m6A Methyltransferase KIAA1429 Regulates the Cisplatin Sensitivity of Gastric Cancer Cells via Stabilizing FOXM1 mRNA
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Antibodies
2.2. Plasmid Overexpression Experiment
2.3. Tissue Culture
2.4. Short Hairpin RNA (shRNA) Knockdown
2.5. Cell Viability
2.6. m6A RNA Methylation Quantification
2.7. Quantitative Reverse Transcription-PCR (QRT-PCR) Experiment
2.8. mRNA Stability Assay
2.9. Western Blotting
2.10. Chromatin Immunoprecipitation (ChIP) Assay
2.11. RNA-Immunoprecipitation (RIP) and MeRIP
2.12. Mouse Studies
2.13. Statistical Analysis
3. Results
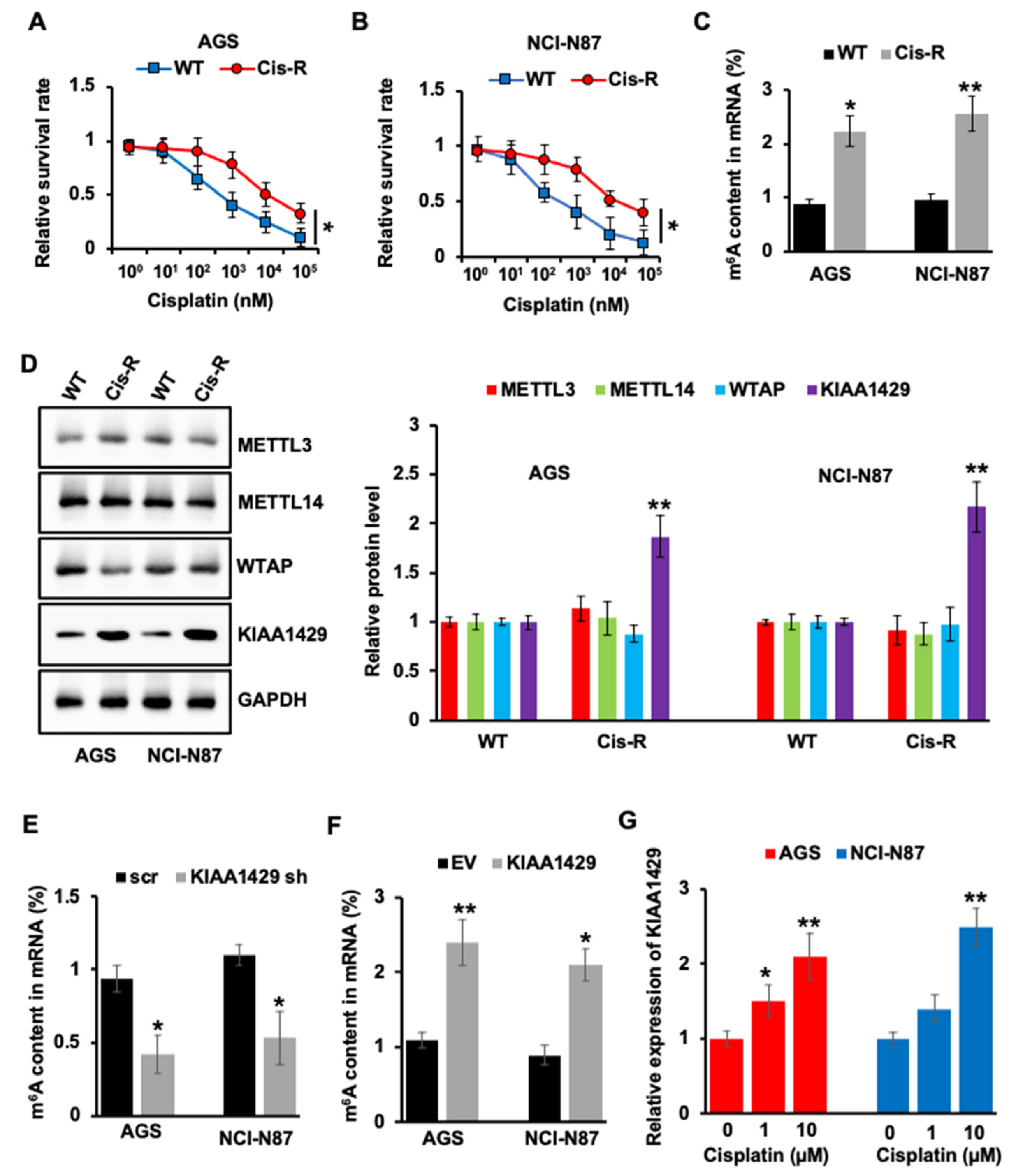
3.1. KIAA1429 Was Highly Expressed in Cisplatin Resistant GC Cells
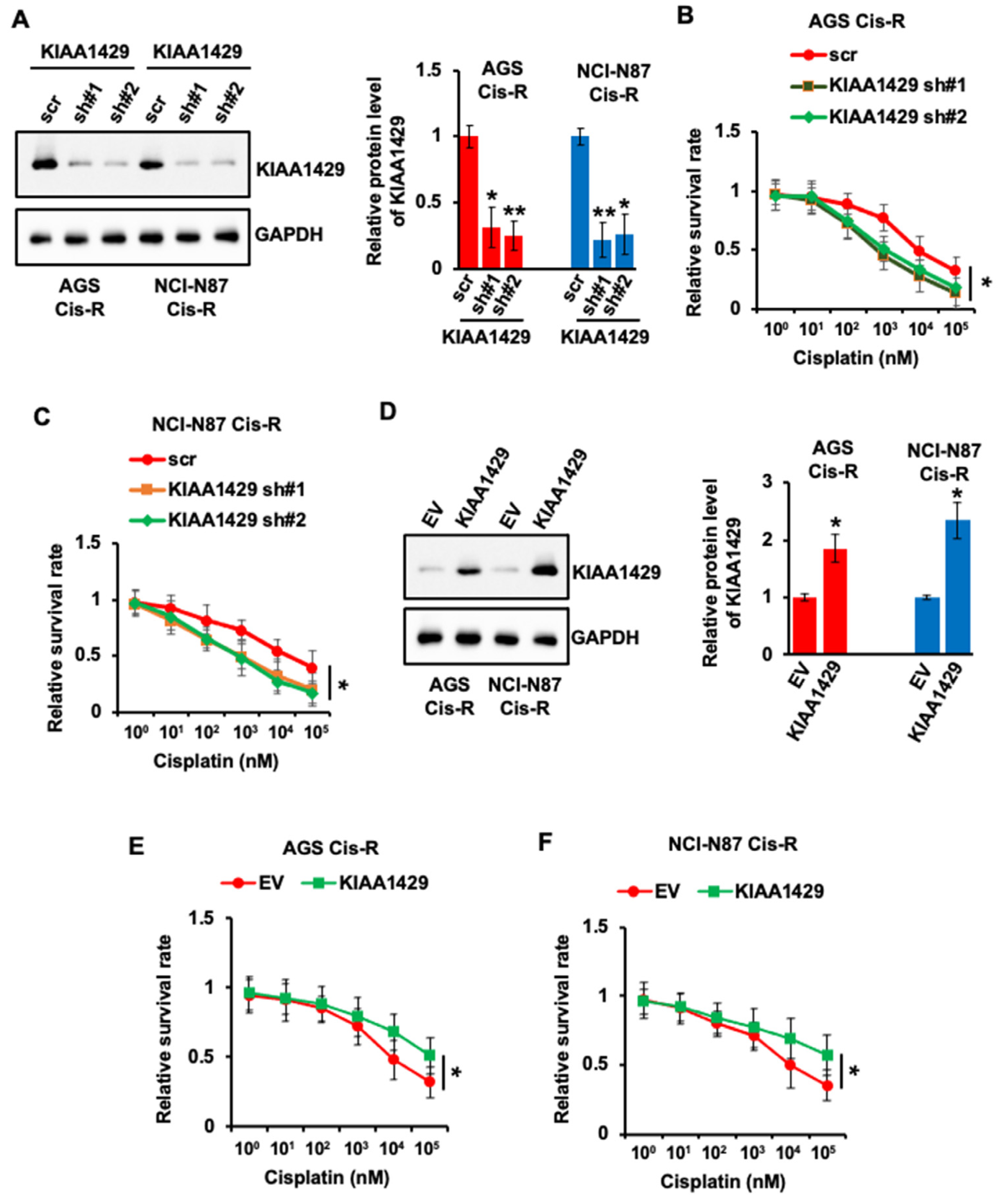
3.2. KIAA1429 Regulated the Sensitivity of GC Cells to Cisplatin
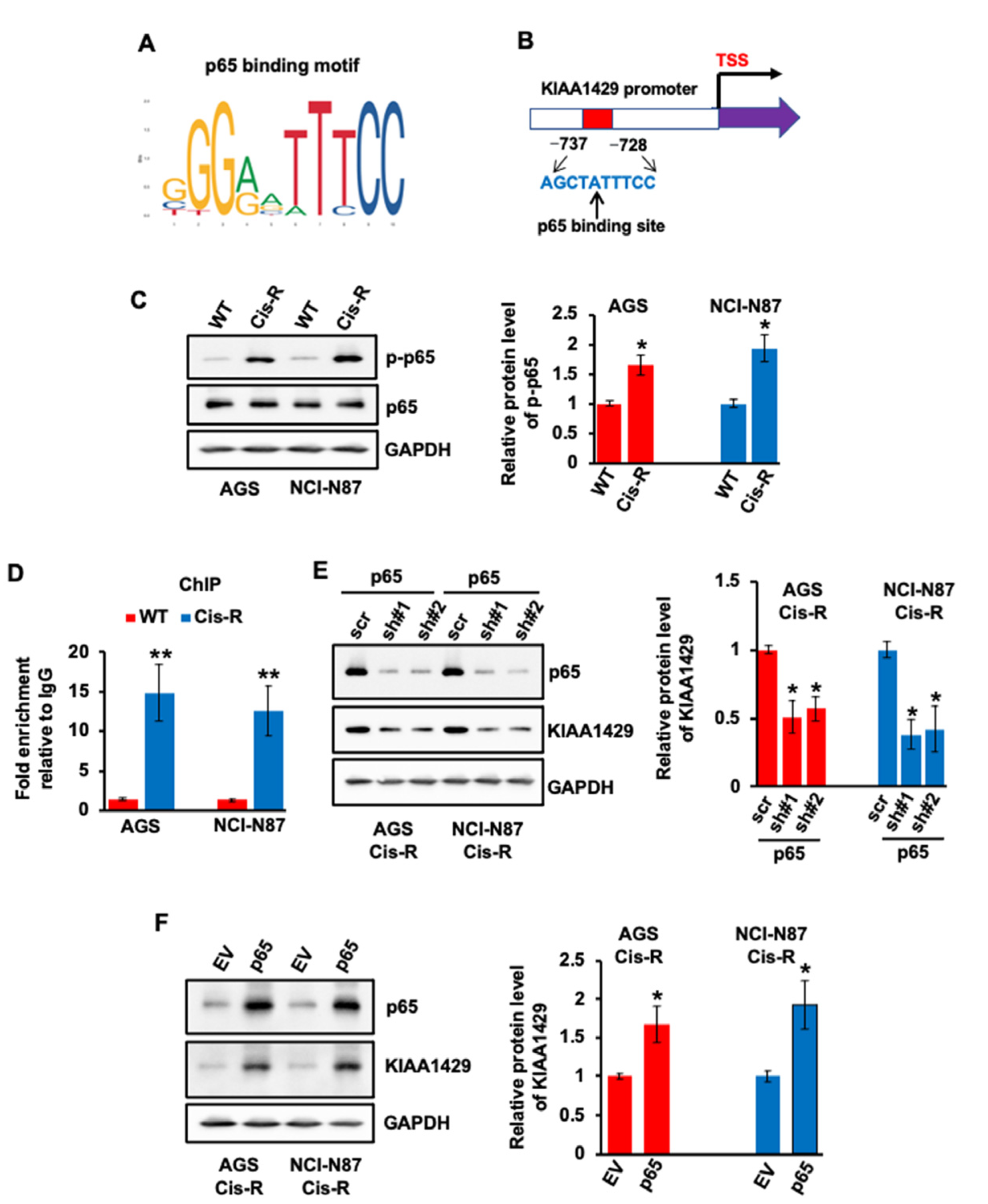
3.3. Expression of KIAA1429 Was Regulated by Transcriptional Factor p65 in GC Cells
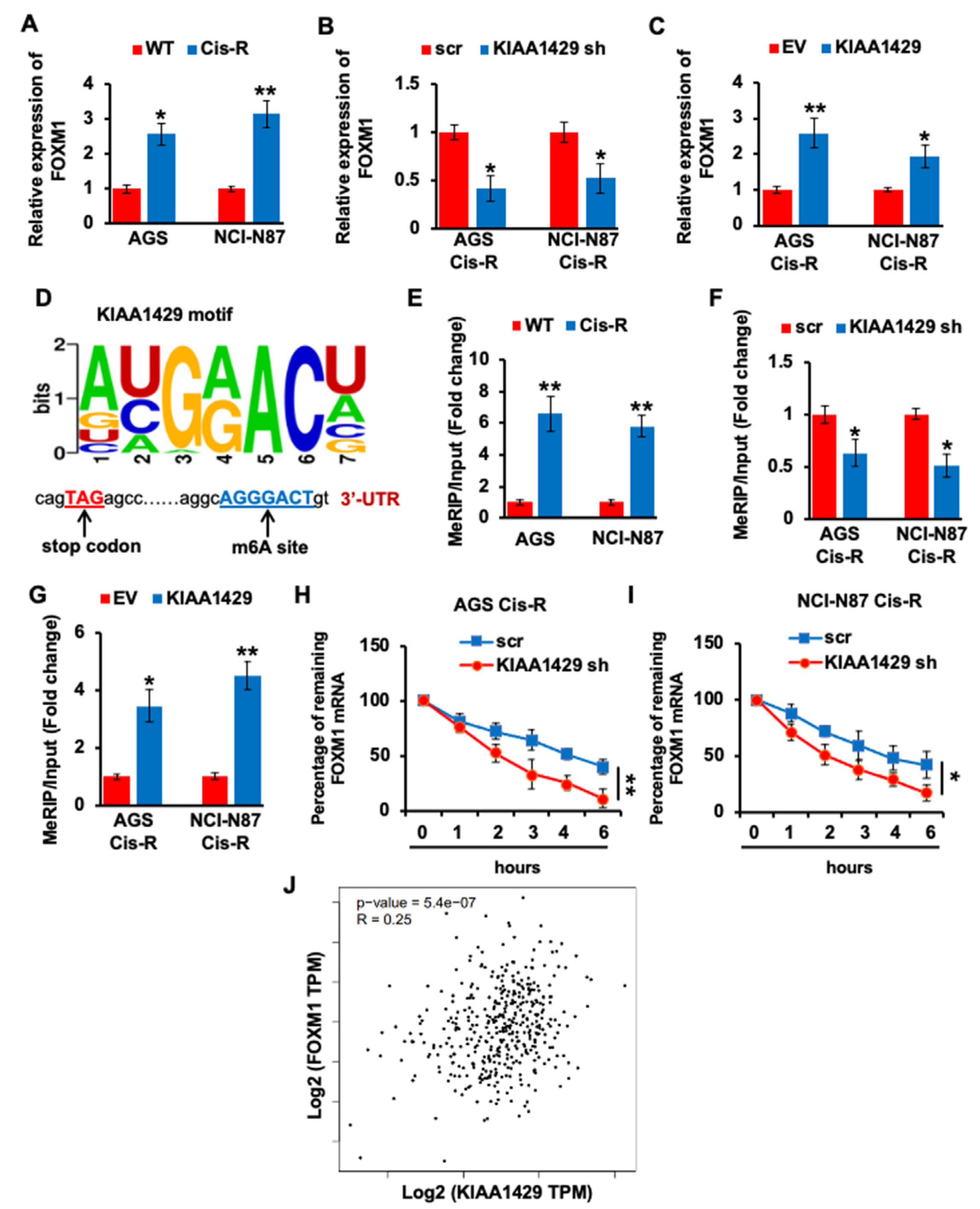
3.4. KIAA1429 Maintained the FOXM1 mRNA Stability in Gastric Cancer Cells
3.5. YTHDF1 Regulated the FOXM1 mRNA Stability in Gastric Cancer Cells
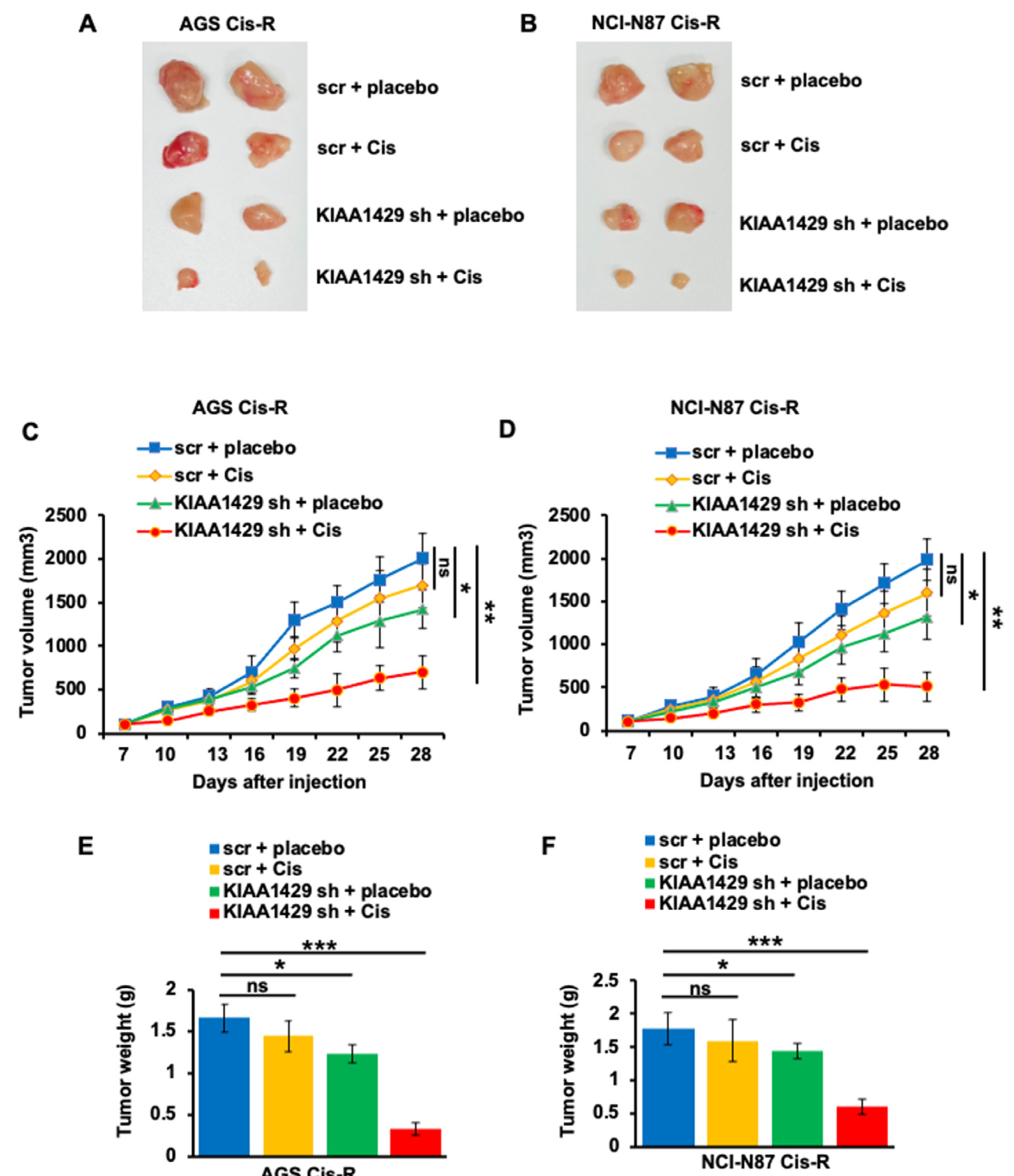
3.6. KIAA1429 Depletion Increased Cisplatin Sensitivity In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zong, L.; Abe, M.; Seto, Y.; Ji, J. The challenge of screening for early gastric cancer in China. Lancet 2016, 388, 2606. [Google Scholar] [CrossRef]
