EGCG Prevents the Transcriptional Reprogramming of an Inflammatory and Immune-Suppressive Molecular Signature in Macrophage-like Differentiated Human HL60 Promyelocytic Leukemia Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture
2.3. Total RNA Isolation, cDNA Synthesis, and Real-Time Quantitative PCR
2.4. Total RNA Library Preparation
2.5. RNA Sequencing
2.6. Reads Alignment and Differential Expression Analysis
2.7. Gene Set Enrichment Analysis
2.8. Human Cancer Inflammation and Immunity Crosstalk PCR Array
2.9. Western Blot
2.10. Gelatin Zymography
2.11. Statistical Data Analysis
2.12. Code Availability
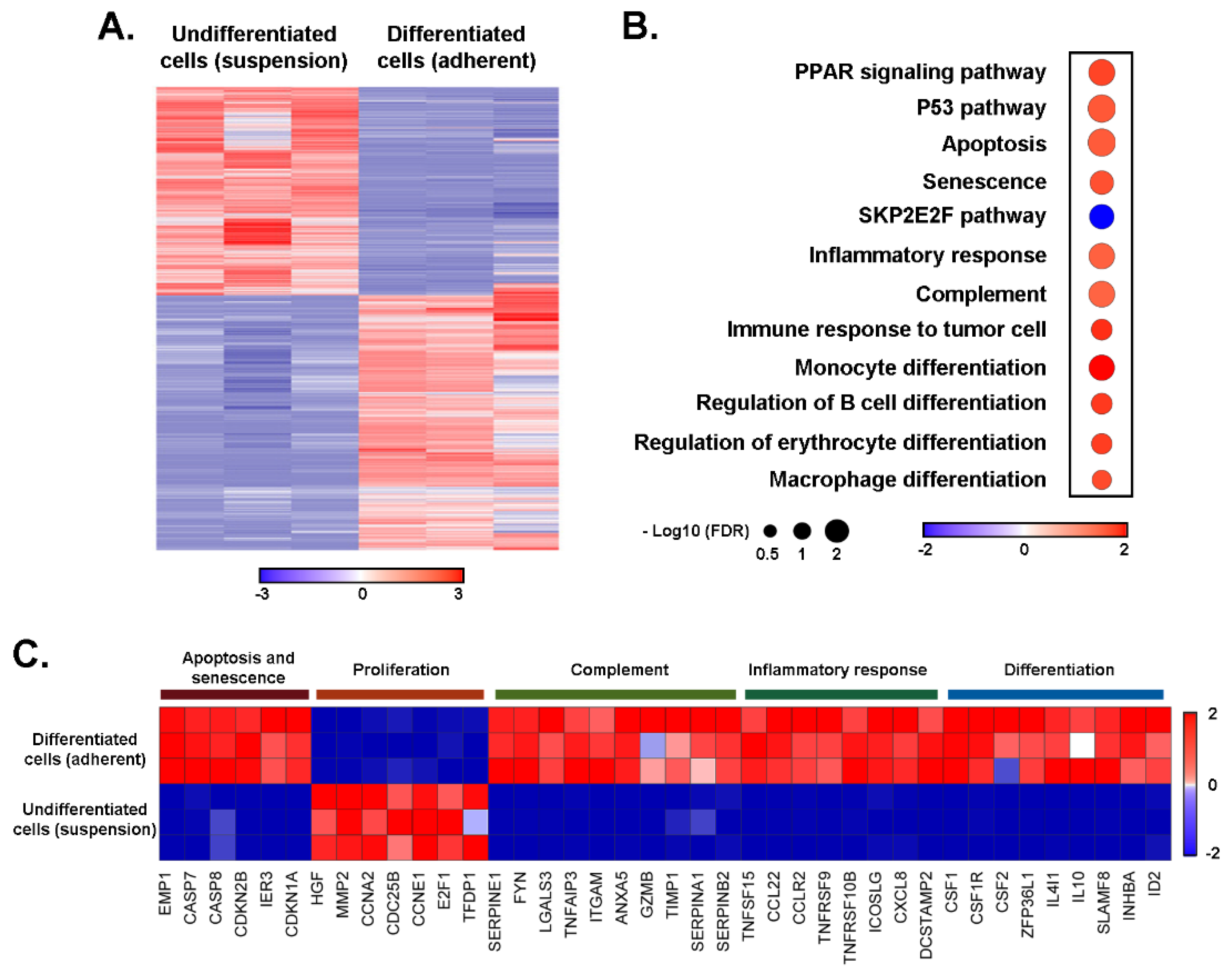
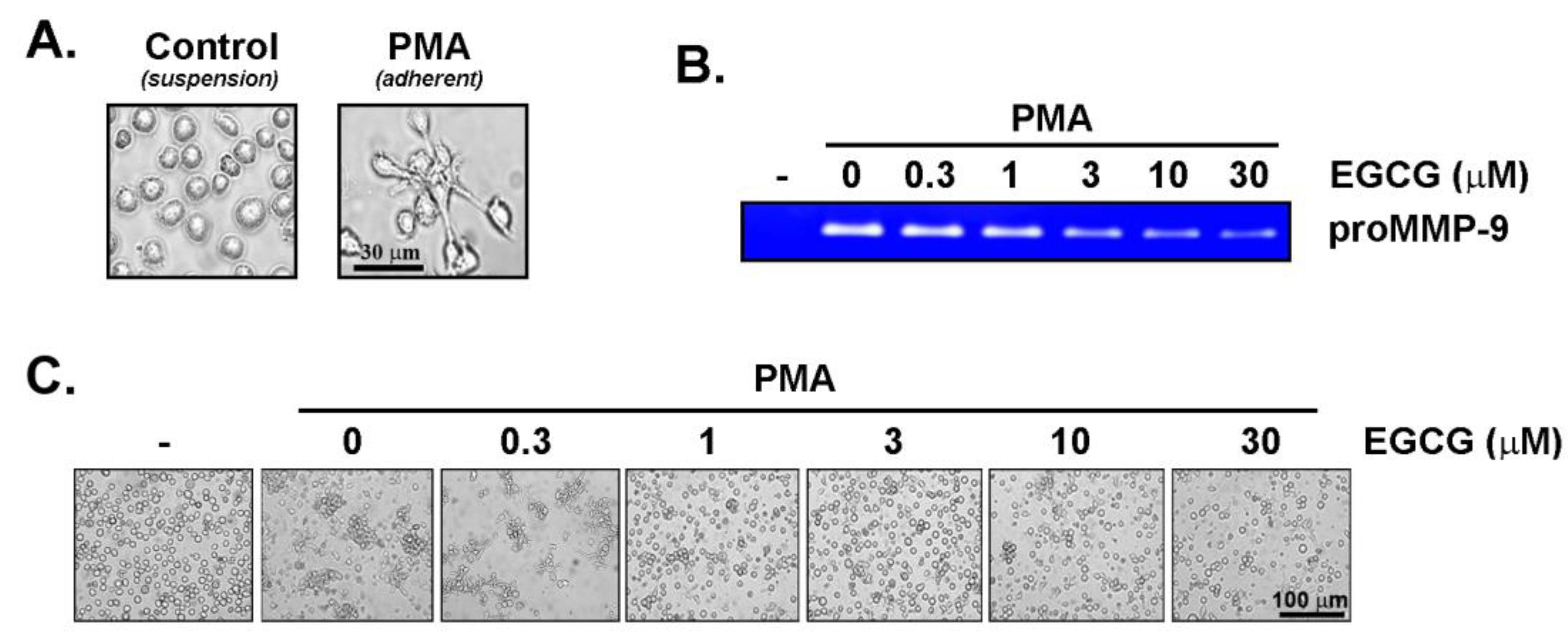
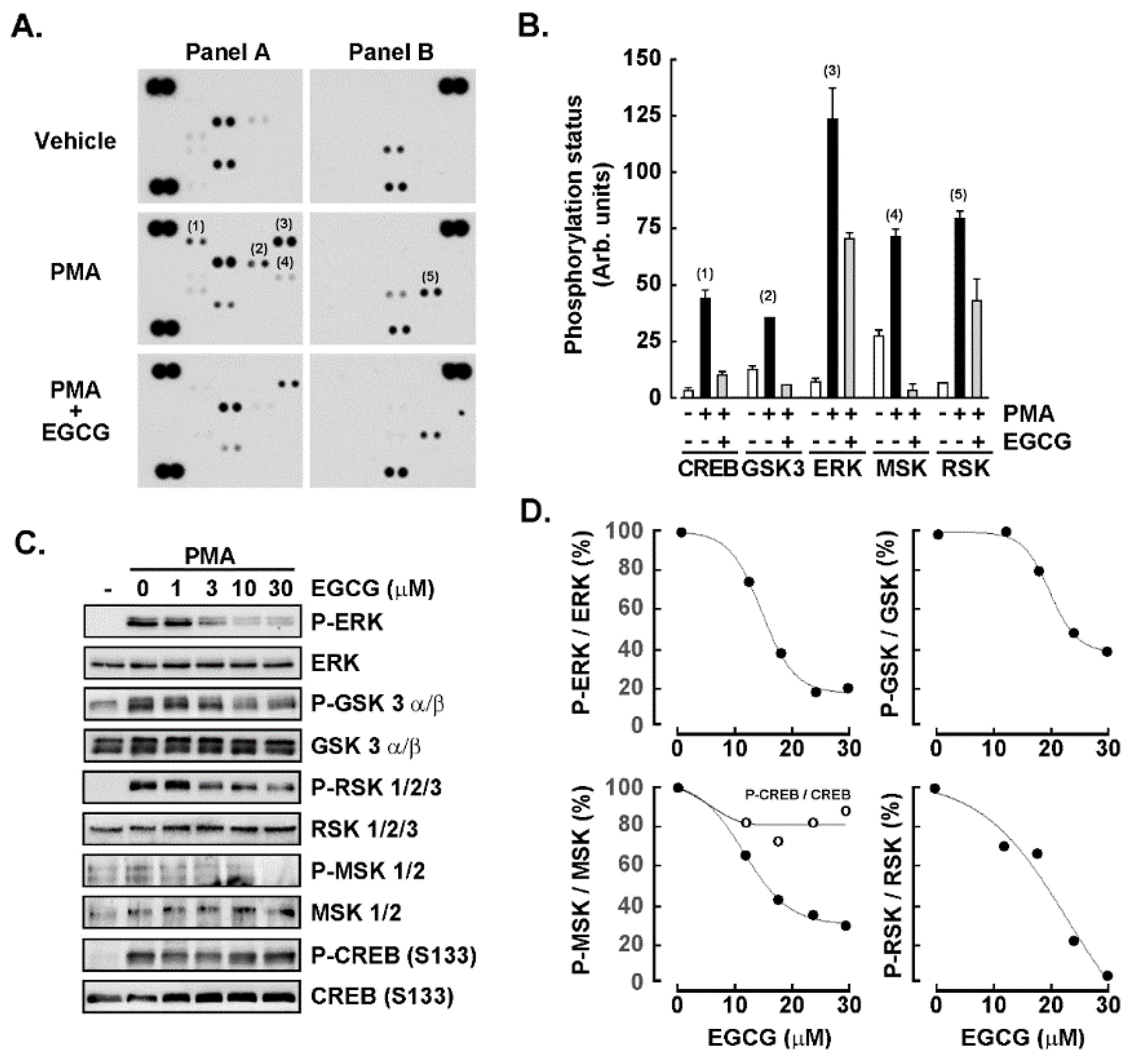
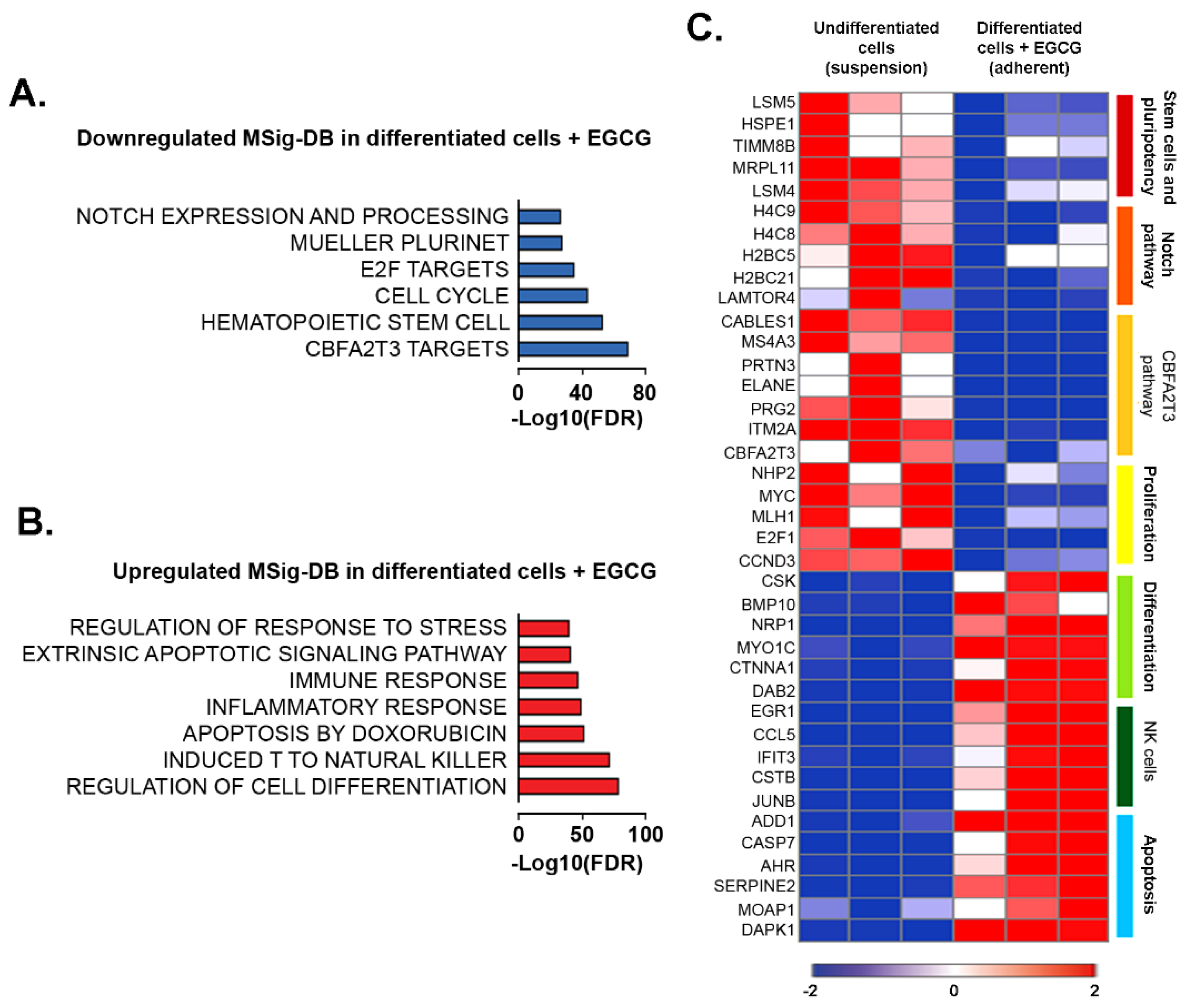
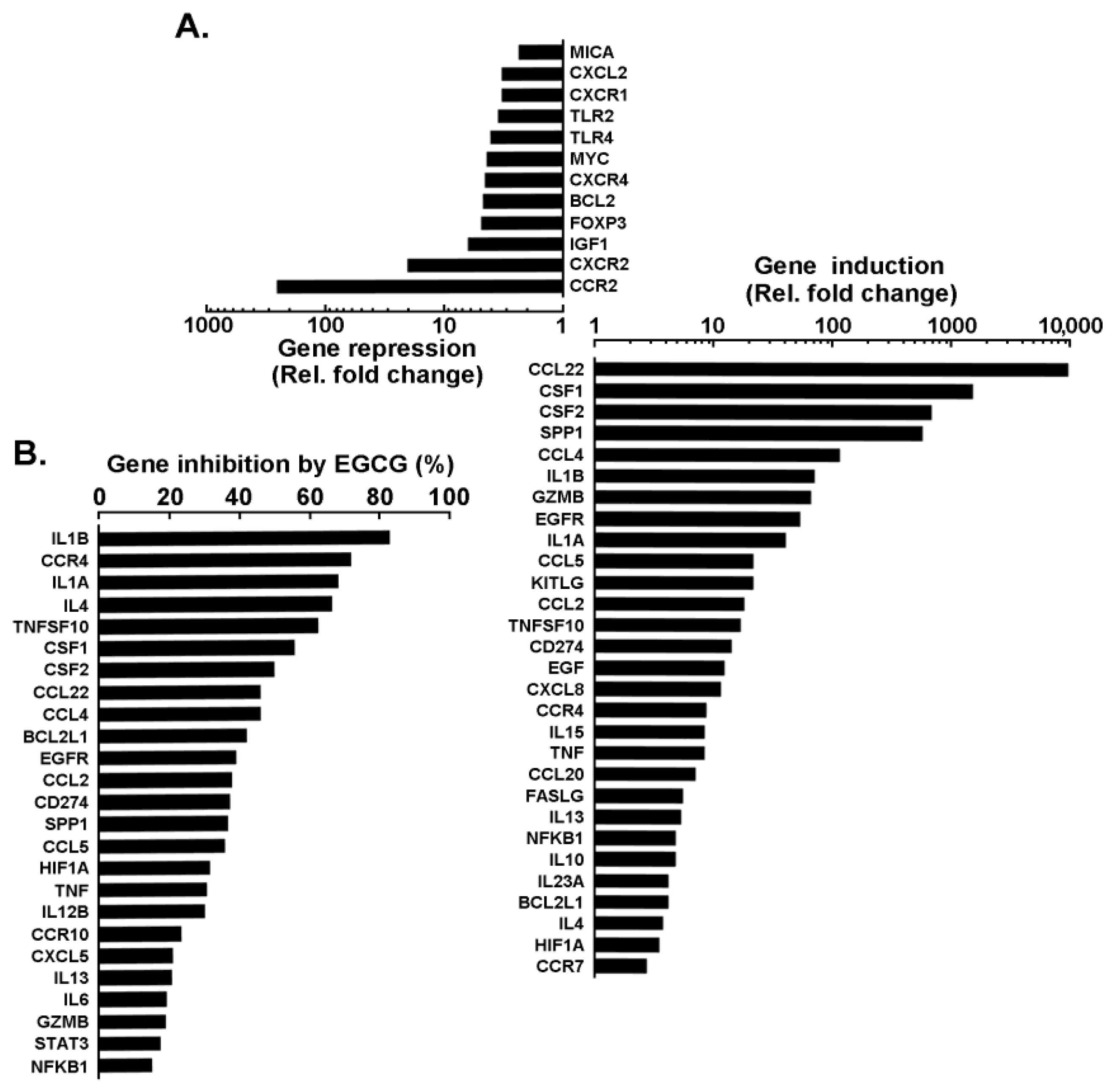
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Coussens, L.M.; Werb, Z. Inflammation and cancer. Nature 2002, 420, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Multhoff, G.; Molls, M.; Radons, J. Chronic Inflammation in Cancer Development. Front. Immunol. 2012, 2, 98. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wu, L.; Yan, G.; Chen, Y.; Zhou, M.; Wu, Y.; Li, Y. Inflammation and tumor progression: Signaling pathways and targeted intervention. Signal Transduct. Target. Ther. 2021, 6, 263. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, D.; Cang, H.; Guo, B. Crosstalk between cancer and immune cells: Role of tumor-associated macrophages in the tumor microenvironment. Cancer Med. 2019, 8, 4709–4721. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Song, Y.; Du, W.; Gong, L.; Chang, H.; Zou, Z. Tumor-associated macrophages: An accomplice in solid tumor progression. J. Biomed. Sci. 2019, 26, 78. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yung, M.; Ngan, H.; Chan, K.; Chan, D. The Impact of the Tumor Microenvironment on Macrophage Polarization in Cancer Metastatic Progression. Int. J. Mol. Sci. 2021, 22, 6560. [Google Scholar] [CrossRef] [PubMed]
