Placental Mesenchymal Dysplasia and Beckwith–Wiedemann Syndrome
Abstract
Simple Summary
Abstract
1. Introduction
2. Incidence and Pathology of PMD
2.1. Incidence
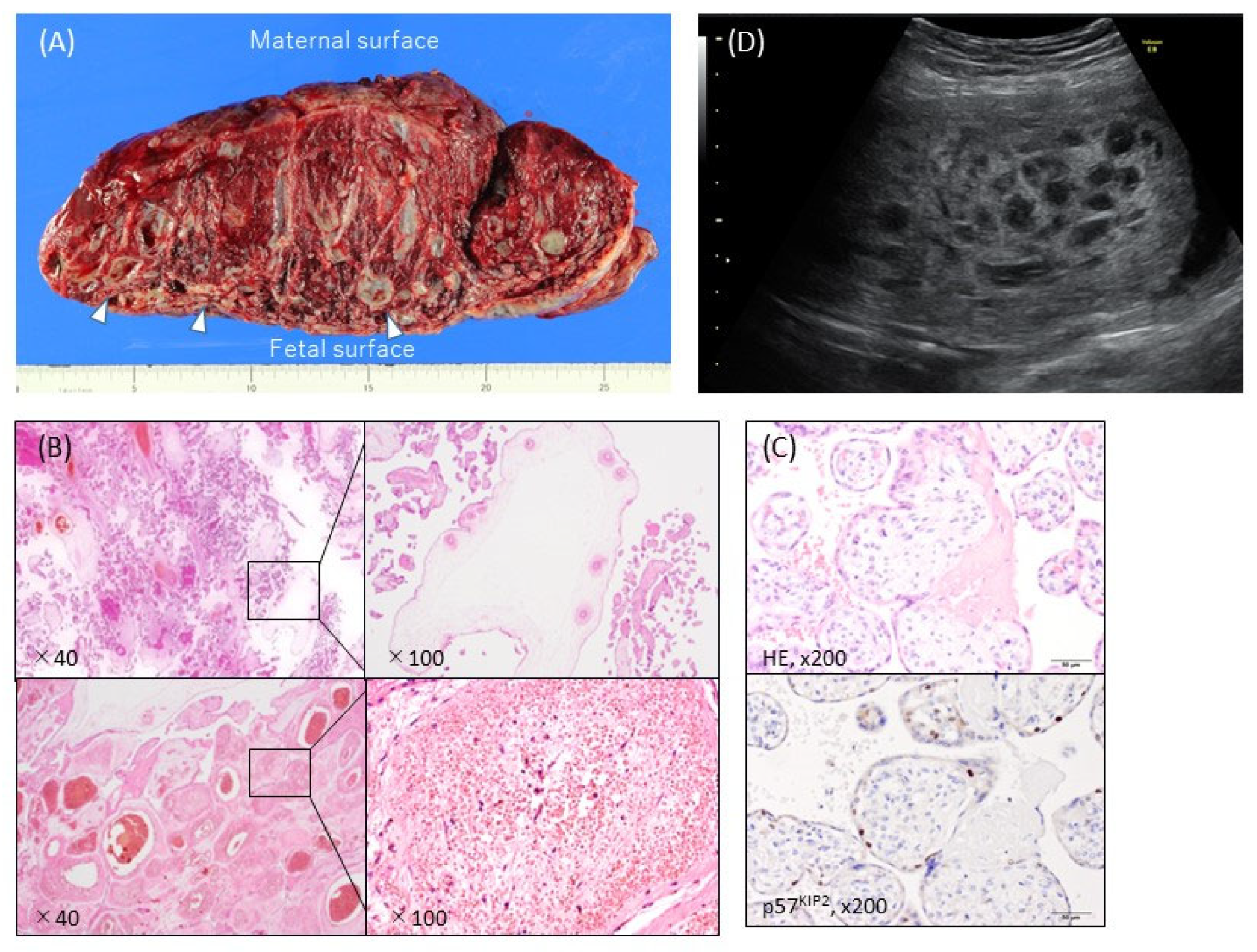
2.2. Pathology
3. Clinical Findings in PMD
3.1. Ultrasound Findings
3.2. Maternal Complications and Pregnancy and Neonatal Outcome
4. Complications of PMD
4.1. BWS
4.2. Hepatic Mesenchymal Hamartoma
5. Etiology
5.1. Etiology of BWS
5.2. Etiology of PMD
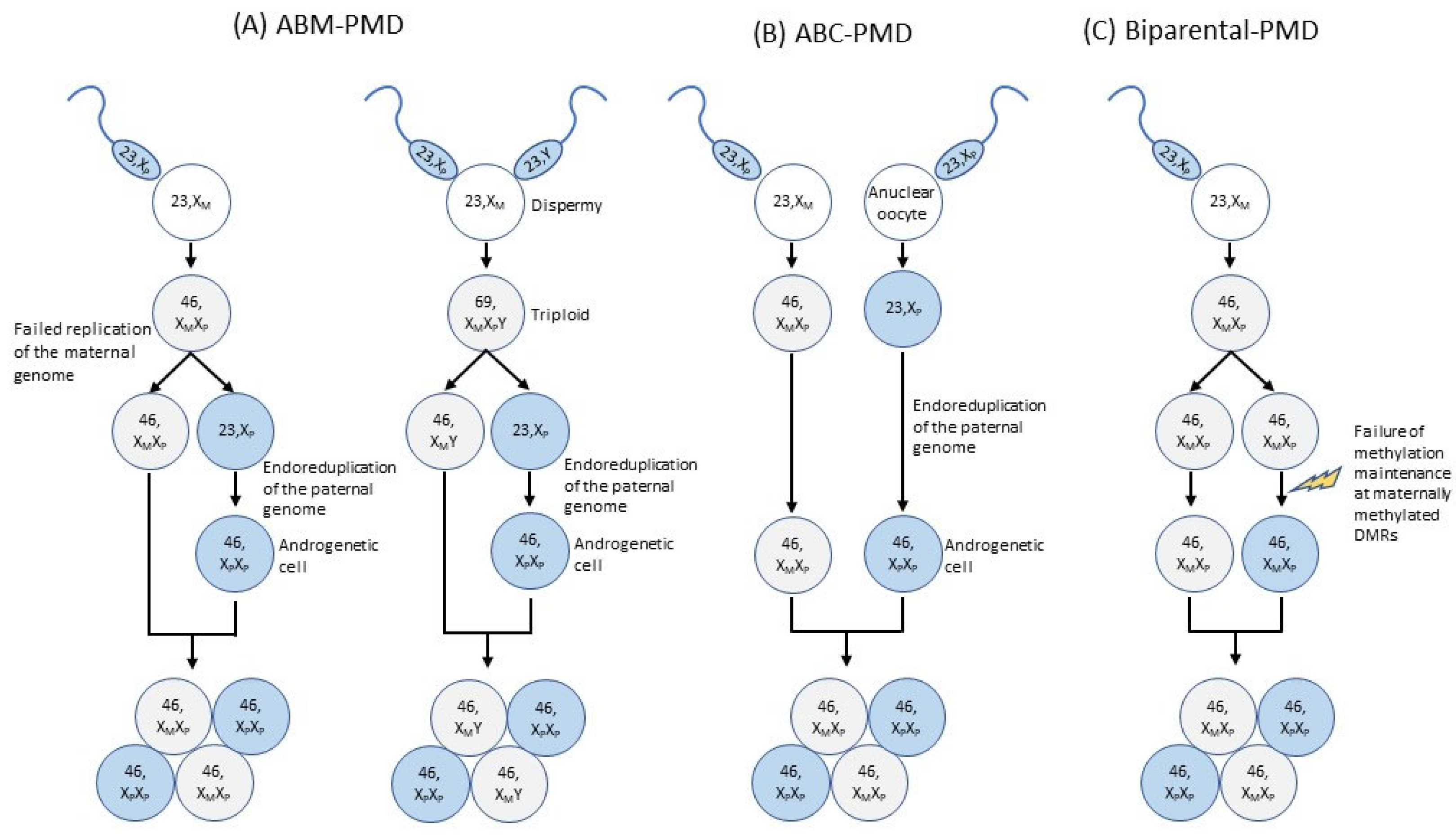
5.2.1. Etiology of PMD with ABM or ABC
5.2.2. Etiology of Biparental PMD
5.2.3. Other Possible Etiologies
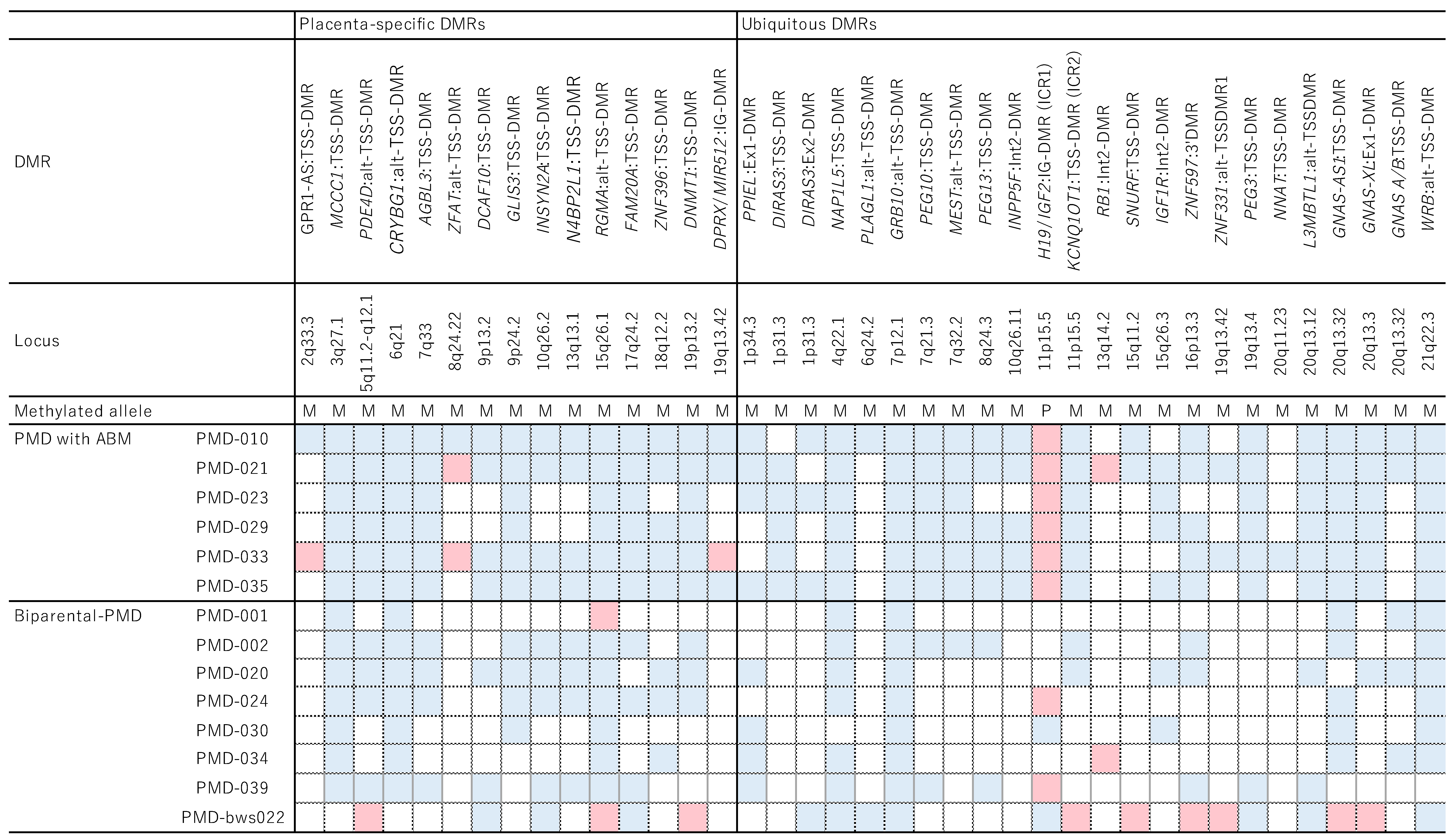
5.3. Molecular Characteristics of BWS and PMD
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Moscoso, G.; Jauniaux, E.; Hustin, J. Placental Vascular Anomaly with Diffuse Mesenchymal Stem Villous Hyperplasia. A New Clinico-Pathological Entity? Pathol. Res. Pract. 1991, 187, 324–328. [Google Scholar] [CrossRef]
- Guenot, C.; Kingdom, J.; De Rham, M.; Osterheld, M.; Keating, S.; Vial, Y.; Van Mieghem, T.; Jastrow, N.; Raio, L.; Spinelli, M.; et al. Placental Mesenchymal Dysplasia: An Underdiagnosed Placental Pathology with Various Clinical Outcomes. Eur. J. Obstet. Gynecol. Reprod. Biol. 2019, 234, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Colpaert, R.M.; Ramseyer, A.M.; Luu, T.; Quick, C.M.; Frye, L.T.; Magann, E.F. Diagnosis and Management of Placental Mesenchymal Disease. A Review of the Literature. Obstet. Gynecol. Surv. 2019, 74, 611–622. [Google Scholar] [PubMed]
- Pham, T.; Stayboldt, C.; Steele, J.; Chan, L.; Benirschke, K. Placental Mesenchymal Dysplasia Is Associated with High Rates of Intrauterine Growth Restriction and Fetal Demise: A Report of 11 New Cases and a Review of the Literature. Am. J. Clin. Pathol. 2006, 126, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Nayeri, U.A.; West, A.B.; Nardini, H.K.G.; Copel, J.A.; Sfakianaki, A.K. Systematic Review of Sonographic Findings of Placental Mesenchymal Dysplasia and Subsequent Pregnancy Outcome. Ultrasound Obstet. Gynecol. 2013, 41, 366–374. [Google Scholar] [CrossRef]
- Kodera, C.; Aoki, S.; Ohba, T.; Higashimoto, K.; Mikami, Y.; Fukunaga, M.; Soejima, H.; Katabuchi, H. Clinical Manifestations of Placental Mesenchymal Dysplasia in Japan: A Multicenter Case Series. J. Obstet. Gynaecol. Res. 2021, 47, 1118–1125. [Google Scholar] [CrossRef]
- Surti, U.; Hill, L.M.; Dunn, J.; Prosen, T.; Hoffner, L. Twin Pregnancy with a Chimeric Androgenetic and Biparental Placenta in One Twin Displaying Placental Mesenchymal Dysplasia Phenotype. Prenat. Diagn. 2005, 25, 1048–1056. [Google Scholar] [CrossRef]
- Kaiser-Rogers, K.A.; McFadden, D.E.; Livasy, C.A.; Dansereau, J.; Jiang, R.; Knops, J.F.; Lefebvre, L.; Rao, K.W.; Robinson, W.P. Androgenetic/Biparental Mosaicism Causes Placental Mesenchymal Dysplasia. J. Med. Genet. 2006, 43, 187–192. [Google Scholar] [CrossRef]
- Robinson, W.P.; Lauzon, J.L.; Innes, A.; Lim, K.; Arsovska, S.; McFadden, D.E. Origin and Outcome of Pregnancies Affected by Androgenetic/Biparental Chimerism. Hum. Reprod. 2007, 22, 1114–1122. [Google Scholar] [CrossRef]
