Development and Validation of Novel Deep-Learning Models Using Multiple Data Types for Lung Cancer Survival
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Study Design and Data Source
2.2. Cohort Selection
2.3. Outcome Measurement
2.4. Feature Selection
- Demographic information: age, gender, body mass index (BMI), smoking, drinking;
- Cancer conditions: tumor size and cancer stage;
- Comorbidities: cardiovascular problems (i.e., myocardial infarction (MI), congestive heart failure (CHF), peripheral vascular disease (PVD), and cardiovascular disease (CVD)), dementia, chronic obstructive pulmonary disease (COPD), rheumatic disease, peptic ulcer disease (PUD), renal disease, liver disease, diabetes, anemia, depression, hyperlipidemia, hypertension, Parkinson’s disease, and Charlson Comorbidity Index (CCI) score. These conditions were considered if they were diagnosed in at least two outpatient claims or one hospitalization over a year before the cancer diagnosis date.
- Medications: alimentary tract and metabolism, blood and blood-forming organs, cardiovascular system, genitourinary system and hormones, musculoskeletal system, nervous system, and respiratory system. We measured patients who had used medications by receiving them for more than a month (i.e., 30 days) during a year (i.e., 360 days) before the index date.
- Laboratory tests: basophil, blood urea nitrogen (BUN), calcium, cholesterol, chloride, creatinine, eosinophil, ferritin, glucose AC, HbA1c, HCT, HGB, potassium, lymphocyte, MCH, MCHC, MCV, monocyte, sodium, neutrophil, platelet (PLT), RBC, triglyceride, and WBC. We only selected laboratory tests with a missing rate of less than 70% values a year before or a month after the index date.
- Genomic tests: ALK, EGFR, KRAS, PDL1, and ROS1. We collected genomic tests if patients had ever taken one a month after the cancer diagnosis date.
2.5. Development of the Algorithms
- The primary mode (e.g., Mode 1) included demographic information, cancer conditions, comorbidities, and medications.