- Elinav, E.; Nowarski, R.; Thaiss, C.A.; Hu, B.; Jin, C.; Flavell, R.A. Inflammation-induced cancer: Crosstalk between tumours, immune cells and microorganisms. Nat. Rev. Cancer 2013, 13, 759–771. [Google Scholar] [CrossRef]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, inflammation, and cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef]
- De Kouchkovsky, I.; Abdul-Hay, M. Acute myeloid leukemia: A comprehensive review and 2016 update. Blood Cancer J. 2016, 6, e441. [Google Scholar] [CrossRef]
- Zhong, Y.; Chiou, Y.-S.; Pan, M.-H.; Shahidi, F. Anti-inflammatory activity of lipophilic epigallocatechin gallate (EGCG) derivatives in LPS-stimulated murine macrophages. Food Chem. 2012, 134, 742–748. [Google Scholar] [CrossRef]
- Gallazzi, M.; Ucciero, M.A.M.; Faraci, D.G.; Mahmoud, A.M.; Al Essa, W.; Gaidano, G.; Mouhssine, S.; Crisà, E. New Frontiers in Monoclonal Antibodies for the Targeted Therapy of Acute Myeloid Leukemia and Myelodysplastic Syndromes. Int. J. Mol. Sci. 2022, 23, 7542. [Google Scholar] [CrossRef]
- Fleck, R.A.; Romero-Steiner, S.; Nahm, M.H. Use of HL-60 Cell Line To Measure Opsonic Capacity of Pneumococcal Antibodies. Clin. Diagn. Lab. Immunol. 2005, 12, 19–27. [Google Scholar] [CrossRef]
- Al-Otaibi, N.A.S.; Cassoli, J.S.; Slater, N.K.H.; Rahmoune, H. Molecular Characterization of Human Leukemia 60 (HL-60) Cells as a Model of Acute Myelogenous Leukemia Post Cryopreservation. Methods Mol. Biol. 2019, 1916, 239–247. [Google Scholar]
- Saland, E.; Boutzen, H.; Castellano, R.; Pouyet, L.; Griessinger, E.; Larrue, C.; De Toni, F.; Scotland, S.; David, M.; Danetdesnoyers, G.; et al. A robust and rapid xenograft model to assess efficacy of chemotherapeutic agents for human acute myeloid leukemia. Blood Cancer J. 2015, 5, e297. [Google Scholar] [CrossRef]
- Nalbandian, G.; Kovats, S. Understanding Sex Biases in Immunity: Effects of Estrogen on the Differentiation and Function of Antigen-Presenting Cells. Immunol. Res. 2005, 31, 91–106. [Google Scholar] [CrossRef]
- Cutolo, M.; Brizzolara, R.; Atzeni, F.; Capellino, S.; Straub, R.H.; Puttini, P.C. The immunomodulatory effects of estrogens: Clinical relevance in immune-mediated rheumatic diseases. Ann. N. Y. Acad. Sci. 2010, 1193, 36–42. [Google Scholar] [CrossRef]