- Morales, C.; Soler, A.; Badenas, C.; Rodríguez-Revenga, L.; Nadal, A.; Martínez, J.M.; Mademont-Soler, I.; Borrell, A.; Milà, M.; Sánchez, A. Reproductive Consequences of Genome-Wide Paternal Uniparental Disomy Mosaicism: Description of Two Cases with Different Mechanisms of Origin and Pregnancy Outcomes. Fertil. Steril. 2009, 92, 393.e5–393.e9. [Google Scholar] [CrossRef]
- Beckwith, J.B. Extreme Cytomegaly of the Adrenal Fetal Cortex, Omphalocele, Hyperplasia of Kidneys and Pancreas, and Leydig-Cell Hyperplasia: Another Syndrome? In Proceedings of the Western Society for Pediatric Research, Los Angeles, CA, USA, 11 November 1963. [Google Scholar]
- Wiedemann, H.R. Complexe Malformatif Familial Avec Hernie Ombilicale Et Macroglossia, Un Syndrome Nouveau. J. Genet. Hum. 1964, 13, 223–232. [Google Scholar] [PubMed]
- Brioude, F.; Kalish, J.M.; Mussa, A.; Foster, A.C.; Bliek, J.; Ferrero, G.B.; Boonen, S.E.; Cole, T.; Baker, R.; Bertoletti, M.; et al. Expert Consensus Document: Clinical and Molecular Diagnosis, Screening and Management of Beckwith-Wiedemann Syndrome: An International Consensus Statement. Nat. Rev. Endocrinol. 2018, 14, 229–249. [Google Scholar] [CrossRef]
- Soejima, H.; Higashimoto, K. Epigenetic and Genetic Alterations of the Imprinting Disorder Beckwith-Wiedemann Syndrome and Related Disorders. J. Hum. Genet. 2013, 58, 402–409. [Google Scholar] [CrossRef]
- Sparago, A.; Russo, S.; Cerrato, F.; Ferraiuolo, S.; Castorina, P.; Selicorni, A.; Schwienbacher, C.; Negrini, M.; Ferrero, G.B.; Silengo, M.C.; et al. Mechanisms Causing Imprinting Defects in Familial Beckwith-Wiedemann Syndrome with Wilms’ Tumour. Hum. Mol. Genet. 2007, 16, 254–264. [Google Scholar] [CrossRef]
- Demars, J.; Shmela, M.E.; Rossignol, S.; Okabe, J.; Netchine, I.; Azzi, S.; Cabrol, S.; Le Caignec, C.; David, A.; Le Bouc, Y.; et al. Analysis of the Igf2/H19 Imprinting Control Region Uncovers New Genetic Defects, Including Mutations of Oct-Binding Sequences, in Patients with 11p15 Fetal Growth Disorders. Hum. Mol. Genet. 2010, 19, 803–814. [Google Scholar] [CrossRef]
- Sheppard, S.E.; Lalonde, E.; Adzick, N.S.; Beck, A.E.; Bhatti, T.; De Leon, D.D.; Duffy, K.A.; Ganguly, A.; Hathaway, E.; Ji, J.; et al. Androgenetic Chimerism as an Etiology for Beckwith-Wiedemann Syndrome: Diagnosis and Management. Genet. Med. 2019, 21, 2644–2649. [Google Scholar] [CrossRef]
- Valente, F.M.; Sparago, A.; Freschi, A.; Hill-Harfe, K.; Maas, S.M.; Frints, S.G.M.; Alders, M.; Pignata, L.; Franzese, M.; Angelini, C.; et al. Transcription Alterations of Kcnq1 Associated with Imprinted Methylation Defects in the Beckwith-Wiedemann Locus. Genet. Med. 2019, 21, 1808–1820. [Google Scholar] [CrossRef]
- Eßinger, C.; Karch, S.; Moog, U.; Fekete, G.; Lengyel, A.; Pinti, E.; Eggermann, T.; Begemann, M. Frequency of Kcnq1 Variants Causing Loss of Methylation of Imprinting Centre 2 in Beckwith-Wiedemann Syndrome. Clin. Epigenetics 2020, 12, 63. [Google Scholar] [CrossRef]
- Cohen, M.C.; Roper, E.C.; Sebire, N.; Stanek, J.; Anumba, D.O.C. Placental Mesenchymal Dysplasia Associated with Fetal Aneuploidy. Prenat. Diagn. 2005, 25, 187–192. [Google Scholar] [CrossRef]
- Paradinas, F.J.; Sebire, N.J.; Fisher, R.A.; Rees, H.C.; Foskett, M.; Seckl, M.J.; Newlands, E.S. Pseudo-Partial Moles: Placental Stem Vessel Hydrops and the Association with Beckwith-Wiedemann Syndrome and Complete Moles. Histopathology 2001, 39, 447–454. [Google Scholar] [CrossRef]