- The second mode (Mode 2) included the data from Mode 1 and the laboratory tests.
- The third mode (Mode 3) included the data from Mode 1 and genomic tests.
- The fourth mode (Mode 4) considered all the above features.
2.6. Evaluating the Algorithms
3. Results
3.1. Baseline Characteristics of Patients
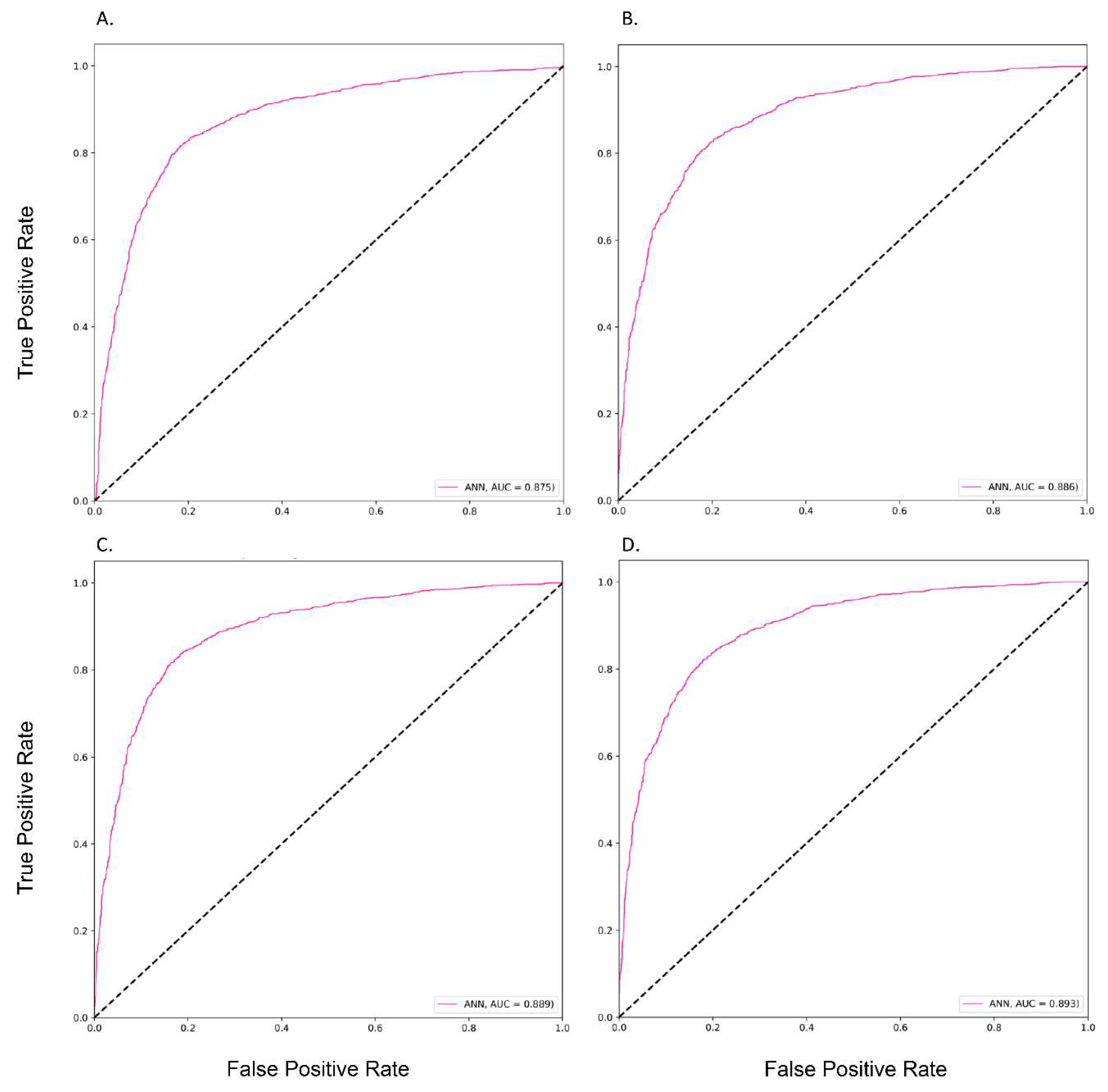
3.2. The Performances of Different Prediction Models
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| NSCLC | Non-small cell lung cancer |
| SCLC | Small cell lung cancer |
| AI | Artificial intelligence |
| TCR | Taiwan Cancer Registry |
| TDR | Taiwan Death Registry |
| TMUCRD | Taipei Medical University Clinical Research Database |
| TMUH | Taipei Medical University Hospital |
| WFH | Wan-Fang Hospital |
| SHH | Shuang-Ho Hospital |
| BMI | Body mass index |
| MI | Myocardial infarction |
| CHF | Congestive heart failure |
| PVD | Peripheral vascular disease |
| CVD | Cardiovascular disease |
| COPD | Chronic obstructive pulmonary disease |
| PUD | Peptic ulcer disease |
| CCI | Charlson Comorbidity Index |
| BUN | Blood urea nitrogen |
| PLT | Platelet |
| LR | Logistic regression |
| LDA | Linear discriminant analysis |
| LGBM | Light gradient boosting machine |
| GBM | Gradient boosting machine |
| XGBoost | Extreme gradient boosting |
| RF | Random forest |
| SVC | Support vector machine |
| ANN | Artificial neural network |
| AUC | The area under the receiver operating characteristic curve |
| PPV | Positive predictive value |
| NPV | Negative predictive value |
| SHAP | Shapley additive explanations |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Cancer Fact Sheets. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 1 November 2022).
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- World Health Organization. Lung Cancer Statistics. Available online: https://www.wcrf.org/cancer-trends/lung-cancer-statistics/ (accessed on 1 November 2022).