- Wood, N.J. Cancer: Integrated epigenomic analysis sheds light on role of BMP4 in regulating cisplatin sensitivity in gastric cancer. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 301. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Qi, X.; Liu, L.; Ma, S.; Liu, J.; Wu, J. Epigenetic Regulation of m(6)A Modifications in Human Cancer. Mol Ther Nucleic Acids 2020, 19, 405–412. [Google Scholar] [CrossRef]
- Chelmicki, T.; Roger, E.; Teissandier, A.; Dura, M.; Bonneville, L.; Rucli, S.; Dossin, F.; Fouassier, C.; Lameiras, S.; Bourc’his, D. m(6)A RNA methylation regulates the fate of endogenous retroviruses. Nature 2021, 591, 312–316. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.S.; Hao, Y.J.; Sun, B.F.; Sun, H.Y.; Li, A.; Ping, X.L.; Lai, W.Y.; et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 2016, 61, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Wang, X.; Lu, Z.; Zhao, B.S.; Ma, H.; Hsu, P.J.; Liu, C.; He, C. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017, 27, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Chen, Y.S.; Ping, X.L.; Yang, X.; Xiao, W.; Yang, Y.; Sun, H.Y.; Zhu, Q.; Baidya, P.; Wang, X.; et al. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 2017, 27, 444–447. [Google Scholar] [CrossRef] [PubMed]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Liu, J.; Zhao, Y.; He, R.; Xu, X.; Guo, X.; Li, X.; Xu, S.; Miao, J.; Guo, J.; et al. Upregulation of METTL14 mediates the elevation of PERP mRNA N(6) adenosine methylation promoting the growth and metastasis of pancreatic cancer. Mol. Cancer 2020, 19, 130. [Google Scholar] [CrossRef]