- Vézina, A.; Chokor, R.; Annabi, B. EGCG targeting efficacy of NF-κB downstream gene products is dictated by the monocytic/macrophagic differentiation status of promyelocytic leukemia cells. Cancer Immunol. Immunother. 2012, 61, 2321–2331. [Google Scholar] [CrossRef]
- Chokor, R.; Lamy, S.; Annabi, B. Transcriptional targeting of sphingosine-1-phosphate receptor S1P2 by epigallocatechin-3-gallate prevents sphingosine-1-phosphate-mediated signaling in macrophage-differentiated HL-60 promyelocytic leukemia cells. OncoTargets Ther. 2014, 7, 667–677. [Google Scholar]
- Annabi, B.; Currie, J.-C.; Moghrabi, A.; Béliveau, R. Inhibition of HuR and MMP-9 expression in macrophage-differentiated HL-60 myeloid leukemia cells by green tea polyphenol EGCg. Leuk. Res. 2007, 31, 1277–1284. [Google Scholar] [CrossRef]
- Annabi, B.; Rojas-Sutterlin, S.; Laroche, M.; Lachambre, M.-P.; Moumdjian, R.; Béliveau, R. The diet-derived sulforaphane inhibits matrix metalloproteinase-9-activated human brain microvascular endothelial cell migration and tubulogenesis. Mol. Nutr. Food Res. 2008, 52, 692–700. [Google Scholar] [CrossRef]
- Nunes, C.D.R.; Arantes, M.B.; de Faria Pereira, S.M.; Da Cruz, L.L.; de Souza Passos, M.; De Moraes, L.P.; Vieira, I.J.C.; de Oliveira, D.B. Plants as Sources of Anti-Inflammatory Agents. Molecules 2020, 25, 3726. [Google Scholar] [CrossRef]
- Farzaei, M.H.; Singh, A.K.; Kumar, R.; Croley, C.R.; Pandey, A.K.; Coy-Barrera, E.; Patra, J.K.; Das, G.; Kerry, R.G.; Annunziata, G.; et al. Targeting Inflammation by Flavonoids: Novel Therapeutic Strategy for Metabolic Disorders. Int. J. Mol. Sci. 2019, 20, 4957. [Google Scholar] [CrossRef]
- Harris, P.; Ralph, P. Human Leukemic Models of Myelomonocytic Development: A Review of the HL-60 and U937 Cell Lines. J. Leukoc. Biol. 1985, 37, 407–422. [Google Scholar] [CrossRef]
