LINC01526 Promotes Proliferation and Metastasis of Gastric Cancer by Interacting with TARBP2 to Induce GNG7 mRNA Decay
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatics Analysis
2.2. Fluorescent In Situ Hybridization (FISH) Analysis
2.3. RNA Extraction and Real-Time Quantitative PCR (RT-qPCR) Assays
- LINC01526-F, 5′-GGAAGGTCCTGCCCTTTGTT-3′; LINC01526-R, 5′-CTGTCCTATTCAGTGGGGGC-3′;
- TARBP2-F, 5′-GCCTGAGGACATTCCGGTTT-3′; TARBP2-R, 5′-TCACTGTGTACTCCGGCAAC-3′;
- GNG7-F, 5′-AAGCTCTCTGAACAACGGGG-3′; GNG7-R, 5′-CGCTCAATCCCGGCTTCTAT-3′;
- CDKN2A-F, 5′-CCAGAGGCAGTAACCATGCC-3′; CDKN2A-R, AAACTACGAAAGCGGGGTGG-3′;
- GSDMD-F, 5′-CATGGTTCTGGAAACCCCGT-3′; GSDMD-R, 5′-CCACACTCAGCGAGTACACA-3′
- POLG-F, 5′-AGGGGCATTGTTGCTTGTTG-3′; POLG-R, 5′-ACTGCCTTGGAGCAGGTTTAT-3′;
- 18srRNA-F, 5′-AAACGGCTACCACATCCAAG-3′; 18srRNA-R, 5′-CCTCCAATGGATCCTCGTTA-3′.
2.4. Cell Culture and Treatments
- si-LINC01526 #1, 5′-GCUGGUUCUAGGACCAUAU-3′;
- si-LINC01526 #2, 5′-GCUCCUAAAGGUUCCUUUA-3′;
- si-TARBP2 #1, 5′-GCUGCCUAGUAUAGAGCAA-3′;
- si-TARBP2 #2, 5′-GCCCACCGCAAAGAAUUCA-3′;
- si-GNG7 #1, 5′-GAGCUACUGUGAGCAACAU-3′;
- si-GNG7 #2, 5′-CCACUAACAACAUAGCCCA-3′;
- si-NC, 5′-UUCUCCGAACGUGUCACGU-3′.
2.5. Cell Proliferation Assays
2.6. Apoptosis Assays
2.7. Transwell Assays
2.8. Xenograft Mouse Model
2.9. Hematoxylin and Eosin (HE) Staining and Immunofluorescence
2.10. RNA Pull-Down Assay
2.11. Western Blotting
2.12. RNA Immunoprecipitation (RIP) Assay
2.13. RNA Sequencing
2.14. RNA Stability Assays
2.15. Statistical Analysis
3. Results
3.1. LINC01526 Is Significantly Upregulated in Human Gastric Cancer Tissues and Associated with a Poor Prognosis
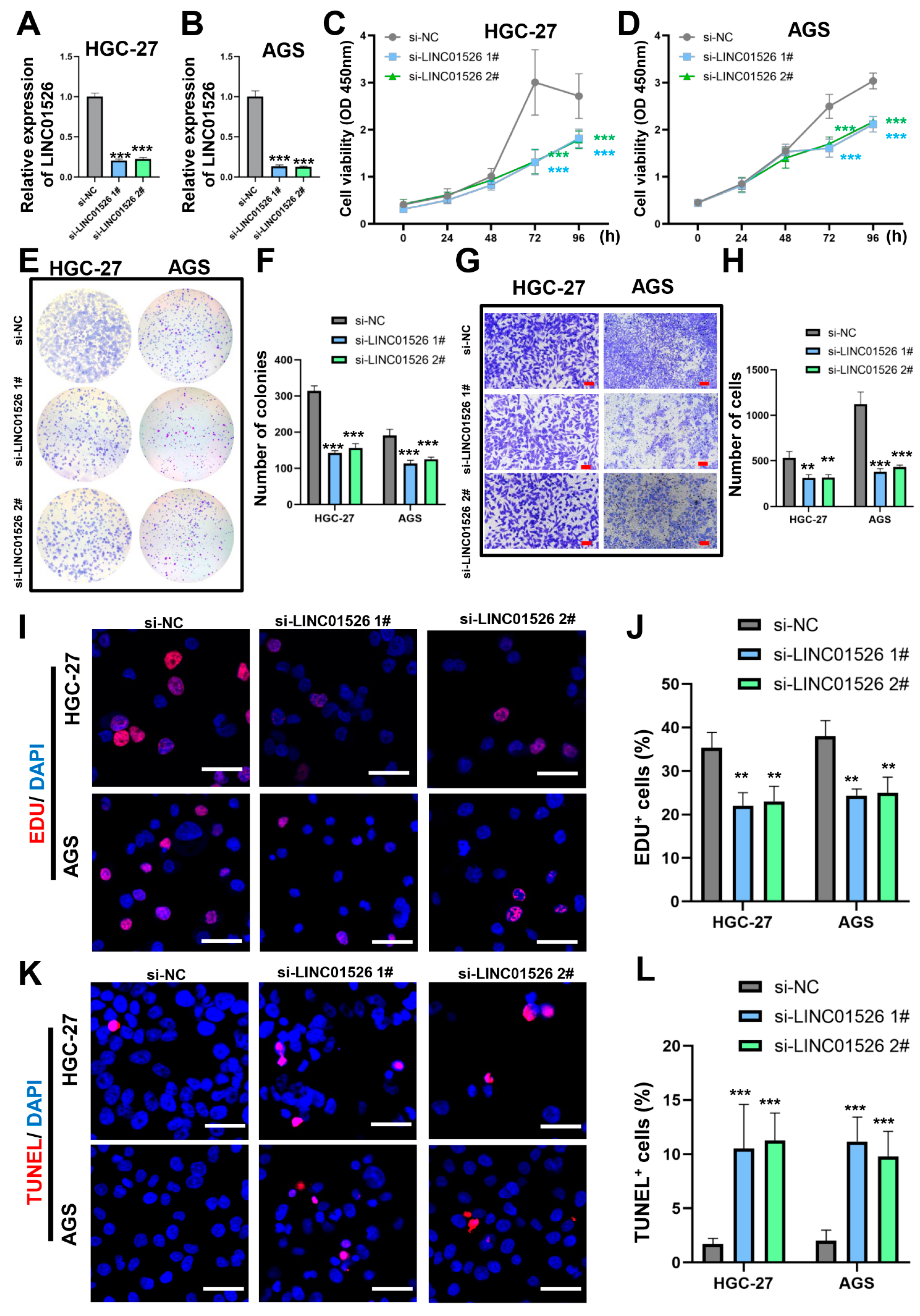
3.2. Knockdown of LINC01526 Suppresses Gastric Cancer Cell Proliferation and Migration and Promotes Apoptosis In Vitro
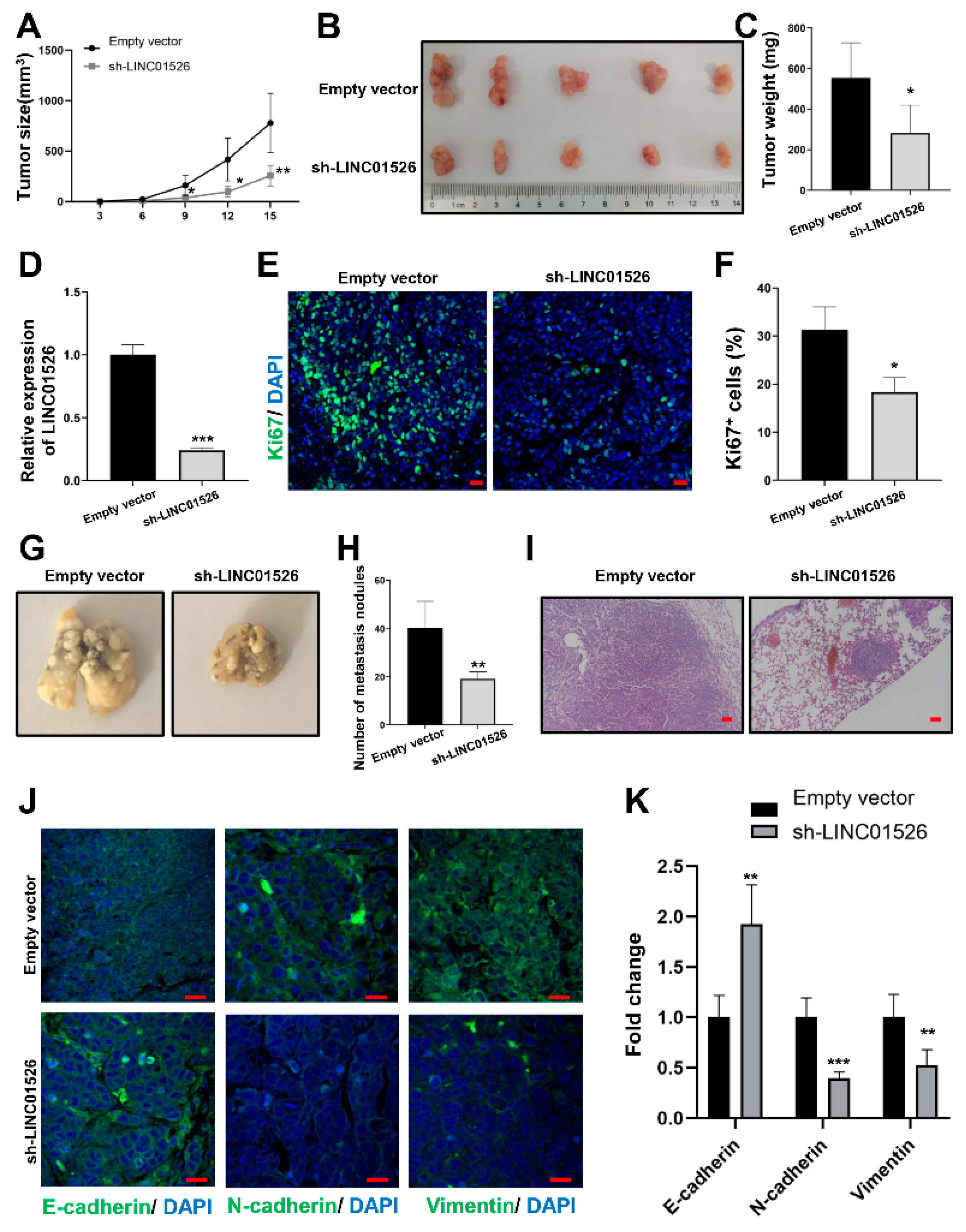
3.3. Knockdown of LINC01526 Suppresses Gastric Cancer Cells Growth and Metastasis In Vivo
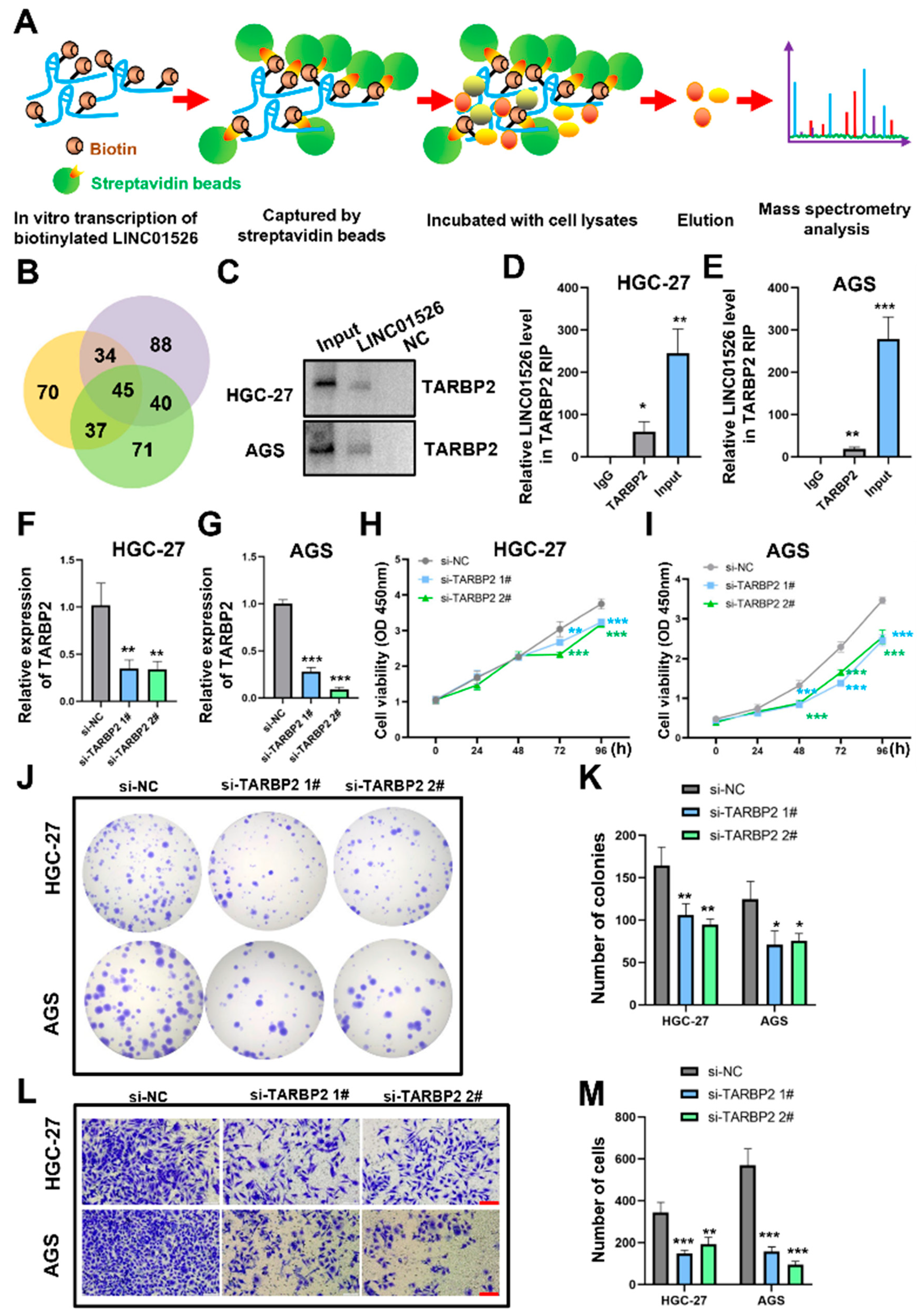
3.4. LINC01526 Interacts with TARBP2, and TARBP2 Functions as an Oncogene in GC Cells
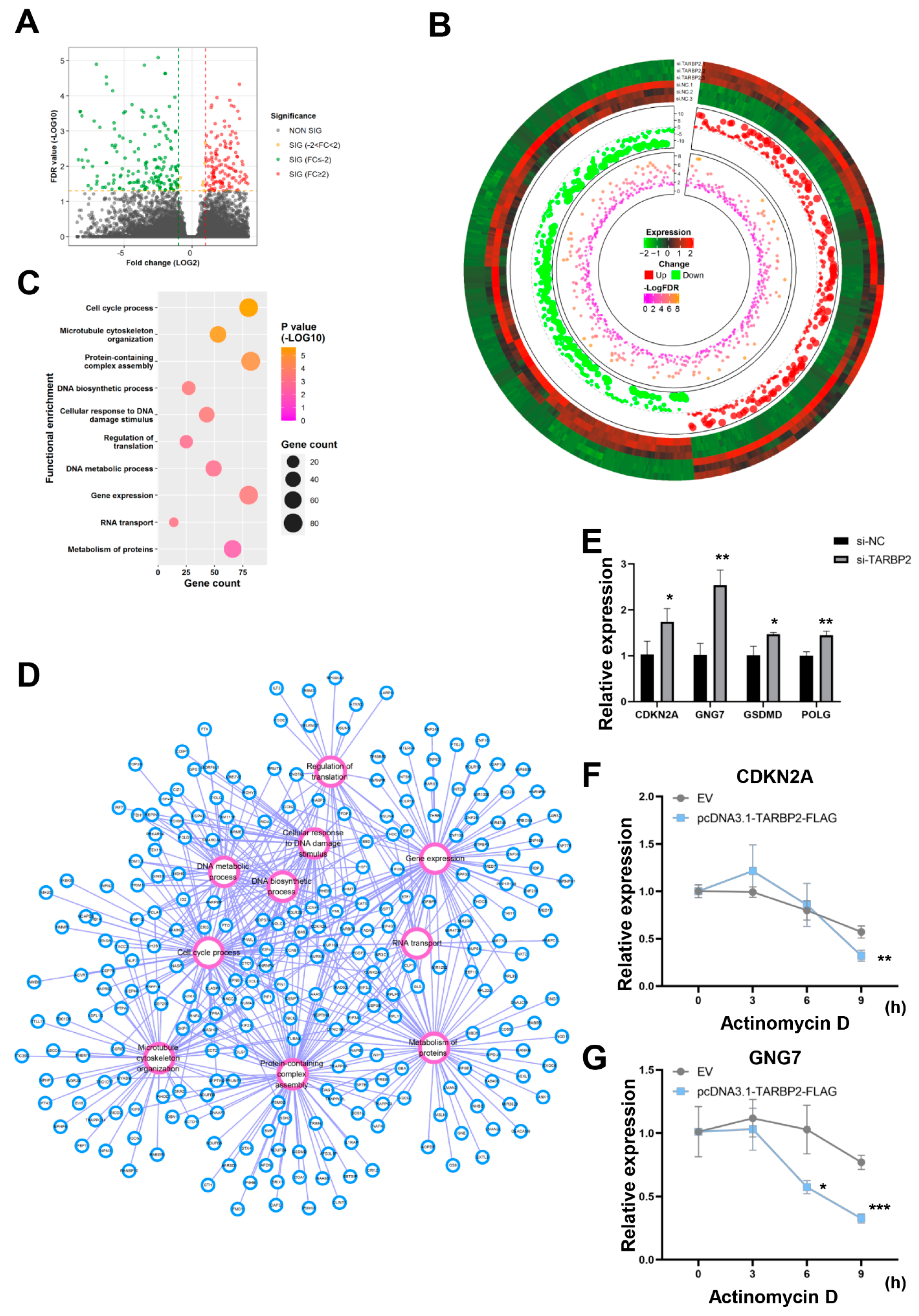
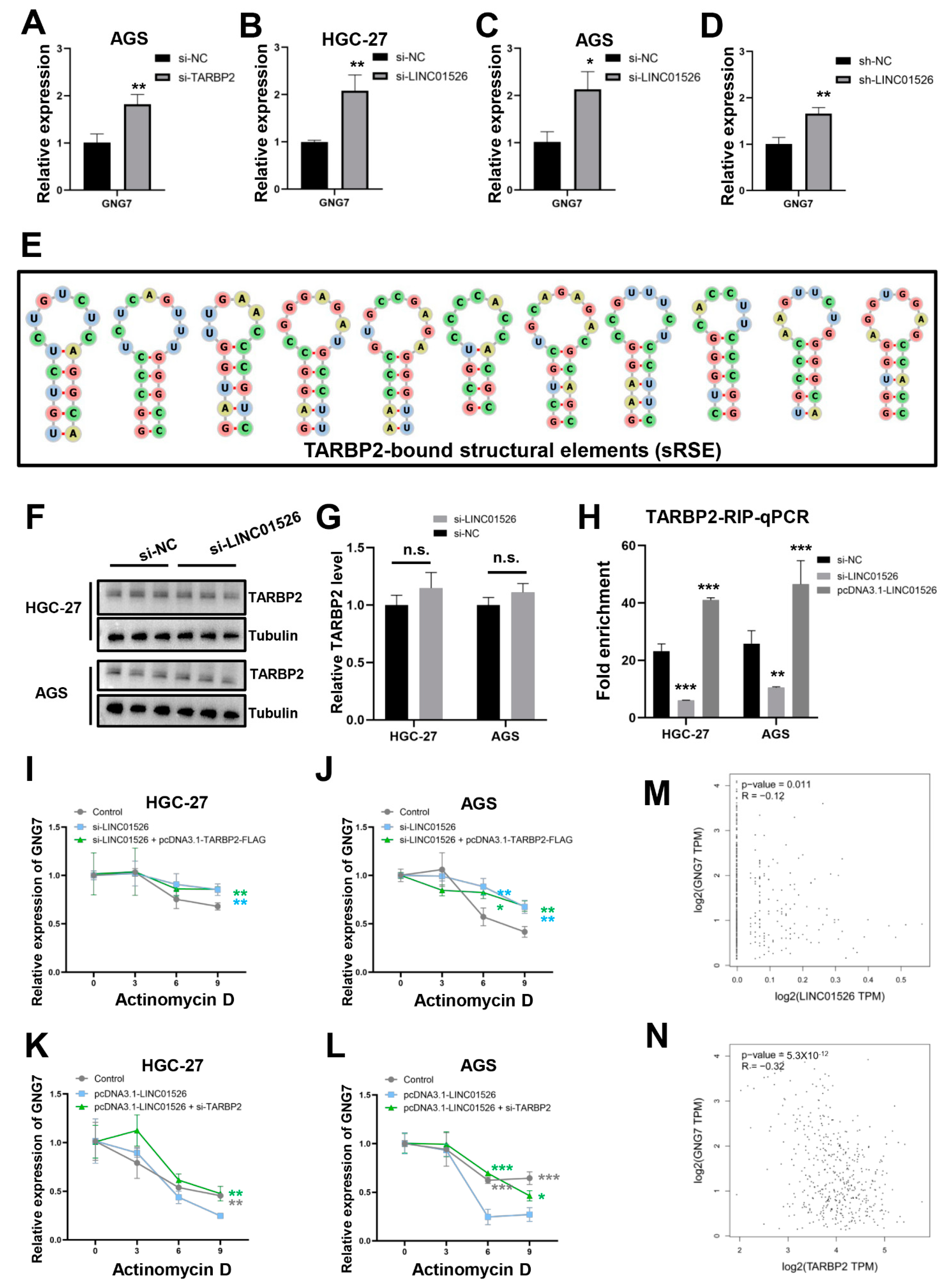
3.5. GNG7 Is the Potential Downstream Target mRNA of TARBP2 in GC Cells
3.6. “LINC01526-TARBP2” Decreased GNG7 mRNAs Stability in GC Cells
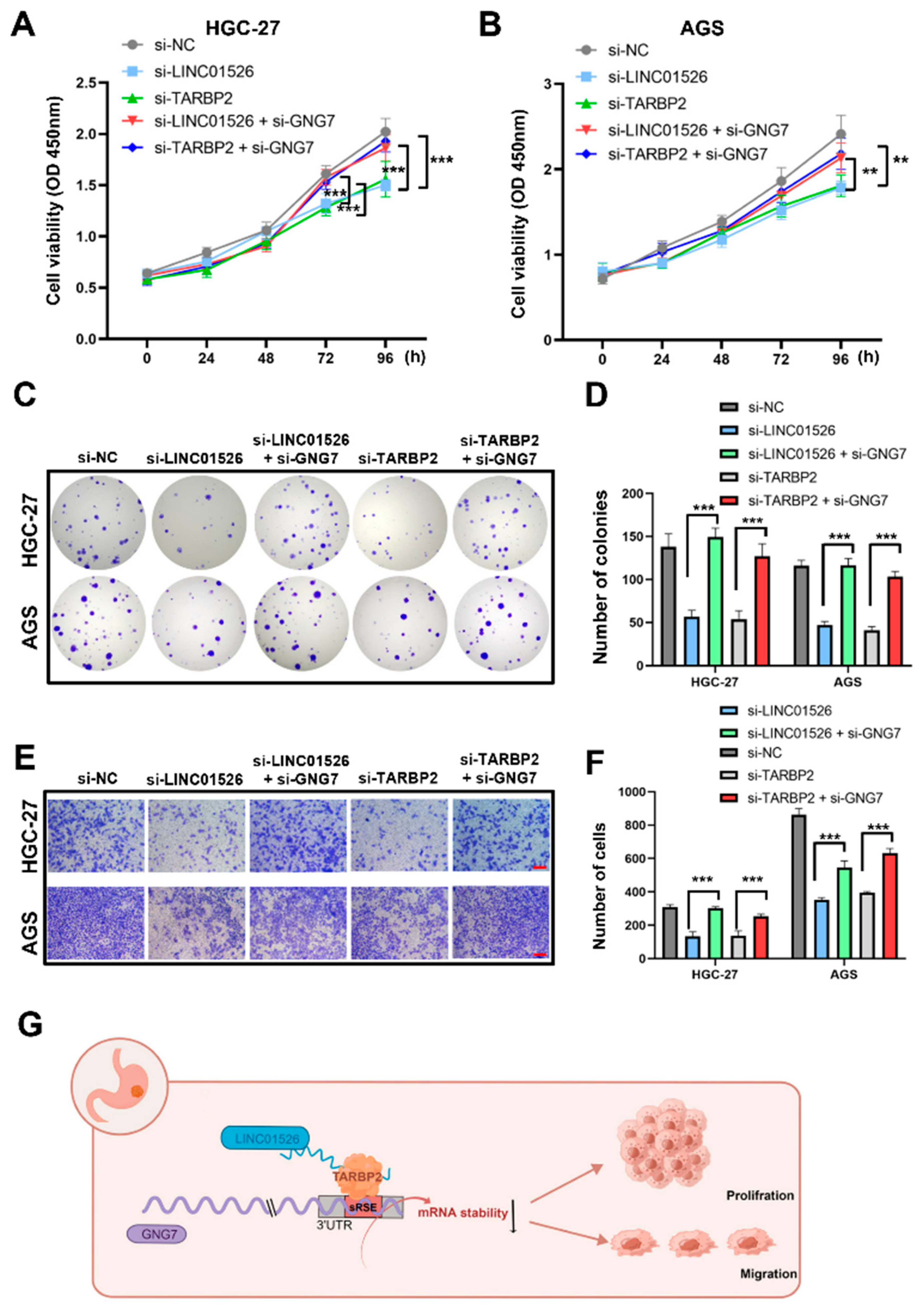