- Zeng, C.; Huang, W.; Li, Y.; Weng, H. Roles of METTL3 in cancer: Mechanisms and therapeutic targeting. J. Hematol. Oncol. 2020, 13, 117. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Liu, J.; Cui, X.; Cao, J.; Luo, G.; Zhang, Z.; Cheng, T.; Gao, M.; Shu, X.; Ma, H.; et al. VIRMA mediates preferential m(6)A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018, 4, 10. [Google Scholar] [CrossRef]
- Schwartz, S.; Mumbach, M.R.; Jovanovic, M.; Wang, T.; Maciag, K.; Bushkin, G.G.; Mertins, P.; Ter-Ovanesyan, D.; Habib, N.; Cacchiarelli, D.; et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 2014, 8, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Li, H.; Zhang, D.; Xu, L.; Liu, H.; Hao, X.; Yan, X.; Liao, H.; Chen, X.; Xie, K.; et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol. Cancer 2019, 18, 186. [Google Scholar]
- Qian, J.Y.; Gao, J.; Sun, X.; Cao, M.D.; Shi, L.; Xia, T.S.; Zhou, W.B.; Wang, S.; Ding, Q.; Wei, J.F. KIAA1429 acts as an oncogenic factor in breast cancer by regulating CDK1 in an N6-methyladenosine-independent manner. Oncogene 2019, 38, 6123–6141. [Google Scholar] [CrossRef] [PubMed]
- Miao, R.; Dai, C.C.; Mei, L.; Xu, J.; Sun, S.W.; Xing, Y.L.; Wu, L.S.; Wang, M.H.; Wei, J.F. KIAA1429 regulates cell proliferation by targeting c-Jun messenger RNA directly in gastric cancer. J. Cell Physiol. 2020, 235, 7420–7432. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Chang, S.; Li, F.; Ma, M.; Yang, J.; Lv, X.; Huangfu, L.; Jia, C. m(6) A transferase KIAA1429-stabilized LINC00958 accelerates gastric cancer aerobic glycolysis through targeting GLUT1. IUBMB Life 2021, 73, 1325–1333. [Google Scholar] [CrossRef] [PubMed]