- Aihara, H.; Asaoka, Y.; Yoshida, K.; Nishizuka, Y. Sustained activation of protein kinase C is essential to HL-60 cell differentiation to macrophage. Proc. Natl. Acad. Sci. USA 1991, 88, 11062–11066. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Gil, J.; Peters, G. Regulation of the INK4b–ARF–INK4a tumour suppressor locus: All for one or one for all. Nat. Rev. Mol. Cell Biol. 2006, 7, 667–677. [Google Scholar] [CrossRef] [PubMed]
- Jang, W.; Park, J.; Kwon, A.; Choi, H.; Kim, J.; Lee, G.D.; Han, E.; Jekarl, D.W.; Chae, H.; Han, K.; et al. CDKN2B downregulation and other genetic characteristics in T-acute lymphoblastic leukemia. Exp. Mol. Med. 2019, 51, 1–15. [Google Scholar] [CrossRef]
- Iwasaki, M.; Liedtke, M.; Gentles, A.J.; Cleary, M.L. CD93 Marks a Non-Quiescent Human Leukemia Stem Cell Population and Is Required for Development of MLL-Rearranged Acute Myeloid Leukemia. Cell Stem Cell 2015, 17, 412–421. [Google Scholar] [CrossRef]
- Morris, L.; Allen, K.E.; La Thangue, N.B. Regulation of E2F transcription by cyclin E-Cdk2 kinase mediated through p300/CBP co-activators. Nat. Cell Biol. 2000, 2, 232–239. [Google Scholar] [CrossRef]
- Dao, T.; Salahuddin, S.; Charfi, C.; Sicard, A.-A.; Jenabian, M.-A.; Annabi, B. Pharmacological targeting of neurotensin response by diet-derived EGCG in macrophage-differentiated HL-60 promyelocytic leukemia cells. PharmaNutrition 2020, 12, 100191. [Google Scholar] [CrossRef]
- Aster, J.C.; Pear, W.S.; Blacklow, S.C. Notch signaling in leukemia. Annu. Rev. Pathol. 2008, 3, 587–613. [Google Scholar] [CrossRef]
- Chyla, B.J.; Moreno-Miralles, I.; Steapleton, M.A.; Thompson, M.A.; Bhaskara, S.; Engel, M.; Hiebert, S.W. Deletion of Mtg16, a Target of t(16;21), Alters Hematopoietic Progenitor Cell Proliferation and Lineage Allocation. Mol. Cell. Biol. 2008, 28, 6234–6247. [Google Scholar] [CrossRef]
- Boutilier, A.; Elsawa, S. Macrophage Polarization States in the Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 6995. [Google Scholar] [CrossRef]
- Li, C.; Kuemmerle, J. Phenotypic alterations of macrophage contributes to extracellular matrix deposition in cultured macrophage RAW 264.7. Gastroenterology 2021, 160, S49. [Google Scholar] [CrossRef]