3.7. LINC01526 Interacts with TARBP2, thereby Regulating GNG7 and Promoting GC Cells’ Proliferation and Migration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Howson, C.P.; Hiyama, T.; Wynder, E.L. The decline in gastric cancer: Epidemiology of an unplanned triumph. Epidemiol. Rev. 1986, 8, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Chen, H.D.; Yu, Y.W.; Li, N.; Chen, W.Q. Changing profiles of cancer burden worldwide and in China: A secondary analysis of the global cancer statistics 2020. Chin. Med. J. 2021, 134, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Abbas, M.; Faggian, A.; Sintali, D.N.; Khan, G.J.; Naeem, S.; Shi, M.; Dingding, C. Current and future biomarkers in gastric cancer. Biomed. Pharmacother. Biomed. Pharmacother. 2018, 103, 1688–1700. [Google Scholar] [CrossRef] [PubMed]
- Durães, C.; Almeida, G.M.; Seruca, R.; Oliveira, C.; Carneiro, F. Biomarkers for gastric cancer: Prognostic, predictive or targets of therapy. Virchows Arch. 2014, 464, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.S.; Badgwell, B.D. Current treatment and recent progress in gastric cancer. CA Cancer J. Clin. 2021, 71, 264–279. [Google Scholar] [CrossRef] [PubMed]
- Necula, L.; Matei, L.; Dragu, D.; Neagu, A.I.; Mambet, C.; Nedeianu, S.; Bleotu, C.; Diaconu, C.C.; Chivu-Economescu, M. Recent advances in gastric cancer early diagnosis. World J. Gastroenterol. 2019, 25, 2029–2044. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, W.; Liu, Y.; Li, S.; Guo, D.; Sun, Q.; Jin, J.; Rao, X.; Li, M.; Sun, M.; Jiang, M.; et al. Long Noncoding RNA GMAN, Up-regulated in Gastric Cancer Tissues, Is Associated with Metastasis in Patients and Promotes Translation of Ephrin A1 by Competitively Binding GMAN-AS. Gastroenterology 2019, 156, 676–691.e11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, E.; He, X.; Zhang, C.; Su, J.; Lu, X.; Si, X.; Chen, J.; Yin, D.; Han, L.; De, W. A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1. Genome Biol. 2018, 19, 154. [Google Scholar] [CrossRef] [PubMed]
- Liao, Q.; Shen, J.; Liu, J.; Sun, X.; Zhao, G.; Chang, Y.; Xu, L.; Li, X.; Zhao, Y.; Zheng, H.; et al. Genome-wide identification and functional annotation of Plasmodium falciparum long noncoding RNAs from RNA-seq data. Parasitol. Res. 2014, 113, 1269–1281. [Google Scholar] [CrossRef] [PubMed]