- Jian, D.; Wang, Y.; Jian, L.; Tang, H.; Rao, L.; Chen, K.; Jia, Z.; Zhang, W.; Liu, Y.; Chen, X.; et al. METTL14 aggravates endothelial inflammation and atherosclerosis by increasing FOXO1 N6-methyladeosine modifications. Theranostics 2020, 10, 8939–8956. [Google Scholar] [CrossRef]
- Ren, Z.; Hu, Y.; Sun, J.; Kang, Y.; Li, G.; Zhao, H. N(6)-methyladenosine methyltransferase WTAP-stabilized FOXD2-AS1 promotes the osteosarcoma progression through m(6)A/FOXM1 axis. Bioengineered 2022, 13, 7963–7973. [Google Scholar] [CrossRef] [PubMed]
- Kalinichenko, V.V.; Major, M.L.; Wang, X.; Petrovic, V.; Kuechle, J.; Yoder, H.M.; Dennewitz, M.B.; Shin, B.; Datta, A.; Raychaudhuri, P.; et al. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004, 18, 830–850. [Google Scholar] [CrossRef]
- Kalin, T.V.; Wang, I.C.; Ackerson, T.J.; Major, M.L.; Detrisac, C.J.; Kalinichenko, V.V.; Lyubimov, A.; Costa, R.H. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res. 2006, 66, 1712–1720. [Google Scholar] [CrossRef] [PubMed]
- Bektas, N.; Haaf, A.; Veeck, J.; Wild, P.J.; Luscher-Firzlaff, J.; Hartmann, A.; Knuchel, R.; Dahl, E. Tight correlation between expression of the Forkhead transcription factor FOXM1 and HER2 in human breast cancer. BMC Cancer 2008, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.M.; Ackerson, T.; Ramakrishna, S.; Tretiakova, M.; Wang, I.C.; Kalin, T.V.; Major, M.L.; Gusarova, G.A.; Yoder, H.M.; Costa, R.H.; et al. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 2006, 66, 2153–2161. [Google Scholar] [CrossRef]