- Arizawa, M.; Nakayama, M. Suspected Involvement of the X Chromosome in Placental Mesenchymal Dysplasia. Congenit. Anom. 2002, 42, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Chen, M.F.; Bureau, Y.-A.; Brown, R. Placental Mesenchymal Dysplasia and an Estimation of the Population Incidence. Acta Obstet. Gynecol. Scand. 2012, 91, 754–757. [Google Scholar] [CrossRef] [PubMed]
- Ohira, S.; Ookubo, N.; Tanaka, K.; Takatsu, A.; Kobara, H.; Kikuchi, N.; Ohya, A.; Kanai, M.; Shiozawa, T. Placental Mesenchymal Dysplasia: Chronological Observation of Placental Images During Gestation and Review of the Literature. Gynecol. Obstet. Investig. 2013, 75, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.L.; Walters-Sen, L.C.; Stanek, J.W. Placental Pathology in Placental Mesenchymal Dysplasia with 13q12.11 Deletion and a 25-Week Gestation Female Infant. Am. J. Case Rep. 2018, 19, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Müngen, E.; Dundar, O.; Muhcu, M.; Haholu, A.; Tunca, Y. Placental Mesenchymal Dysplasia Associated with Trisomy 13: Sonographic Findings. J. Clin. Ultrasound 2008, 36, 454–456. [Google Scholar] [CrossRef]
- Lokan, J.; Chan, Y.F.; Agnesta, F. Placental Mesenchymal Dysplasia. Pathology 2002, 34, 375–378. [Google Scholar] [CrossRef]
- Aoki, S.; Higashimoto, K.; Hidaka, H.; Ohtsuka, Y.; Aoki, S.; Mishima, H.; Yoshiura, K.-I.; Nakabayashi, K.; Hata, K.; Yatsuki, H.; et al. Aberrant Hypomethylation at Imprinted Differentially Methylated Regions Is Involved in Biparental Placental Mesenchymal Dysplasia. Clin. Epigenetics 2022, 14, 64. [Google Scholar] [CrossRef]
- Chilosi, M.; Piazzola, E.; Lestani, M.; Benedetti, A.; Guasparri, I.; Granchelli, G.; Aldovini, D.; Leonardi, E.; Pizzolo, G.; Doglioni, C.; et al. Differential Expression of P57kip2, a Maternally Imprinted Cdk Inhibitor, in Normal Human Placenta and Gestational Trophoblastic Disease. Lab. Investig. 1998, 78, 269–276. [Google Scholar]
- Chan, Y.F.; Sampson, A. Placental Mesenchymal Dysplasia: A Report of Four Cases with Differentiation from Partial Hydatidiform Mole. Aust. N. Z. J. Obstet. Gynaecol. 2003, 43, 475–479. [Google Scholar] [CrossRef]
- Fisher, R.; Hodges, M.D.J.; Rees, H.C.; Sebire, N.; Seckl, M.J.; Newlands, E.S.; Genest, D.R.; Castrillon, D.H. The Maternally Transcribed Gene P57(Kip2) (Cdnk1c) Is Abnormally Expressed in Both Androgenetic and Biparental Complete Hydatidiform Moles. Hum. Mol. Genet. 2002, 11, 3267–3272. [Google Scholar] [CrossRef]
- Armes, J.E.; McGown, I.; Williams, M.; Broomfield, A.; Gough, K.; Lehane, F.; Lourie, R. The Placenta in Beckwith-Wiedemann Syndrome: Genotype-Phenotype Associations, Excessive Extravillous Trophoblast and Placental Mesenchymal Dysplasia. Pathology 2012, 44, 519–527. [Google Scholar] [CrossRef]
- Sun, Y.; Zheng, J.; Jiang, Z.; Zhang, L.; Ding, S.; Liu, X. Placental Mesenchymal Dysplasia in a Normal Female Infant: A Rare Case Report with Follow-Up. Int. J. Clin. Exp. Pathol. 2020, 13, 896–900. [Google Scholar]
- Gaillot-Durand, L.; Brioude, F.; Beneteau, C.; Le Breton, F.; Massardier, J.; Michon, L.; Devouassoux-Shisheboran, M.; Allias, F. Placental Pathology in Beckwith-Wiedemann Syndrome According to Genotype/Epigenotype Subgroups. Fetal Pediatr. Pathol. 2018, 37, 387–399. [Google Scholar] [CrossRef]