- Johnsson, P.; Lipovich, L.; Grandér, D.; Morris, K.V. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim. Biophys. Acta 2014, 1840, 1063–1071. [Google Scholar] [CrossRef] [PubMed]
- Ye, D.; Jian, W.; Feng, J.; Liao, X. Role of long noncoding RNA ZFAS1 in proliferation, apoptosis and migration of chondrocytes in osteoarthritis. Biomed. Pharmacother. Biomed. Pharmacother. 2018, 104, 825–831. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Shao, L.; Zhang, J.; Zhang, H.; Wen, A.; Zhang, P. Knockdown of ZFAS1 Inhibits Hippocampal Neurons Apoptosis and Autophagy by Activating the PI3K/AKT Pathway via Up-regulating miR-421 in Epilepsy. Neurochem. Res. 2020, 45, 2433–2441. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.J.; Li, Y.; Yang, M.S.; Chen, R.; Cen, C.Q. SP1-induced ZFAS1 aggravates sepsis-induced cardiac dysfunction via miR-590-3p/NLRP3-mediated autophagy and pyroptosis. Arch. Biochem. Biophys. 2020, 695, 108611. [Google Scholar] [CrossRef]
- Wu, T.; Wu, D.; Wu, Q.; Zou, B.; Huang, X.; Cheng, X.; Wu, Y.; Hong, K.; Li, P.; Yang, R.; et al. Knockdown of Long Non-Coding RNA-ZFAS1 Protects Cardiomyocytes Against Acute Myocardial Infarction Via Anti-Apoptosis by Regulating miR-150/CRP. J. Cell. Biochem. 2017, 118, 3281–3289. [Google Scholar] [CrossRef]
- Wang, Y.; He, L.; Du, Y.; Zhu, P.; Huang, G.; Luo, J.; Yan, X.; Ye, B.; Li, C.; Xia, P.; et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell 2015, 16, 413–425. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, B.; Zhang, M.; Guo, W.; Wu, Z.; Wang, Y.; Jia, L.; Li, S.; Xie, W.; Yang, D. lncRNA Epigenetic Landscape Analysis Identifies EPIC1 as an Oncogenic lncRNA that Interacts with MYC and Promotes Cell-Cycle Progression in Cancer. Cancer Cell 2018, 33, 706–720.e9. [Google Scholar] [CrossRef]
- Olivero, C.E.; Martínez-Terroba, E.; Zimmer, J.; Liao, C.; Tesfaye, E.; Hooshdaran, N.; Schofield, J.A.; Bendor, J.; Fang, D.; Simon, M.D.; et al. p53 Activates the Long Noncoding RNA Pvt1b to Inhibit Myc and Suppress Tumorigenesis. Mol. Cell 2020, 77, 761–774.e8. [Google Scholar] [CrossRef]
- Ma, Z.; Gao, X.; Shuai, Y.; Wu, X.; Yan, Y.; Xing, X.; Ji, J. EGR1-mediated linc01503 promotes cell cycle progression and tumorigenesis in gastric cancer. Cell Prolif. 2021, 54, e12922. [Google Scholar] [CrossRef]