- Uddin, S.; Ahmed, M.; Hussain, A.; Abubaker, J.; Al-Sanea, N.; AbdulJabbar, A.; Ashari, L.H.; Alhomoud, S.; Al-Dayel, F.; Jehan, Z.; et al. Genome-wide expression analysis of Middle Eastern colorectal cancer reveals FOXM1 as a novel target for cancer therapy. Am. J. Pathol. 2011, 178, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, N.; Jia, Z.; Le, X.; Dai, B.; Wei, D.; Huang, S.; Tan, D.; Xie, K. Critical role and regulation of transcription factor FoxM1 in human gastric cancer angiogenesis and progression. Cancer Res. 2009, 69, 3501–3509. [Google Scholar] [CrossRef]
- Okada, K.; Fujiwara, Y.; Takahashi, T.; Nakamura, Y.; Takiguchi, S.; Nakajima, K.; Miyata, H.; Yamasaki, M.; Kurokawa, Y.; Mori, M.; et al. Overexpression of forkhead box M1 transcription factor (FOXM1) is a potential prognostic marker and enhances chemoresistance for docetaxel in gastric cancer. Ann. Surg. Oncol. 2013, 20, 1035–1043. [Google Scholar] [CrossRef]
- Qu, K.; Xu, X.; Liu, C.; Wu, Q.; Wei, J.; Meng, F.; Zhou, L.; Wang, Z.; Lei, L.; Liu, P. Negative regulation of transcription factor FoxM1 by p53 enhances oxaliplatin-induced senescence in hepatocellular carcinoma. Cancer Lett. 2013, 331, 105–114. [Google Scholar] [CrossRef]
- Feng, Y.; Wang, L.; Zeng, J.; Shen, L.; Liang, X.; Yu, H.; Liu, S.; Liu, Z.; Sun, Y.; Li, W.; et al. FoxM1 is overexpressed in Helicobacter pylori-induced gastric carcinogenesis and is negatively regulated by miR-370. Mol. Cancer Res. 2013, 11, 834–844. [Google Scholar] [CrossRef]
- Ballard, D.W.; Dixon, E.P.; Peffer, N.J.; Bogerd, H.; Doerre, S.; Stein, B.; Greene, W.C. The 65-kDa subunit of human NF-kappa B functions as a potent transcriptional activator and a target for v-Rel-mediated repression. Proc. Natl. Acad. Sci. USA 1992, 89, 1875–1879. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Guide for the Care and Use of Laboratory Animals, 3rd ed.; National Academies Press: Washington, DC, USA, 2011.