- Duffy, K.A.; Cielo, C.M.; Cohen, J.L.; Gonzalez-Gandolfi, C.X.; Griff, J.R.; Hathaway, E.R.; Kupa, J.; Taylor, J.A.; Wang, K.H.; Ganguly, A.; et al. Characterization of the Beckwith-Wiedemann Spectrum: Diagnosis and Management. Am. J. Med. Genet. C Semin. Med. Genet. 2019, 181, 693–708. [Google Scholar] [CrossRef]
- Jimbo, T.; Fujita, Y.; Yumoto, Y.; Fukushima, K.; Kato, K. Rare Fetal Complications Associated with Placental Mesenchymal Dysplasia: A Report of Two Cases. J. Obstet. Gynaecol. Res. 2015, 41, 304–308. [Google Scholar] [CrossRef]
- Meyers, R.L. Tumors of the Liver in Children. Surg. Oncol. 2007, 16, 195–203. [Google Scholar] [CrossRef]
- Foucar, E.; Williamson, R.; Yiu-Chiu, V.; Varner, M.; Kay, B. Mesenchymal Hamartoma of the Liver Identified by Fetal Sonography. AJR Am. J. Roentgenol. 1983, 140, 970–972. [Google Scholar] [CrossRef]
- Alwaidh, M.H.; Woodhall, C.R.; Carty, H.T. Mesenchymal Hamartoma of the Liver: A Case Report. Pediatr. Radiol. 1997, 27, 247–249. [Google Scholar] [CrossRef]
- Tovbin, J.; Segal, M.; Tavori, I.; Lotan, G.; Maymon, R. Hepatic Mesenchymal Hamartoma: A Pediatric Tumor That May Be Diagnosed Prenatally. Ultrasound Obstet. Gynecol. 1997, 10, 63–65. [Google Scholar] [CrossRef]
- Kitano, Y.; Ruchelli, E.; Weiner, S.; Adzick, N.S. Hepatic Mesenchymal Hamartoma Associated with Mesenchymal Stem Villous Hyperplasia of the Placenta. Fetal Diagn. Ther. 2000, 15, 134–138. [Google Scholar] [CrossRef]
- Carta, M.; Maresi, E.; Giuffrè, M.; Catalano, G.; Piro, E.; Siracusa, F.; Corsello, G. Congenital Hepatic Mesenchymal Hamartoma Associated with Mesenchymal Stem Villous Hyperplasia of the Placenta: Case Report. J. Pediatr. Surg. 2005, 40, e37–e39. [Google Scholar] [CrossRef] [PubMed]
- Laberge, J.-M.; Patenaude, Y.; Desilets, V.; Cartier, L.; Khalife, S.; Jutras, L.; Chen, M.-F. Chen. Large Hepatic Mesenchymal Hamartoma Leading to Mid-Trimester Fetal Demise. Fetal Diagn. Ther. 2005, 20, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Francis, B.; Hallam, L.; Kecskes, Z.; Croaker, D.; Ellwood, D.; Kent, A. Placental Mesenchymal Dysplasia Associated with Hepatic Mesenchymal Hamartoma in the Newborn. Pediatr. Dev. Pathol. 2007, 10, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Reed, R.C.; Beischel, L.; Schoof, J.; Johnson, J.; Raff, M.L.; Kapur, R.P. Androgenetic/Biparental Mosaicism in an Infant with Hepatic Mesenchymal Hamartoma and Placental Mesenchymal Dysplasia. Pediatr. Dev. Pathol. 2008, 11, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Cornette, J.; Festen, S.; Hoonaard, T.V.D.; Steegers, E. Mesenchymal Hamartoma of the Liver: A Benign Tumor with Deceptive Prognosis in the Perinatal Period. Case Report and Review of the Literature. Fetal Diagn. Ther. 2009, 25, 196–202. [Google Scholar] [CrossRef]
- Tortoledo, M.; Galindo, A.; Ibarrola, C. Placental Mesenchymal Dysplasia Associated with Hepatic and Pulmonary Hamartoma. Fetal Pediatr. Pathol. 2010, 29, 261–270. [Google Scholar] [CrossRef]
- Mack-Detlefsen, B.; Boemers, T.M.; Groneck, P.; Bald, R. Multiple Hepatic Mesenchymal Hamartomas in a Premature Associated with Placental Mesenchymal Dysplasia. J. Pediatr. Surg. 2011, 46, e23–e25. [Google Scholar] [CrossRef]
- Ruhland, B.; Schröer, A.; Gembruch, U.; Noack, F.; Weichert, J. Prenatal Imaging and Postnatal Pathologic Work-up in a Case of Fetal Hepatic Hamartoma and Placental Mesenchymal Dysplasia. Ultrasound Obstet. Gynecol. 2011, 38, 360–362. [Google Scholar] [CrossRef]