- Mao, X.; Ji, T.; Liu, A.; Weng, Y. ELK4-mediated lncRNA SNHG22 promotes gastric cancer progression through interacting with EZH2 and regulating miR-200c-3p/Notch1 axis. Cell Death Dis. 2021, 12, 957. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Zheng, S.; Wu, Q.; Wu, J.; Zhou, R.; Wang, C.; Wu, Z.; Rong, X.; Huang, N.; Sun, L.; et al. Long noncoding RNA (lncRNA) EIF3J-DT induces chemoresistance of gastric cancer via autophagy activation. Autophagy 2021, 17, 4083–4101. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Liang, X.; Liu, L.; Guo, Y.; Shen, S.; Liang, J.; Dong, Z. MiR-6872 host gene SEMA3B and its antisense lncRNA SEMA3B-AS1 function synergistically to suppress gastric cardia adenocarcinoma progression. Gastric Cancer 2019, 22, 705–722. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Wang, Q.; Zhu, M.; Liu, K.; Zhang, Z. Integrated analysis reveals potential long non-coding RNA biomarkers and their potential biological functions for disease free survival in gastric cancer patients. Cancer Cell Int. 2019, 19, 123. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, J.; Qian, C.; Qiu, F.; Shen, Q.; Tong, R.; Yang, Q.; Xu, J.; Zheng, B.; Lv, J.; et al. LINC00624/TEX10/NF-κB axis promotes proliferation and migration of human prostate cancer cells. Biochem. Biophys. Res. Commun. 2022, 601, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, H.; Najafabadi, H.S.; Oikonomou, P.; Greco, T.M.; Fish, L.; Salavati, R.; Cristea, I.M.; Tavazoie, S. Systematic discovery of structural elements governing stability of mammalian messenger RNAs. Nature 2012, 485, 264–268. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Wang, J.; Xu, J.; Li, S.; Zhang, R.; Shen, C.; Xie, M.; Zheng, B.; Gu, M. Long non-coding RNA DEPDC1-AS1 promotes proliferation and migration of human gastric cancer cells HGC-27 via the human antigen R-F11R pathway. J. Int. Med. Res. 2022, 50, 1–15. [Google Scholar] [CrossRef]
- Zheng, B.; Yu, J.; Guo, Y.; Gao, T.; Shen, C.; Zhang, X.; Li, H.; Huang, X. Cellular nucleic acid-binding protein is vital to testis development and spermatogenesis in mice. Reproduction 2018, 156, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Guo, Y.; Zheng, B.; Shao, B.; Jiang, M.; Wang, G.; Zhou, T.; Wang, L.; Zhou, Z.; Guo, X.; et al. Establishment of a proteome profile and identification of molecular markers for mouse spermatogonial stem cells. J. Cell. Mol. Med. 2015, 19, 521–534. [Google Scholar] [CrossRef]