- Yan, F.; Al-Kali, A.; Zhang, Z.; Liu, J.; Pang, J.; Zhao, N.; He, C.; Litzow, M.R.; Liu, S. A dynamic N(6)-methyladenosine methylome regulates intrinsic and acquired resistance to tyrosine kinase inhibitors. Cell Res. 2018, 28, 1062–1076. [Google Scholar] [CrossRef] [PubMed]
- Xiang, M.; Liu, W.; Tian, W.; You, A.; Deng, D. RNA N-6-methyladenosine enzymes and resistance of cancer cells to chemotherapy and radiotherapy. Epigenomics 2020, 12, 801–809. [Google Scholar] [CrossRef]
- Hu, J.; Li, P.; Shi, B.; Tie, J. Effects and Mechanisms of Saikosaponin D Improving the Sensitivity of Human Gastric Cancer Cells to Cisplatin. ACS Omega 2021, 6, 18745–18755. [Google Scholar] [CrossRef]
- Wang, Z.; Li, F.; He, S.; Zhao, L.; Wang, F. Period circadian regulator 2 suppresses drug resistance to cisplatin by PI3K/AKT pathway and improves chronochemotherapeutic efficacy in cervical cancer. Gene 2022, 809, 146003. [Google Scholar] [CrossRef]
- Zhang, C.; Ou, S.; Zhou, Y.; Liu, P.; Zhang, P.; Li, Z.; Xu, R.; Li, Y. m(6)A Methyltransferase METTL14-Mediated Upregulation of Cytidine Deaminase Promoting Gemcitabine Resistance in Pancreatic Cancer. Front. Oncol. 2021, 11, 696371. [Google Scholar] [CrossRef]
- Raychaudhuri, P.; Park, H.J. FoxM1: A master regulator of tumor metastasis. Cancer Res 2011, 71, 4329–4333. [Google Scholar] [CrossRef]