- Harris, K.; Carreon, C.K.; Vohra, N.; Williamson, A.; Dolgin, S.; Rochelson, B. Placental Mesenchymal Dysplasia with Hepatic Mesenchymal Hamartoma: A Case Report and Literature Review. Fetal Pediatr. Pathol. 2013, 32, 448–453. [Google Scholar] [CrossRef]
- Kalish, J.M.; Conlin, L.K.; Bhatti, T.R.; Dubbs, H.A.; Harris, M.C.; Izumi, K.; Mostoufi-Moab, S.; Mulchandani, S.; Saitta, S.; States, L.J.; et al. Clinical Features of Three Girls with Mosaic Genome-Wide Paternal Uniparental Isodisomy. Am. J. Med. Genet. A 2013, 161A, 1929–1939. [Google Scholar] [CrossRef]
- Kapur, R.P.; Berry, J.E.; Tsuchiya, K.D.; Opheim, K.E. Activation of the Chromosome 19q Microrna Cluster in Sporadic and Androgenetic-Biparental Mosaicism-Associated Hepatic Mesenchymal Hamartoma. Pediatr. Dev. Pathol. 2014, 17, 75–84. [Google Scholar] [CrossRef]
- Keller, R.B.; El Demellawy, D.; Quaglia, A.; Finegold, M.; Kapur, R.P. Methylation Status of the Chromosome Arm 19q Microrna Cluster in Sporadic and Androgenetic-Biparental Mosaicism-Associated Hepatic Mesenchymal Hamartoma. Pediatr. Dev. Pathol. 2015, 18, 218–227. [Google Scholar] [CrossRef]
- Gurram, D.; Joung, S.J.S.; Ryder, L.; Nayyar, R. Late Diagnosis of Hepatic Mesenchymal Hamartoma and Placental Mesenchymal Dysplasia. Australas J. Ultrasound Med. 2016, 19, 123–125. [Google Scholar] [CrossRef]
- Repnikova, E.; Roberts, J.; Kats, A.; Habeebu, S.; Schwager, C.; Joyce, J.; Manalang, M.; Amudhavalli, S. Biparental/Androgenetic Mosaicism in a Male with Features of Overgrowth and Placental Mesenchymal Dysplasia. Clin. Genet. 2018, 94, 564–568. [Google Scholar] [CrossRef]
- Noguer-Dance, M.; Abu-Amero, S.; Al-Khtib, M.; Lefèvre, A.; Coullin, P.; Moore, G.E.; Cavaillé, J. The Primate-Specific Microrna Gene Cluster (C19mc) Is Imprinted in the Placenta. Hum. Mol. Genet. 2010, 19, 3566–3582. [Google Scholar] [CrossRef]
- Higashimoto, K.; Jozaki, K.; Kosho, T.; Matsubara, K.; Fuke, T.; Yamada, D.; Yatsuki, H.; Maeda, T.; Ohtsuka, Y.; Nishioka, K.; et al. A Novel De Novo Point Mutation of the Oct-Binding Site in the Igf2/H19-Imprinting Control Region in a Beckwith-Wiedemann Syndrome Patient. Clin. Genet. 2014, 86, 539–544. [Google Scholar] [CrossRef]
- Sun, F.; Higashimoto, K.; Awaji, A.; Ohishi, K.; Nishizaki, N.; Tanoue, Y.; Aoki, S.; Watanabe, H.; Yatsuki, H.; Soejima, H. The Extent of DNA Methylation Anticipation Due to a Genetic Defect in Icr1 in Beckwith-Wiedemann Syndrome. J. Hum. Genet. 2019, 64, 937–943. [Google Scholar] [CrossRef]
- Ohtsuka, Y.; Higashimoto, K.; Sasaki, K.; Jozaki, K.; Yoshinaga, H.; Okamoto, N.; Takama, Y.; Kubota, A.; Nakayama, M.; Yatsuki, H.; et al. Autosomal Recessive Cystinuria Caused by Genome-Wide Paternal Uniparental Isodisomy in a Patient with Beckwith-Wiedemann Syndrome. Clin. Genet. 2015, 88, 261–266. [Google Scholar] [CrossRef]
- Ohtsuka, Y.; Higashimoto, K.; Oka, T.; Yatsuki, H.; Jozaki, K.; Maeda, T.; Kawahara, K.; Hamasaki, Y.; Matsuo, M.; Nishioka, K.; et al. Identification of Consensus Motifs Associated with Mitotic Recombination and Clinical Characteristics in Patients with Paternal Uniparental Isodisomy of Chromosome 11. Hum. Mol. Genet. 2016, 25, 1406–1419. [Google Scholar] [CrossRef]