- Hu, Y.; Gu, J.; Lin, J.; Wang, Y.; Yang, F.; Yin, J.; Yu, Z.; Wu, S.; Lv, H.; Ji, X.; et al. (-)-Epigallocatechin-3-gallate (EGCG) modulates polarized macrophages to suppress M1 phenotype and promote M2 polarization in vitro and in vivo. J. Funct. Foods 2021, 87, 104743. [Google Scholar] [CrossRef]
- Niedzwiecki, A.; Roomi, M.W.; Kalinovsky, T.; Rath, M. Anticancer Efficacy of Polyphenols and Their Combinations. Nutrients 2016, 8, 552. [Google Scholar] [CrossRef]
- Nagarsheth, N.; Wicha, M.S.; Zou, W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 2017, 17, 559–572. [Google Scholar] [CrossRef]
- Taniguchi, K.; Karin, M. NF-κB, inflammation, immunity and cancer: Coming of age. Nat. Rev. Immunol. 2018, 18, 309–324. [Google Scholar] [CrossRef]
- Luster, A.D.; Alon, R.; Von Andrian, U.H. Immune cell migration in inflammation: Present and future therapeutic targets. Nat. Immunol. 2005, 6, 1182–1190. [Google Scholar] [CrossRef]
- Mandic, M.; Marjanovic, M.M.; Vucicevic, L.; Jovanovic, M.; Bosnjak, M.; Perovic, V.; Ristic, B.; Ciric, D.; Harhaji-Trajkovic, L.; Trajkovic, V. MAP kinase-dependent autophagy controls phorbol myristate acetate-induced macrophage differentiation of HL-60 leukemia cells. Life Sci. 2022, 297, 120481. [Google Scholar] [CrossRef]
- Saqib, U.; Sarkar, S.; Suk, K.; Mohammad, O.; Baig, M.S.; Savai, R. Phytochemicals as modulators of M1-M2 macrophages in inflammation. Oncotarget 2018, 9, 17937–17950. [Google Scholar] [CrossRef]
- Zhang, J.R.; Lu, F.; Lu, T.; Dong, W.H.; Li, P.; Liu, N.; Ma, D.X.; Ji, C.Y. Inactivation of FoxM1 transcription factor contributes to curcumin-induced inhibition of survival, angiogenesis, and chemosensitivity in acute myeloid leukemia cells. J. Mol. Med. 2014, 92, 1319–1330. [Google Scholar] [CrossRef]
- Momtazi-Borojeni, A.A.; Abdollahi, E.; Nikfar, B.; Chaichian, S.; Ekhlasi-Hundrieser, M. Curcumin as a potential modulator of M1 and M2 macrophages: New insights in atherosclerosis therapy. Heart Fail. Rev. 2019, 24, 399–409. [Google Scholar] [CrossRef]
- Abdollahi, E.; Johnston, T.P.; Ghaneifar, Z.; Vahedi, P.; Goleij, P.; Azhdari, S.; Moghaddam, A.S. Immunomodulatory therapeutic effects of curcumin on M1/M2 macrophage polarization in inflammatory diseases. Curr. Mol. Pharmacol. 2022; In press. [Google Scholar]