- Weng, J.; Wu, A.; Ying, J. Chemosensitivity of gastric cancer: Analysis of key pathogenic transcription factors. J. Gastrointest. Oncol. 2022, 13, 977–984. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Jin, X.; Jiang, M.L.; Wu, Z.H.; Fan, Y. Progress of Individualized Chemotherapy for Gastric Carcinoma Under the Guidance of Genetic Testing. Curr. Med. Chem. 2020, 27, 2322–2334. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, P.Y.; Kim, G.; Poudel, M.; Lim, S.C.; Choi, H.S. METTL3 induces PLX4032 resistance in melanoma by promoting m(6)A-dependent EGFR translation. Cancer Lett. 2021, 522, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Han, T.; Tong, W.; Zhao, J.; Wang, W. N(6)-methyladenosine (m(6)A) methyltransferase KIAA1429 accelerates the gefitinib resistance of non-small-cell lung cancer. Cell Death Discov. 2021, 7, 108. [Google Scholar] [CrossRef] [PubMed]
- Myatt, S.S.; Lam, E.W. Targeting FOXM1. Nat. Rev. Cancer 2008, 8, 242. [Google Scholar] [CrossRef]
- Li, X.; Liang, J.; Liu, Y.X.; Wang, Y.; Yang, X.H.; Bao, H.; Zhang, G.L.; Du, J.; Wu, X.H. Knockdown of the FoxM1 enhances the sensitivity of gastric cancer cells to cisplatin by targeting Mcl-1. Pharmazie 2016, 71, 345–348. [Google Scholar] [PubMed]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Chen, E.R.; Nilsen, T.W. Kaposi’s Sarcoma-Associated Herpesvirus Utilizes and Manipulates RNA N(6)-Adenosine Methylation To Promote Lytic Replication. J. Virol. 2017, 91, e00466-17. [Google Scholar] [CrossRef]
- Hsu, P.J.; Zhu, Y.; Ma, H.; Guo, Y.; Shi, X.; Liu, Y.; Qi, M.; Lu, Z.; Shi, H.; Wang, J.; et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017, 27, 1115–1127. [Google Scholar] [CrossRef]