- Papulino, C.; Chianese, U.; Nicoletti, M.M.; Benedetti, R.; Altucci, L. Preclinical and Clinical Epigenetic-Based Reconsideration of Beckwith-Wiedemann Syndrome. Front. Genet. 2020, 11, 563718. [Google Scholar] [CrossRef]
- Postema, F.A.; Bliek, J.; Noesel, C.J.; Zutven, L.J.; Oosterwijk, J.C.; Hopman, S.M.J.; Merks, J.H.M.; Hennekam, R.C. Multiple Tumors Due to Mosaic Genome-Wide Paternal Uniparental Disomy. Pediatr. Blood Cancer 2019, 66, e27715. [Google Scholar] [CrossRef] [PubMed]
- Heide, S.; Chantot-Bastaraud, S.; Keren, B.; Harbison, M.D.; Azzi, S.; Rossignol, S.; Michot, C.; Lys, M.L.-P.; Demeer, B.; Heinrichs, C.; et al. Chromosomal Rearrangements in the 11p15 Imprinted Region: 17 New 11p15.5 Duplications with Associated Phenotypes and Putative Functional Consequences. J. Med. Genet. 2018, 55, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Delgado, M.; Riccio, A.; Eggermann, T.; Maher, E.R.; Lapunzina, P.; Mackay, D.; Monk, D. Causes and Consequences of Multi-Locus Imprinting Disturbances in Humans. Trends Genet. 2016, 32, 444–455. [Google Scholar] [CrossRef] [PubMed]
- Begemann, M.; Rezwan, F.I.; Beygo, J.; Docherty, L.E.; Kolarova, J.; Schroeder, C.; Buiting, K.; Chokkalingam, K.; Degenhardt, F.; Wakeling, E.L.; et al. Maternal Variants in Nlrp and Other Maternal Effect Proteins Are Associated with Multilocus Imprinting Disturbance in Offspring. J. Med. Genet. 2018, 55, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Elbracht, M.; Mackay, D.J.; Begemann, M.; Kagan, K.O.; Eggermann, T. Disturbed Genomic Imprinting and Its Relevance for Human Reproduction: Causes and Clinical Consequences. Hum. Reprod. Update 2020, 26, 197–213. [Google Scholar] [CrossRef]
- Monk, D.; Sanchez-Delgado, M.; Fisher, R. Fisher. Nlrps, the Subcortical Maternal Complex and Genomic Imprinting. Reproduction 2017, 154, R161–R170. [Google Scholar] [CrossRef]
- Robinson, W.; Slee, J.; Smith, N.; Murch, A.; Watson, S.K.; Lam, W.; McFadden, D. Placental Mesenchymal Dysplasia Associated with Fetal Overgrowth and Mosaic Deletion of the Maternal Copy of 11p15.5. Am. J. Med. Genet. A 2007, 143A, 1752–1759. [Google Scholar] [CrossRef]
- Drut, R.M.; Drut, R. Nonimmune Fetal Hydrops and Placentomegaly: Diagnosis of Familial Wiedemann-Beckwith Syndrome with Trisomy 11p15 Using Fish. Am. J. Med. Genet. 1996, 62, 145–149. [Google Scholar] [CrossRef]
- Caspary, T.; Cleary, M.A.; Perlman, E.J.; Zhang, P.; Elledge, S.J.; Tilghman, S.M. Oppositely Imprinted Genes P57(Kip2) and Igf2 Interact in a Mouse Model for Beckwith-Wiedemann Syndrome. Genes Dev. 1999, 13, 3115–3124. [Google Scholar] [CrossRef]
- Court, F.; Tayama, C.; Romanelli, V.; Martin-Trujillo, A.; Iglesias-Platas, I.; Okamura, K.; Sugahara, N.; Simón, C.; Moore, H.; Harness, J.V.; et al. Genome-Wide Parent-of-Origin DNA Methylation Analysis Reveals the Intricacies of Human Imprinting and Suggests a Germline Methylation-Independent Mechanism of Establishment. Genome Res. 2014, 24, 554–569. [Google Scholar] [CrossRef]
- HaHanna, C.W.; Peñaherrera, M.S.; Saadeh, H.; Andrews, S.; McFadden, D.E.; Kelsey, G.; Robinson, W.P. Pervasive Polymorphic Imprinted Methylation in the Human Placenta. Genome Res. 2016, 26, 756–767. [Google Scholar] [CrossRef]
