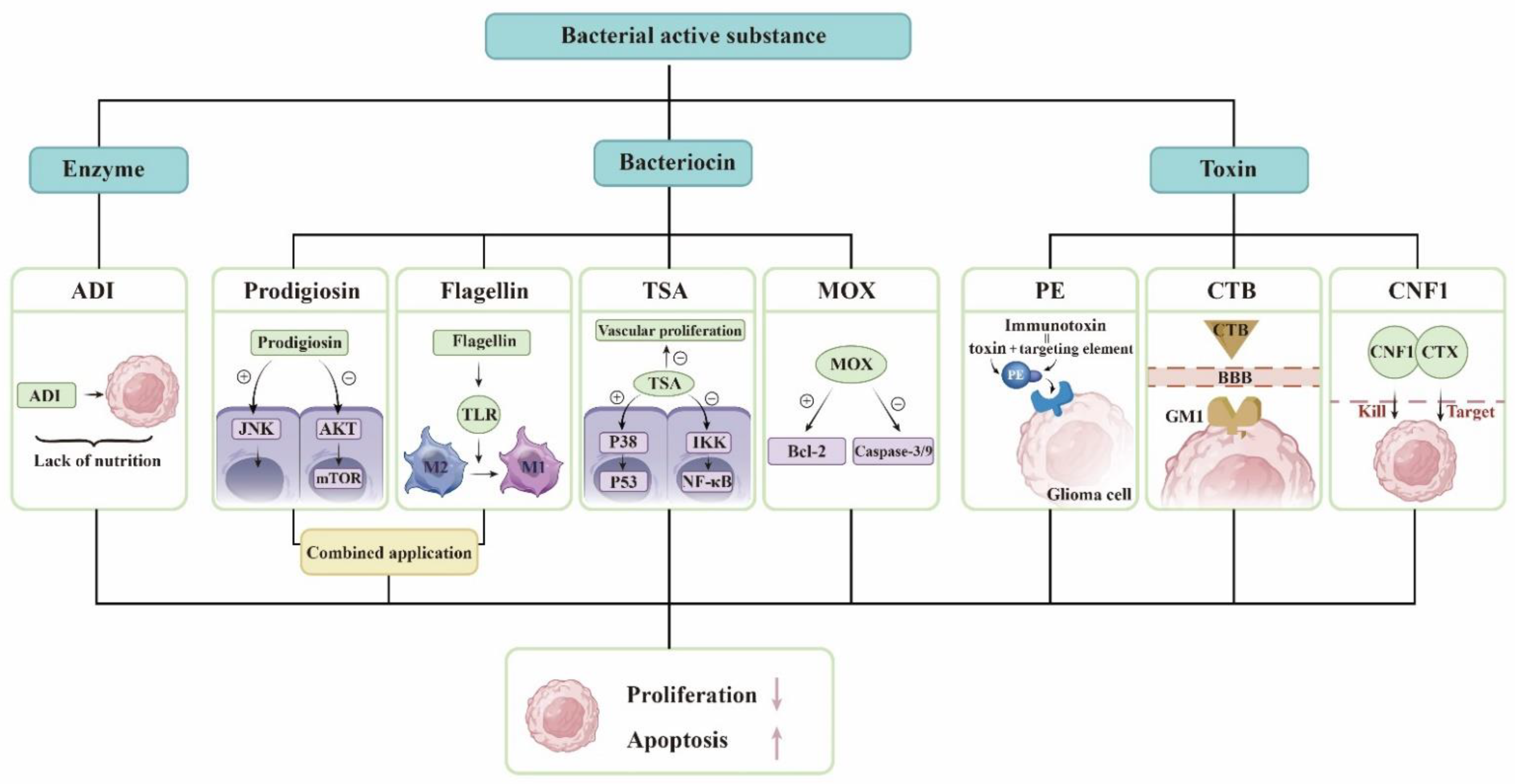
| Derivatives | Serratia marcescens | Prodigiosin induces autophagy and apoptosis in glioma cells [18]. Flagellin breaks immune tolerance [19]. |
| Pseudomonas aeruginosa | PE is used to synthesize fusion toxin to kill glioma [20]. | |
| Streptomyces | As a deacetylase inhibitor, TSA can kill glioma cells [21]. MOX induces apoptosis in glioma cells [22]. | |
| Marine Bacteria | Marine bacteria produce antiglioma substances [23]. | |
| Vibrio cholerae | CTB facilitates glioma-targeted drug delivery [24]. | |
| E. coli. | CNF1 affects the state of neurons and fights brain tumors [25]. Binding CNF1 to CTX forms a protein that prolongs the survival time of glioma mice [26]. | |
| Streptococcus pyogenes | They can produce ADI [27]. | |
| Microorganisms | HSV-1 | The virus is able to specifically infect tumor cells and induce tumor lysis by releasing viral progeny [28]. |
| Salmonella | Glioma xenografts can be targeted by injecting genetically engineered Salmonella typhimurium in the tail vein of mice [29]. Glioma mice survived longer by using genetically engineered Salmonella typhimurium that targets delivery and expresses TIMP-2 [30]. | |
| Filamentous phage | Filamentous phages are ideal drug delivery agents [31]. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Liu, Y.; Zhang, A.; Yu, W.; Lei, Q.; Xiao, B.; Luo, Z. Investigational Microbiological Therapy for Glioma. Cancers 2022, 14, 5977. https://doi.org/10.3390/cancers14235977
Wang J, Liu Y, Zhang A, Yu W, Lei Q, Xiao B, Luo Z. Investigational Microbiological Therapy for Glioma. Cancers. 2022; 14(23):5977. https://doi.org/10.3390/cancers14235977
Chicago/Turabian StyleWang, Jing, Yaxue Liu, Aohan Zhang, Wenxin Yu, Qian Lei, Bo Xiao, and Zhaohui Luo. 2022. "Investigational Microbiological Therapy for Glioma" Cancers 14, no. 23: 5977. https://doi.org/10.3390/cancers14235977
APA StyleWang, J., Liu, Y., Zhang, A., Yu, W., Lei, Q., Xiao, B., & Luo, Z. (2022). Investigational Microbiological Therapy for Glioma. Cancers, 14(23), 5977. https://doi.org/10.3390/cancers14235977