- Siddiqui, F.; Vaqar, S.; Siddiqui, A.H. Lung Cancer. In StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2022. [Google Scholar]
- Testa, U.; Castelli, G.; Pelosi, E. Lung Cancers: Molecular Characterization, Clonal Heterogeneity and Evolution, and Cancer Stem Cells. Cancers 2018, 10, 248. [Google Scholar] [CrossRef] [PubMed]
- Ryan, C.; Burke, L. Pathology of lung tumours. Surgery 2017, 35, 234–242. [Google Scholar] [CrossRef]
- Liang, C.-W.; Yang, H.-C.; Islam, M.M.; Nguyen, P.A.A.; Feng, Y.-T.; Hou, Z.Y.; Huang, C.-W.; Poly, T.N.; Li, Y.-C.J. Predicting Hepatocellular Carcinoma With Minimal Features From Electronic Health Records: Development of a Deep Learning Model. JMIR Cancer 2021, 7, e19812. [Google Scholar] [CrossRef] [PubMed]
- Poly, T.N.; Islam, M.M.; Muhtar, M.S.; Yang, H.-C.; Nguyen, P.A.; Li, Y.-C. Machine Learning Approach to Reduce Alert Fatigue Using a Disease Medication–Related Clinical Decision Support System: Model Development and Validation. JMIR Med Inform. 2020, 8, e19489. [Google Scholar] [CrossRef] [PubMed]
- Le, N.Q.K.; Ho, Q.-T. Deep transformers and convolutional neural network in identifying DNA N6-methyladenine sites in cross-species genomes. Methods 2022, 204, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Dang, H.H.; Ta, H.D.K.; Nguyen, T.T.T.; Anuraga, G.; Wang, C.-Y.; Lee, K.-H.; Le, N.Q.K. Prospective role and immunotherapeutic targets of sideroflexin protein family in lung adenocarcinoma: Evidence from bioinformatics validation. Funct. Integr. Genom. 2022, 22, 1057–1072. [Google Scholar] [CrossRef] [PubMed]
- Lynch, C.M.; Abdollahi, B.; Fuqua, J.D.; de Carlo, A.R.; Bartholomai, J.A.; Balgemann, R.N.; van Berkel, V.H.; Frieboes, H.B. Prediction of lung cancer patient survival via supervised machine learning classification techniques. Int. J. Med Inform. 2017, 108, 1–8. [Google Scholar] [CrossRef]
- Siah, K.W.; Khozin, S.; Wong, C.H.; Lo, A.W. Machine-Learning and Stochastic Tumor Growth Models for Predicting Outcomes in Patients With Advanced Non-Small-Cell Lung Cancer. JCO Clin. Cancer Inform. 2019, 3, 1–11. [Google Scholar] [CrossRef]
- Cui, L.; Li, H.; Hui, W.; Chen, S.; Yang, L.; Kang, Y.; Bo, Q.; Feng, J. A deep learning-based framework for lung cancer survival analysis with biomarker interpretation. BMC Bioinform. 2020, 21, 112. [Google Scholar] [CrossRef]
- She, Y.; Jin, Z.; Wu, J.; Deng, J.; Zhang, L.; Su, H.; Jiang, G.; Liu, H.; Xie, D.; Cao, N.; et al. Development and Validation of a Deep Learning Model for Non–Small Cell Lung Cancer Survival. JAMA Netw. Open 2020, 3, e205842. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.-H.; Chen, W.-N.; Hsu, T.-C.; Lin, C.; Tsao, Y.; Wu, S. Overall survival prediction of non-small cell lung cancer by integrating microarray and clinical data with deep learning. Sci. Rep. 2020, 10, 4679. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Van Zandt, M.; Liu, Y.; Li, J.; Wang, X.; Chen, Y.; Chen, Z.; Cho, J.; Dorajoo, S.R.; Feng, M.; et al. Analysis of Dual Combination Therapies Used in Treatment of Hypertension in a Multinational Cohort. JAMA Netw. Open 2022, 5, e223877. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.-A.; Chang, C.-C.; Galvin, C.J.; Wang, Y.-C.; An, S.Y.; Huang, C.-W.; Wang, Y.-H.; Hsu, M.-H.; Li, Y.-C.; Yang, H.-C. Statins use and its impact in EGFR-TKIs resistance to prolong the survival of lung cancer patients: A Cancer registry cohort study in Taiwan. Cancer Sci. 2020, 111, 2965–2973. [Google Scholar] [CrossRef]
- Dreiseitl, S.; Ohno-Machado, L. Logistic regression and artificial neural network classification models: A methodology review. J. Biomed. Inform. 2002, 35, 352–359. [Google Scholar] [CrossRef]
- Izenman, A.J. Linear discriminant analysis. In Modern Multivariate Statistical Techniques; Springer: Berlin/Heidelberg, Germany, 2013; pp. 237–280. [Google Scholar]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. Lightgbm: A highly efficient gradient boosting decision tree. Adv. Neural Inf. Process. Syst. 2017, 30, 3149–3157. [Google Scholar]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Chen, T.; He, T.; Benesty, M.; Khotilovich, V.; Tang, Y.; Cho, H.; Chen, K. Xgboost: Extreme gradient boosting. R Package Version 0.4-2 2015, 1, 1–4. [Google Scholar]
- Ho, T.K. Random decision forests. In Proceedings of the 3rd International Conference on Document Analysis and Recognition, IEEE, Montreal, QC, Canada, 14–16 August 1995; pp. 278–282. [Google Scholar]
- Hastie, T.; Rosset, S.; Zhu, J.; Zou, H. Multi-class adaboost. Stat. Its Interface 2009, 2, 349–360. [Google Scholar] [CrossRef]
- Gunn, S.R. Support vector machines for classification and regression. ISIS Tech. Rep. 1998, 14, 5–16. [Google Scholar]
- Agatonovic-Kustrin, S.; Beresford, R. Basic concepts of artificial neural network (ANN) modeling and its application in pharmaceutical research. J. Pharm. Biomed. Anal. 2000, 22, 717–727. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.-I. A unified approach to interpreting model predictions. Adv. Neural Inf. Process. Syst. 2017, 30, 4768–4777. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- He, J.; Zhang, J.X.; Chen, C.T.; Ma, Y.; De Guzman, R.; Meng, J.; Pu, Y. The Relative Importance of Clinical and Socio-demographic Variables in Prognostic Prediction in Non-Small Cell Lung Cancer: A Variable Importance Approach. Med Care 2020, 58, 461–467. [Google Scholar] [CrossRef]
- Bartholomai, J.A.; Frieboes, H.B. Lung Cancer Survival Prediction via Machine Learning Regression, Classification, and Statistical Techniques. In Proceedings of the 2018 IEEE International Symposium on Signal Processing and Information Technology (ISSPIT), Louisville, KY, USA, 6–8 December 2018; Volume 2018, pp. 632–637. [Google Scholar] [CrossRef]
- Goussault, H.; Gendarme, S.; Assié, J.B.; Bylicki, O.; Chouaïd, C. Factors associated with early lung cancer mortality: A systematic review. Expert Rev. Anticancer Ther. 2021, 21, 1125–1133. [Google Scholar] [CrossRef]