- Hamada, H.; Okae, H.; Toh, H.; Chiba, H.; Hiura, H.; Shirane, K.; Sato, T.; Suyama, M.; Yaegashi, N.; Sasaki, H.; et al. Allele-Specific Methylome and Transcriptome Analysis Reveals Widespread Imprinting in the Human Placenta. Am. J. Hum. Genet. 2016, 99, 1045–1058. [Google Scholar] [CrossRef]
- Højberg, K.-E.; Aagaard, J.; Henriques, U.; Sunde, L. Placental Vascular Malformation with Mesenchymal Hyperplasia and a Localized Chorioangioma. A Rarity Simulating Partial Mole. Pathol. Res. Pract. 1994, 190, 808–813, discussion 14. [Google Scholar] [CrossRef]
- Chen, C.-P.; Chern, S.-R.; Wang, T.-Y.; Huang, Z.-D.; Huang, M.-C.; Chuang, C.-Y. Pregnancy with Concomitant Chorangioma and Placental Vascular Malformation with Mesenchymal Hyperplasia. Hum. Reprod. 1997, 12, 2553–2556. [Google Scholar] [CrossRef][Green Version]
- Kotani, T.; Sumigama, S.; Tsuda, H.; Mano, Y.; Yamamoto, E.; Iwase, A.; Shimoyama, Y.; Nagasaka, T.; Hayakawa, H.; Ino, K.; et al. A Case Report of Placental Mesenchymal Dysplasia with an Increased Vegf-D Expression. Placenta 2012, 33, 888–891. [Google Scholar] [CrossRef]
- Huang, T.-C.; Chang, K.-C.; Chang, J.-Y.; Tsai, Y.-S.; Yang, Y.-J.; Chang, W.-C.; Mo, C.-F.; Yu, P.-H.; Chiang, C.-T.; Lin, S.-P.; et al. Variants in Maternal Effect Genes and Relaxed Imprinting Control in a Special Placental Mesenchymal Dysplasia Case with Mild Trophoblast Hyperplasia. Biomedicines 2021, 9, 544. [Google Scholar] [CrossRef]
- Hoffner, L.; Dunn, J.; Esposito, N.; Macpherson, T.; Surti, U. P57kip2 Immunostaining and Molecular Cytogenetics: Combined Approach Aids in Diagnosis of Morphologically Challenging Cases with Molar Phenotype and in Detecting Androgenetic Cell Lines in Mosaic/Chimeric Conceptions. Hum. Pathol. 2008, 39, 63–72. [Google Scholar] [CrossRef]
- Beechey, C.V. A Reassessment of Imprinting Regions and Phenotypes on Mouse Chromosome 6: Nap1l5 Locates within the Currently Defined Sub-Proximal Imprinting Region. Cytogenet. Genome Res. 2004, 107, 108–114. [Google Scholar] [CrossRef]
- Litzky, J.F.; Deyssenroth, M.A.; Everson, T.M.; Armstrong, D.A.; Lambertini, L.; Chen, J.; Marsit, C.J. Placental Imprinting Variation Associated with Assisted Reproductive Technologies and Subfertility. Epigenetics 2017, 12, 653–661. [Google Scholar] [CrossRef]
- Parry, D.; Logan, C.; Hayward, B.E.; Shires, M.; Landolsi, H.; Diggle, C.; Carr, I.; Rittore, C.; Touitou, I.; Philibert, L.; et al. Mutations Causing Familial Biparental Hydatidiform Mole Implicate C6orf221 as a Possible Regulator of Genomic Imprinting in the Human Oocyte. Am. J. Hum. Genet. 2011, 89, 451–458. [Google Scholar] [CrossRef]
- Rezaei, M.; Suresh, B.; Bereke, E.; Hadipour, Z.; Aguinaga, M.; Qian, J.; Bagga, R.; Fardaei, M.; Hemida, R.; Jagadeesh, S.; et al. Novel Pathogenic Variants in Nlrp7, Nlrp5, and Padi6 in Patients with Recurrent Hydatidiform Moles and Reproductive Failure. Clin. Genet. 2021, 99, 823–828. [Google Scholar] [CrossRef] [PubMed]
- El-Maarri, O.; Seoud, M.; Coullin, P.; Herbiniaux, U.; Oldenburg, J.; Rouleau, G.; Slim, R. Maternal Alleles Acquiring Paternal Methylation Patterns in Biparental Complete Hydatidiform Moles. Hum. Mol. Genet. 2003, 12, 1405–1413. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sanchez-Delgado, M.; Martin-Trujillo, A.; Tayama, C.; Vidal, E.; Esteller, M.; Iglesias-Platas, I.; Deo, N.; Barney, O.; Maclean, K.; Hata, K.; et al. Absence of Maternal Methylation in Biparental Hydatidiform Moles from Women with Nlrp7 Maternal-Effect Mutations Reveals Widespread Placenta-Specific Imprinting. PLoS Genet. 2015, 11, e1005644. [Google Scholar] [CrossRef] [PubMed]