- Zhang, K.; Xu, J.; Ding, Y.; Shen, C.; Lin, M.; Dai, X.; Zhou, H.; Huang, X.; Xue, B.; Zheng, B. BMI1 promotes spermatogonia proliferation through epigenetic repression of Ptprm. Biochem. Biophys. Res. Commun. 2021, 583, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wu, Y.; Lin, M.; Wang, G.; Liu, J.; Xie, M.; Zheng, B.; Shen, C.; Shen, J. BMI1 promotes osteosarcoma proliferation and metastasis by repressing the transcription of SIK1. Cancer Cell Int. 2022, 22, 136. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Chen, X.; Wu, Y.; Zheng, Q.; Chen, W.; Yan, Y.; Luan, X.; Shen, C.; Fang, J.; Zheng, B.; et al. RpS13 controls the homeostasis of germline stem cell niche through Rho1-mediated signals in the Drosophila testis. Cell Prolif. 2020, 53, e12899. [Google Scholar] [CrossRef]
- Liu, Y.; Yu, X.; Huang, A.; Zhang, X.; Wang, Y.; Geng, W.; Xu, R.; Li, S.; He, H.; Zheng, B.; et al. INTS7-ABCD3 Interaction Stimulates the Proliferation and Osteoblastic Differentiation of Mouse Bone Marrow Mesenchymal Stem Cells by Suppressing Oxidative Stress. Front. Physiol. 2021, 12, 758607. [Google Scholar] [CrossRef]
- Kozak, C.A.; Gatignol, A.; Graham, K.; Jeang, K.T.; McBride, O.W. Genetic mapping in human and mouse of the locus encoding TRBP, a protein that binds the TAR region of the human immunodeficiency virus (HIV-1). Genomics 1995, 25, 66–72. [Google Scholar] [CrossRef]
- Goodarzi, H.; Zhang, S.; Buss, C.G.; Fish, L.; Tavazoie, S.; Tavazoie, S.F. Metastasis-suppressor transcript destabilization through TARBP2 binding of mRNA hairpins. Nature 2014, 513, 256–260. [Google Scholar] [CrossRef] [PubMed]
- Lima, E.M.; Leal, M.F.; Burbano, R.R.; Khayat, A.S.; Assumpção, P.P.; Bello, M.J.; Rey, J.A.; Smith, M.A.; Casartelli, C. Methylation status of ANAPC1, CDKN2A and TP53 promoter genes in individuals with gastric cancer. Braz. J. Med. Biol. Res. Rev. Bras. Pesqui. Med. Biol. 2008, 41, 539–543. [Google Scholar] [CrossRef]
- Xu, J.; Li, N.; Deng, W.; Luo, S. Discovering the mechanism and involvement of the methylation of cyclin-dependent kinase inhibitor 2A (CDKN2A) gene and its special locus region in gastric cancer. Bioengineered 2021, 12, 1286–1298. [Google Scholar] [CrossRef]