- Kang, J.; Chang, Y.; Ahn, J.; Oh, S.; Koo, D.H.; Lee, Y.G.; Shin, H.; Ryu, S. Neutrophil-to-lymphocyte ratio and risk of lung cancer mortality in a low-risk population: A cohort study. Int. J. Cancer 2019, 145, 3267–3275. [Google Scholar] [CrossRef]
- Spicer, J.D.; McDonald, B.; Cools-Lartigue, J.J.; Chow, S.C.; Giannias, B.; Kubes, P.; Ferri, L.E. Neutrophils promote liver metastasis via Mac-1-mediated interactions with circulating tumor cells. Cancer Res. 2012, 72, 3919–3927. [Google Scholar] [CrossRef]
- Powell, D.R.; Huttenlocher, A. Neutrophils in the Tumor Microenvironment. Trends Immunol. 2016, 37, 41–52. [Google Scholar] [CrossRef]
- Contursi, A.; Grande, R.; Dovizio, M.; Bruno, A.; Fullone, R.; Patrignani, P. Platelets in cancer development and diagnosis. Biochem. Soc. Trans. 2018, 46, 1517–1527. [Google Scholar] [CrossRef]
- Wang, J.; Xu, H.; Zhou, S.; Wang, D.; Zhu, L.; Hou, J.; Tang, J.; Zhao, J.; Zhong, S. Body mass index and mortality in lung cancer patients: A systematic review and meta-analysis. Eur. J. Clin. Nutr. 2018, 72, 4–17. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Toyazaki, T.; Chiba, N.; Ueda, Y.; Gotoh, M. Prognostic value of body mass index and change in body weight in postoperative outcomes of lung cancer surgery. Interact. Cardiovasc. Thorac. Surg. 2016, 23, 560–566. [Google Scholar] [CrossRef] [PubMed]


| Features | Overall n = 3714 | Training Set a n = 2280 | Testing Set b n = 1434 |
|---|---|---|---|
| Male, N (%) | 2136 (57.5) | 1258 (55.2) | 878 (61.2) |
| Age, Mean (SD), yrs. | 68.0 (13.7) | 67.9 (13.8) | 68.0 (13.4) |
| BMI, Mean (SD), kg/m2 | 23.4 (4.33) | 23.4 (3.93) | 23.4 (4.81) |
| Smoking, N (%) | |||
| No | 1170 (31.5) | 710 (31.1) | 460 (32.1) |
| Yes | 993 (26.7) | 523 (22.9) | 470 (32.8) |
| Unknown | 1551 (41.8) | 1047 (45.9) | 504 (35.1) |
| Drinking, N (%) | |||
| No | 1750 (47.1) | 983 (43.1) | 767 (53.5) |
| Yes | 408 (11.0) | 247 (10.8) | 161 (11.2) |
| Unknown | 1556 (41.9) | 1050 (46.1) | 506 (35.3) |
| Tumor size, cm | |||
| Mean (SD) | 4.23 (2.45) | 4.11 (2.39) | 4.46 (2.55) |
| Median [IQR] | 3.8 [2.4–5.5] | 3.6 [2.3–5.5] | 4.0 [2.5–5.7] |
| Cancer stage, N (%) | |||
| 0 | 11 (0.3) | 10 (0.4) | 1 (0.1) |
| I | 533 (14.4) | 348 (15.3) | 185 (12.9) |
| II | 139 (3.7) | 88 (3.9) | 51 (3.6) |
| III | 527 (14.1) | 330 (14.5) | 197 (13.7) |
| IV | 2034 (54.8) | 1207 (52.9) | 827 (57.7) |
| Missing | 470 (12.7) | 297 (13.0) | 173 (12.1) |
| Genomic Test | |||
| ALK, N (%) | |||
| Negative | 681 (18.3) | 457 (20.0) | 224 (15.6) |
| Positive | 39 (1.1) | 21 (0.9) | 18 (1.3) |
| Unknown | 2994 (80.6) | 1802 (79.0) | 1192 (83.1) |
| EGFR, N (%) | |||
| Negative | 842 (22.7) | 473 (20.7) | 369 (25.7) |
| Positive | 787 (21.2) | 467 (20.5) | 320 (22.3) |
| Unknown | 2085 (56.1) | 1340 (58.8) | 745 (52.0) |
| KRAS, N (%) | |||
| Negative | 45 (1.2) | 32 (1.4) | 13 (0.9) |
| Positive | 5 (0.1) | 2 (0.1) | 3 (0.2) |
| Unknown | 3664 (98.7) | 2246 (98.5) | 1418 (98.9) |
| PDL1, N (%) | |||
| Negative | 269 (7.2) | 149 (6.5) | 120 (8.4) |
| Positive | 66 (1.8) | 42 (1.8) | 24 (1.7) |