- Demond, H.; Anvar, Z.; Jahromi, B.N.; Sparago, A.; Verma, A.; Davari, M.; Calzari, L.; Russo, S.; Jahromi, M.A.; Monk, D.; et al. A Khdc3l Mutation Resulting in Recurrent Hydatidiform Mole Causes Genome-Wide DNA Methylation Loss in Oocytes and Persistent Imprinting Defects Post-Fertilisation. Genome Med. 2019, 11, 84. [Google Scholar] [CrossRef]
- Takahashi, H.; Matsubara, S.; Kuwata, T.; Saruyama, M.; Usui, R.; Ohkuchi, A.; Takizawa, T.; Suzuki, M. Changes in Expression of Vascular Endothelial Growth Factor D-Related Genes in Placental Mesenchymal Dysplasia. J. Obstet. Gynaecol. Res. 2014, 40, 1145–1149. [Google Scholar] [CrossRef]
- Aviram, R.; Kidron, D.; Silverstein, S.; Lerer, I.; Abeliovich, D.; Tepper, R.; Dolfin, Z.; Markovitch, O.; Arnon, S. Placental Mesenchymal Dysplasia Associated with Transient Neonatal Diabetes Mellitus and Paternal Upd6. Placenta 2008, 29, 646–649. [Google Scholar] [CrossRef]
- H’Mida, D.; Gribaa, M.; Yacoubi, T.; Chaieb, A.; Adala, L.; Elghezal, H.; Saad, A. Placental Mesenchymal Dysplasia with Beckwith-Wiedemann Syndrome Fetus in the Context of Biparental and Androgenic Cell Lines. Placenta 2008, 29, 454–460. [Google Scholar] [CrossRef]
| BWS | PMD | |||||||
|---|---|---|---|---|---|---|---|---|
| Sex | Karyotype | Molecular Characteristics | Karyotype | PMD Genotype | DNA methylation of DMRs | Suspected Fertilization | Case no. or ID in the Publication | Reference |
| Female | 46,XX | ABM (probable heterodisomy) | 46,XX | ABM (probable heterodisomy) | NA | 2 sperms | one case report | H’mida et al. [88] |
| Male | 46,XY [17]/46,XX [2] | ABM (isodisomy) | NA | ABM (isodisomy) | NA | 2 sperms | PMD-bws027 | Aoki et al. [28] |
| Female | NA | Biparental with pUPD11 (limited to 11p) | NA | Biparental with pUPD11 (limited to 11p) | NA | 1 sperm | 4 | Armes et al. [32] |
| Female | 46,XX | No alteration | NA | ABM (isodisomy) | NA | 1 sperm | PMD-022 | Aoki et al. [28] |
| Male | NA | ICR2-LOM | NA | Biparental | Multiple aberrant methylation of both placenta-specific DMRs and ubiquitous DMRs | 1 sperm | PMD-020 * | Aoki et al. [28] |
| Female | 46,XX | pUPD11 (limited to 11p) | NA | Biparental | Multiple aberrant methylation of both placenta-specific DMRs and ubiquitous DMRs | 1 sperm | PMD-bws022 * | Aoki et al. [28] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soejima, H.; Hara, S.; Ohba, T.; Higashimoto, K. Placental Mesenchymal Dysplasia and Beckwith–Wiedemann Syndrome. Cancers 2022, 14, 5563. https://doi.org/10.3390/cancers14225563
Soejima H, Hara S, Ohba T, Higashimoto K. Placental Mesenchymal Dysplasia and Beckwith–Wiedemann Syndrome. Cancers. 2022; 14(22):5563. https://doi.org/10.3390/cancers14225563
Chicago/Turabian StyleSoejima, Hidenobu, Satoshi Hara, Takashi Ohba, and Ken Higashimoto. 2022. "Placental Mesenchymal Dysplasia and Beckwith–Wiedemann Syndrome" Cancers 14, no. 22: 5563. https://doi.org/10.3390/cancers14225563
APA StyleSoejima, H., Hara, S., Ohba, T., & Higashimoto, K. (2022). Placental Mesenchymal Dysplasia and Beckwith–Wiedemann Syndrome. Cancers, 14(22), 5563. https://doi.org/10.3390/cancers14225563