- Mantovani, A.; Sozzani, S.; Locati, M.; Schioppa, T.; Saccani, A.; Allavena, P.; Sica, A. Infiltration of tumours by macrophages and dendritic cells: Tumour-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Novartis Found. Symp. 2004, 256, 137–148. [Google Scholar] [PubMed]
- van Dalen, F.J.; van Stevendaal, M.H.M.E.; Fennemann, F.L.; Verdoes, M.; Ilina, O. Molecular repolarisation of tumour-associated macrophages. Molecules 2018, 24, 9. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Schioppa, T.; Porta, C.; Allavena, P.; Sica, A. Role of tumor-associated macrophages in tumor progression and invasion. Cancer Metastasis Rev. 2006, 25, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Luan, B.; Yoon, Y.-S.; Le Lay, J.; Kaestner, K.H.; Hedrick, S.; Montminy, M. CREB pathway links PGE2 signaling with macrophage polarization. Proc. Natl. Acad. Sci. USA 2015, 112, 15642–15647. [Google Scholar] [CrossRef]
- Ruffell, D.; Mourkioti, F.; Gambardella, A.; Kirstetter, P.; Lopez, R.G.; Rosenthal, N.; Nerlov, C. A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc. Natl. Acad. Sci. USA 2009, 106, 17475–17480. [Google Scholar] [CrossRef]
- Jiang, H.; Wei, H.; Wang, H.; Wang, Z.; Li, J.; Ou, Y.; Xiao, X.; Wang, W.; Chang, A.; Sun, W.; et al. Zeb1-induced metabolic reprogramming of glycolysis is essential for macrophage polarization in breast cancer. Cell Death Dis. 2022, 13, 206. [Google Scholar] [CrossRef]
- Nath, N.; Kashfi, K. Tumor associated macrophages and ‘NO’. Biochem. Pharmacol. 2020, 176, 113899. [Google Scholar] [CrossRef]
- Carter, N.; Stamper, B.; Elbarbry, F.; Nguyen, V.; Lopez, S.; Kawasaki, Y.; Poormohamadian, R.; Roberts, S. Natural Products That Target the Arginase in Leishmania Parasites Hold Therapeutic Promise. Microorganisms 2021, 9, 267. [Google Scholar] [CrossRef]
- Diskin, C.; Pålsson-McDermott, E.M. Metabolic Modulation in Macrophage Effector Function. Front. Immunol. 2018, 9, 270. [Google Scholar] [CrossRef]