| Unknown | 3379 (91.0) | 2089 (91.6) | 1290 (90.0) |
| ROS1, N (%) | |||
| Negative | 288 (7.8) | 287 (12.6) | 1 (0.1) |
| Positive | 29 (0.8) | 27 (1.2) | 2 (0.1) |
| Unknown | 3397 (91.4) | 1966 (86.2) | 1431 (99.8) |
| Comorbidity, N (%) | |||
| CVD problems | 432 (11.6) | 296 (13.0) | 136 (9.5) |
| Dementia | 124 (3.3) | 71 (3.1) | 53 (3.7) |
| COPD | 599 (16.1) | 391 (17.1) | 208 (14.5) |
| Rheumatic disease | 28 (0.75) | 16 (0.7) | 12 (0.8) |
| PUD | 365 (9.8) | 246 (10.8) | 119 (8.3) |
| Renal disease | 128 (3.4) | 92 (4.0) | 31 (2.2) |
| Liver disease | 211 (5.7) | 147 (6.4) | 64 (4.5) |
| DM | 372 (10.0) | 248 (10.9) | 124 (8.6) |
| Anemia | 107 (2.9) | 76 (3.3) | 31 (2.2) |
| Depression | 245 (6.6) | 175 (7.7) | 70 (4.9) |
| Hyperlipidemia | 516 (13.9) | 385 (16.9) | 131 (9.1) |
| Hypertension | 736 (19.8) | 503 (22.1) | 233 (16.2) |
| Parkinson’s disease | 50 (1.3) | 29 (1.3) | 21 (1.5) |
| Charlson Comorbidity Index (CCI) | |||
| Mean (SD) | 3.08 (2.07) | 3.13 (2.19) | 2.97 (1.86) |
| Median [IQR] | 3.0 [2.0–4.0] | 3.0 [2.0–4.0] | 3.0 [2.0–4.0] |
| Follow-up, yrs. | |||
| Mean (SD) | 2.25 (2.47) | 2.44 (2.61) | 1.96 (2.19) |
| Median [IQR] | 1.41 [0.46–3.04] | 1.51 [0.53–3.36] | 1.24 [0.38–2.64] |
| Medications, N (%) | |||
| Alimentary tract and metabolism | 591 (15.9) | 394 (17.3) | 197 (14.7) |
| Blood and blood-forming organs | 446 (12.0) | 293 (12.9) | 153 (11.3) |
| Cardiovascular system | 675 (18.2) | 448 (19.6) | 227 (16.9) |
| Genitourinary system and hormones | 132 (3.6) | 74 (3.2) | 58 (4.3) |
| Musculoskeletal system | 252 (6.8) | 141 (6.2) | 111 (8.3) |
| Nervous system | 391 (10.5) | 254 (11.1) | 137 (10.2) |
| Respiratory system | 319 (8.6) | 226 (9.9) | 93 (6.9) |
| Laboratory Test, Mean (SD) | |||
| Basophil | 0.50 (0.40) | 0.53 (0.42) | 0.48 (0.39) |
| BUN | 19.4 (14.9) | 18.8 (13.1) | 20.5 (17.6) |
| Creatinine | 1.05 (0.98) | 1.02 (0.90) | 1.10 (1.07) |
| Eosinophil | 1.89 (2.31) | 2.03 (2.59) | 1.76 (1.97) |
| HCT | 38.3 (5.69) | 38.5 (5.61) | 37.9 (5.80) |
| HGB | 12.9 (1.97) | 13.0 (1.91) | 12.7 (2.05) |
| K | 3.99 (0.56) | 4.02 (0.53) | 3.95 (0.60) |
| Lymphocyte | 18.7 (9.98) | 19.6 (9.55) | 17.8 (10.3) |
| MCH | 29.9 (3.02) | 29.9 (3.03) | 29.8 (3.00) |
| MCHC | 33.6 (0.95) | 33.7 (0.96) | 33.6 (0.94) |
| MCV | 88.6 (7.61) | 88.5 (7.64) | 88.7 (7.57) |
| Monocyte | 7.45 (2.90) | 7.42 (2.93) | 7.48 (2.87) |
| Na | 137 (4.46) | 137 (4.39) | 137 (4.53) |
| Neutrophil | 71.3 (11.9) | 70.2 (11.4) | 72.3 (12.2) |
| PLT | 263 (109) | 258 (100) | 269 (121) |
| RBC | 4.35 (0.68) | 4.38 (0.67) | 4.29 (0.69) |
| WBC | 9.72 (5.38) | 9.16 (4.16) | 10.6 (6.80) |
| Modes | Models | AUC Training | AUC Testing | Accuracy | Precision | Recall | F1-score |
|---|---|---|---|---|---|---|---|
| Mode 1 | LR | 0.70 | 0.72 | 0.65 | 0.88 | 0.64 | 0.75 |
| LDA | 0.78 | 0.78 | 0.71 | 0.90 | 0.70 | 0.80 | |
| LGBM | 0.98 | 0.81 | 0.73 | 0.92 | 0.72 | 0.81 | |
| GBM | 0.96 | 0.83 | 0.75 | 0.91 | 0.76 | 0.84 | |