- Ni, S.J.; Zhao, L.Q.; Wang, X.F.; Wu, Z.H.; Hua, R.X.; Wan, C.H.; Zhang, J.Y.; Zhang, X.W.; Huang, M.Z.; Gan, L.; et al. CBX7 regulates stem cell-like properties of gastric cancer cells via p16 and AKT-NF-κB-miR-21 pathways. J. Hematol. Oncol. 2018, 11, 17. [Google Scholar] [CrossRef]
- Dong, Z.; Liu, Z.; Liang, M.; Pan, J.; Lin, M.; Lin, H.; Luo, Y.; Zhou, X.; Yao, W. Identification of circRNA-miRNA-mRNA networks contributes to explore underlying pathogenesis and therapy strategy of gastric cancer. J. Transl. Med. 2021, 19, 226. [Google Scholar] [CrossRef]
- Wang, Y.; Yin, B.; Li, D.; Wang, G.; Han, X.; Sun, X. GSDME mediates caspase-3-dependent pyroptosis in gastric cancer. Biochem. Biophys. Res. Commun. 2018, 495, 1418–1425. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.J.; Chen, D.; Jiang, M.Z.; Xu, B.; Li, X.W.; Chu, Y.; Zhang, Y.J.; Mao, R.; Liang, J.; Fan, D.M. Downregulation of gasdermin D promotes gastric cancer proliferation by regulating cell cycle-related proteins. J. Dig. Dis. 2018, 19, 74–83. [Google Scholar] [CrossRef]
- Saeki, N.; Usui, T.; Aoyagi, K.; Kim, D.H.; Sato, M.; Mabuchi, T.; Yanagihara, K.; Ogawa, K.; Sakamoto, H.; Yoshida, T.; et al. Distinctive expression and function of four GSDM family genes (GSDMA-D) in normal and malignant upper gastrointestinal epithelium. Genes Chromosomes Cancer 2009, 48, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Deng, B.B.; Jiao, B.P.; Liu, Y.J.; Li, Y.R.; Wang, G.J. BIX-01294 enhanced chemotherapy effect in gastric cancer by inducing GSDME-mediated pyroptosis. Cell Biol. Int. 2020, 44, 1890–1899. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.; Zhang, S.; Dong, Y.; Cao, L.; Guo, S. PolG Inhibits Gastric Cancer Glycolysis and Viability by Suppressing PKM2 Phosphorylation. Cancer Manag. Res. 2021, 13, 1559–1570. [Google Scholar] [CrossRef] [PubMed]
- Gatignol, A.; Buckler, C.; Jeang, K.T. Relatedness of an RNA-binding motif in human immunodeficiency virus type 1 TAR RNA-binding protein TRBP to human P1/dsI kinase and Drosophila staufen. Mol. Cell. Biol. 1993, 13, 2193–2202. [Google Scholar] [PubMed]
- Smyth, E.C.; Nilsson, M.; Grabsch, H.I.; van Grieken, N.C.; Lordick, F. Gastric cancer. Lancet 2020, 396, 635–648. [Google Scholar] [CrossRef]