- Steinauer, N.; Guo, C.; Zhang, J. Emerging Roles of MTG16 in Cell-Fate Control of Hematopoietic Stem Cells and Cancer. Stem Cells Int. 2017, 2017, 6301385. [Google Scholar] [CrossRef]
- Engel, M.E.; Nguyen, H.N.; Mariotti, J.; Hunt, A.; Hiebert, S.W. Myeloid Translocation Gene 16 (MTG16) Interacts with Notch Transcription Complex Components To Integrate Notch Signaling in Hematopoietic Cell Fate Specification. Mol. Cell. Biol. 2010, 30, 1852–1863. [Google Scholar] [CrossRef][Green Version]
- Salaverria, I.; Akasaka, T.; Gesk, S.; Szczepanowski, M.; Burkhardt, B.; Harder, L.; Damm-Welk, C.; Oschlies, I.; Klapper, W.; Dyer, M.J.; et al. The CBFA2T3/ACSF3 locus is recurrently involved in IGH chromosomal translocation t(14;16)(q32;q24) in pediatric B-cell lymphoma with germinal center phenotype. Genes Chromosomes Cancer 2012, 51, 338–343. [Google Scholar] [CrossRef]
- Thirant, C.; Ignacimouttou, C.; Lopez, C.K.; Diop, M.; Le Mouël, L.; Thiollier, C.; Siret, A.; Dessen, P.; Aid, Z.; Rivière, J.; et al. ETO2-GLIS2 hijacks transcriptional complexes to drive cellular identity and self-renewal in pediatric acute megakaryoblastic leukemia. Cancer Cell 2017, 31, 452–465. [Google Scholar] [CrossRef]
- Steinauer, N.; Guo, C.; Zhang, J. The transcriptional corepressor CBFA2T3 inhibits all-trans-retinoic acid–induced myeloid gene expression and differentiation in acute myeloid leukemia. J. Biol. Chem. 2020, 295, 8887–8900. [Google Scholar] [CrossRef] [PubMed]
- Gow, C.-H.; Guo, C.; Wang, D.; Hu, Q.; Zhang, J. Differential involvement of E2A-corepressor interactions in distinct leukemogenic pathways. Nucleic Acids Res. 2014, 42, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Britschgi, A.; Simon, H.U.; Tobler, A.; Fey, M.F.; Tschan, M.P. Epigallocatechin-3-gallate induces cell death in acute myeloid leukaemia cells and supports all-trans retinoic acid-induced neutrophil differentiation via death-associated protein kinase 2. Br. J. Haematol. 2010, 149, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Montuori, N.; Selleri, C.; Risitano, A.M.; Raiola, A.M.; Ragno, P.; Del Vecchio, L.; Rotoli, B.; Rossi, G. Expression of the 67-kDa laminin receptor in acute myeloid leukemia cells mediates adhesion to laminin and is frequently associated with monocytic differentiation. Clin. Cancer Res. 1999, 5, 1465–1472. [Google Scholar]