| XGBoost | 0.99 | 0.80 | 0.75 | 0.90 | 0.77 | 0.84 | |
| RF | 0.90 | 0.82 | 0.72 | 0.92 | 0.70 | 0.80 | |
| AdaBoost | 0.94 | 0.81 | 0.73 | 0.91 | 0.72 | 0.81 | |
| SVC | 0.78 | 0.78 | 0.71 | 0.89 | 0.72 | 0.79 | |
| ANN * | 0.89 | 0.88 | 0.82 | 0.90 | 0.75 | 0.64 | |
| Mode 2 | LR | 0.74 | 0.75 | 0.60 | 0.93 | 0.53 | 0.67 |
| LDA | 0.81 | 0.79 | 0.71 | 0.90 | 0.70 | 0.80 | |
| LGBM | 0.99 | 0.83 | 0.78 | 0.91 | 0.79 | 0.86 | |
| GBM | 0.96 | 0.84 | 0.78 | 0.91 | 0.80 | 0.87 | |
| XGBoost | 1.00 | 0.81 | 0.78 | 0.90 | 0.81 | 0.86 | |
| RF | 0.92 | 0.83 | 0.69 | 0.94 | 0.64 | 0.76 | |
| AdaBoost | 0.95 | 0.80 | 0.74 | 0.90 | 0.76 | 0.83 | |
| SVC | 0.81 | 0.79 | 0.70 | 0.91 | 0.68 | 0.78 | |
| ANN * | 0.89 | 0.89 | 0.80 | 0.91 | 0.75 | 0.64 | |
| Mode 3 | LR | 0.70 | 0.73 | 0.65 | 0.88 | 0.63 | 0.74 |
| LDA | 0.80 | 0.81 | 0.75 | 0.91 | 0.76 | 0.83 | |
| LGBM | 0.98 | 0.85 | 0.80 | 0.92 | 0.81 | 0.87 | |
| GBM | 0.96 | 0.85 | 0.79 | 0.92 | 0.79 | 0.86 | |
| XGBoost | 1.00 | 0.83 | 0.79 | 0.91 | 0.80 | 0.86 | |
| RF | 0.91 | 0.84 | 0.72 | 0.93 | 0.69 | 0.80 | |
| AdaBoost | 0.95 | 0.83 | 0.79 | 0.91 | 0.80 | 0.86 | |
| SVC | 0.80 | 0.81 | 0.75 | 0.90 | 0.75 | 0.83 | |
| ANN * | 0.89 | 0.89 | 0.83 | 0.89 | 0.81 | 0.64 | |
| Mode 4 | LR | 0.74 | 0.75 | 0.61 | 0.93 | 0.53 | 0.67 |
| LDA | 0.83 | 0.82 | 0.76 | 0.90 | 0.77 | 0.84 | |
| LGBM | 0.99 | 0.86 | 0.81 | 0.92 | 0.83 | 0.88 | |
| GBM | 0.97 | 0.85 | 0.79 | 0.92 | 0.81 | 0.87 | |
| XGBoost | 1.00 | 0.84 | 0.77 | 0.92 | 0.77 | 0.85 | |
| RF | 0.93 | 0.85 | 0.75 | 0.93 | 0.73 | 0.82 | |
| AdaBoost | 0.96 | 0.83 | 0.76 | 0.92 | 0.75 | 0.83 | |
| SVC | 0.83 | 0.81 | 0.75 | 0.90 | 0.76 | 0.84 | |
| ANN * | 0.89 | 0.89 | 0.82 | 0.91 | 0.75 | 0.65 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsu, J.C.; Nguyen, P.-A.; Phuc, P.T.; Lo, T.-C.; Hsu, M.-H.; Hsieh, M.-S.; Le, N.Q.K.; Cheng, C.-T.; Chang, T.-H.; Chen, C.-Y. Development and Validation of Novel Deep-Learning Models Using Multiple Data Types for Lung Cancer Survival. Cancers 2022, 14, 5562. https://doi.org/10.3390/cancers14225562
Hsu JC, Nguyen P-A, Phuc PT, Lo T-C, Hsu M-H, Hsieh M-S, Le NQK, Cheng C-T, Chang T-H, Chen C-Y. Development and Validation of Novel Deep-Learning Models Using Multiple Data Types for Lung Cancer Survival. Cancers. 2022; 14(22):5562. https://doi.org/10.3390/cancers14225562
Chicago/Turabian StyleHsu, Jason C., Phung-Anh Nguyen, Phan Thanh Phuc, Tsai-Chih Lo, Min-Huei Hsu, Min-Shu Hsieh, Nguyen Quoc Khanh Le, Chi-Tsun Cheng, Tzu-Hao Chang, and Cheng-Yu Chen. 2022. "Development and Validation of Novel Deep-Learning Models Using Multiple Data Types for Lung Cancer Survival" Cancers 14, no. 22: 5562. https://doi.org/10.3390/cancers14225562
APA StyleHsu, J. C., Nguyen, P.-A., Phuc, P. T., Lo, T.-C., Hsu, M.-H., Hsieh, M.-S., Le, N. Q. K., Cheng, C.-T., Chang, T.-H., & Chen, C.-Y. (2022). Development and Validation of Novel Deep-Learning Models Using Multiple Data Types for Lung Cancer Survival. Cancers, 14(22), 5562. https://doi.org/10.3390/cancers14225562