- Guo, X.; Lv, X.; Ru, Y.; Zhou, F.; Wang, N.; Xi, H.; Zhang, K.; Li, J.; Chang, R.; Xie, T.; et al. Circulating Exosomal Gastric Cancer-Associated Long Noncoding RNA1 as a Biomarker for Early Detection and Monitoring Progression of Gastric Cancer: A Multiphase Study. JAMA Surg. 2020, 155, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Fattahi, S.; Kosari-Monfared, M.; Golpour, M.; Emami, Z.; Ghasemiyan, M.; Nouri, M.; Akhavan-Niaki, H. LncRNAs as potential diagnostic and prognostic biomarkers in gastric cancer: A novel approach to personalized medicine. J. Cell. Physiol. 2020, 235, 3189–3206. [Google Scholar] [CrossRef]
- Wang, C.Y.; Colognori, D.; Sunwoo, H.; Wang, D.; Lee, J.T. PRC1 collaborates with SMCHD1 to fold the X-chromosome and spread Xist RNA between chromosome compartments. Nat. Commun. 2019, 10, 2950. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Wang, H.; Hu, X.; Cao, X. lncRNA MALAT1 binds chromatin remodeling subunit BRG1 to epigenetically promote inflammation-related hepatocellular carcinoma progression. Oncoimmunology 2019, 8, e1518628. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Hu, W.; Wang, Y.; An, Y.; Song, L.; Shang, P.; Yue, Z. Long non-coding RNA UCA1 promotes malignant phenotypes of renal cancer cells by modulating the miR-182-5p/DLL4 axis as a ceRNA. Mol. Cancer 2020, 19, 18. [Google Scholar] [CrossRef] [PubMed]
- Hou, P.; Meng, S.; Li, M.; Lin, T.; Chu, S.; Li, Z.; Zheng, J.; Gu, Y.; Bai, J. LINC00460/DHX9/IGF2BP2 complex promotes colorectal cancer proliferation and metastasis by mediating HMGA1 mRNA stability depending on m6A modification. J. Exp. Clin. Cancer Res. 2021, 40, 52. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, W.; Liu, W.; Huang, L.; Wang, Y.; Li, D.; Wang, G.; Zhao, Z.; Chi, X.; Xue, Y.; et al. Long Noncoding RNA VESTAR Regulates Lymphangiogenesis and Lymph Node Metastasis of Esophageal Squamous Cell Carcinoma by Enhancing VEGFC mRNA Stability. Cancer Res. 2021, 81, 3187–3199. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; He, F.; Hou, Y.; Tu, G.; Li, Q.; Jin, T.; Zeng, H.; Qin, Y.; Wan, X.; Qiao, Y.; et al. A novel hypoxic long noncoding RNA KB-1980E6.3 maintains breast cancer stem cell stemness via interacting with IGF2BP1 to facilitate c-Myc mRNA stability. Oncogene 2021, 40, 1609–1627. [Google Scholar] [CrossRef]