- Fujimura, Y.; Kumazoe, M.; Tachibana, H. 67-kDa Laminin Receptor-Mediated Cellular Sensing System of Green Tea Polyphenol EGCG and Functional Food Pairing. Molecules 2022, 27, 5130. [Google Scholar] [CrossRef]
- Koji, A.; Yasushi, M.; Daisuke, I.; Masako, I.; Tsushima, H.; Taguchi, J.; Fukushima, T.; Yoshida, S.; Hata, T.; Tomonaga, M. Expression of 67-Kda Laminin Receptor (67LR) Relates to the Aggressiveness and Poor Prognosis of AML: Modulation of the Expression of GM-CSF Receptor by 67LR. Blood 2008, 112, 926. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kassouri, C.; Rodriguez Torres, S.; Gonzalez Suarez, N.; Duhamel, S.; Annabi, B. EGCG Prevents the Transcriptional Reprogramming of an Inflammatory and Immune-Suppressive Molecular Signature in Macrophage-like Differentiated Human HL60 Promyelocytic Leukemia Cells. Cancers 2022, 14, 5065. https://doi.org/10.3390/cancers14205065
Kassouri C, Rodriguez Torres S, Gonzalez Suarez N, Duhamel S, Annabi B. EGCG Prevents the Transcriptional Reprogramming of an Inflammatory and Immune-Suppressive Molecular Signature in Macrophage-like Differentiated Human HL60 Promyelocytic Leukemia Cells. Cancers. 2022; 14(20):5065. https://doi.org/10.3390/cancers14205065
Chicago/Turabian StyleKassouri, Celia, Sahily Rodriguez Torres, Narjara Gonzalez Suarez, Stéphanie Duhamel, and Borhane Annabi. 2022. "EGCG Prevents the Transcriptional Reprogramming of an Inflammatory and Immune-Suppressive Molecular Signature in Macrophage-like Differentiated Human HL60 Promyelocytic Leukemia Cells" Cancers 14, no. 20: 5065. https://doi.org/10.3390/cancers14205065
APA StyleKassouri, C., Rodriguez Torres, S., Gonzalez Suarez, N., Duhamel, S., & Annabi, B. (2022). EGCG Prevents the Transcriptional Reprogramming of an Inflammatory and Immune-Suppressive Molecular Signature in Macrophage-like Differentiated Human HL60 Promyelocytic Leukemia Cells. Cancers, 14(20), 5065. https://doi.org/10.3390/cancers14205065