- Du, H.; Zhao, Y.; He, J.; Zhang, Y.; Xi, H.; Liu, M.; Ma, J.; Wu, L. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 2016, 7, 12626. [Google Scholar] [CrossRef]
- Huo, F.C.; Zhu, Z.M.; Zhu, W.T.; Du, Q.Y.; Liang, J.; Mou, J. METTL3-mediated m(6)A methylation of SPHK2 promotes gastric cancer progression by targeting KLF2. Oncogene 2021, 40, 2968–2981. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Z.; Zhou, Y.; Chen, Y.; Zhou, Z.; Liu, W.; Zheng, L.; Pei, Q.; Tan, F.; Pei, H.; Li, Y. m6A Methyltransferase KIAA1429 Regulates the Cisplatin Sensitivity of Gastric Cancer Cells via Stabilizing FOXM1 mRNA. Cancers 2022, 14, 5025. https://doi.org/10.3390/cancers14205025
Zhu Z, Zhou Y, Chen Y, Zhou Z, Liu W, Zheng L, Pei Q, Tan F, Pei H, Li Y. m6A Methyltransferase KIAA1429 Regulates the Cisplatin Sensitivity of Gastric Cancer Cells via Stabilizing FOXM1 mRNA. Cancers. 2022; 14(20):5025. https://doi.org/10.3390/cancers14205025
Chicago/Turabian StyleZhu, Zhongcheng, Yuan Zhou, Yongheng Chen, Zhongyi Zhou, Wenxue Liu, Linyi Zheng, Qian Pei, Fengbo Tan, Haiping Pei, and Yuqiang Li. 2022. "m6A Methyltransferase KIAA1429 Regulates the Cisplatin Sensitivity of Gastric Cancer Cells via Stabilizing FOXM1 mRNA" Cancers 14, no. 20: 5025. https://doi.org/10.3390/cancers14205025
APA StyleZhu, Z., Zhou, Y., Chen, Y., Zhou, Z., Liu, W., Zheng, L., Pei, Q., Tan, F., Pei, H., & Li, Y. (2022). m6A Methyltransferase KIAA1429 Regulates the Cisplatin Sensitivity of Gastric Cancer Cells via Stabilizing FOXM1 mRNA. Cancers, 14(20), 5025. https://doi.org/10.3390/cancers14205025