- Lee, S.; Kopp, F.; Chang, T.C.; Sataluri, A.; Chen, B.; Sivakumar, S.; Yu, H.; Xie, Y.; Mendell, J.T. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell 2016, 164, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, H.; Tavazoie, S.F.; Tavazoie, S. TARBP2 binding structured RNA elements drives metastasis. Cell Cycle 2014, 13, 2799–2800. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhu, C.; Huang, J.; Zhao, X.; Deng, R.; Zhang, H.; Dou, J.; Chen, Q.; Xu, M.; Yuan, H.; et al. SUMOylation of TARBP2 regulates miRNA/siRNA efficiency. Nat. Commun. 2015, 6, 8899. [Google Scholar] [CrossRef] [PubMed]
- Fish, L.; Navickas, A.; Culbertson, B.; Xu, Y.; Nguyen, H.; Zhang, S.; Hochman, M.; Okimoto, R.; Dill, B.D.; Molina, H.; et al. Nuclear TARBP2 Drives Oncogenic Dysregulation of RNA Splicing and Decay. Mol. Cell 2019, 75, 967–981.e9. [Google Scholar] [CrossRef]
- Wang, M.Y.; Huang, H.Y.; Kuo, Y.L.; Lo, C.; Sun, H.Y.; Lyu, Y.J.; Chen, B.R.; Li, J.N.; Chen, P.S. TARBP2-Enhanced Resistance during Tamoxifen Treatment in Breast Cancer. Cancers 2019, 11, 156. [Google Scholar] [CrossRef]
- Sand, M.; Skrygan, M.; Georgas, D.; Arenz, C.; Gambichler, T.; Sand, D.; Altmeyer, P.; Bechara, F.G. Expression levels of the microRNA maturing microprocessor complex component DGCR8 and the RNA-induced silencing complex (RISC) components argonaute-1, argonaute-2, PACT, TARBP1, and TARBP2 in epithelial skin cancer. Mol. Carcinog. 2012, 51, 916–922. [Google Scholar] [CrossRef]
- Lai, H.H.; Li, C.W.; Hong, C.C.; Sun, H.Y.; Chiu, C.F.; Ou, D.L.; Chen, P.S. TARBP2-mediated destabilization of Nanog overcomes sorafenib resistance in hepatocellular carcinoma. Mol. Oncol. 2019, 13, 928–945. [Google Scholar] [CrossRef] [PubMed]
- Caramuta, S.; Lee, L.; Ozata, D.M.; Akçakaya, P.; Xie, H.; Höög, A.; Zedenius, J.; Bäckdahl, M.; Larsson, C.; Lui, W.O. Clinical and functional impact of TARBP2 over-expression in adrenocortical carcinoma. Endocr. Relat. Cancer 2013, 20, 551–564. [Google Scholar] [CrossRef]
- Caramuta, S.; Lee, L.; Ozata, D.M.; Akçakaya, P.; Georgii-Hemming, P.; Xie, H.; Amini, R.M.; Lawrie, C.H.; Enblad, G.; Larsson, C.; et al. Role of microRNAs and microRNA machinery in the pathogenesis of diffuse large B-cell lymphoma. Blood Cancer J. 2013, 3, e152. [Google Scholar] [CrossRef]
- Chen, G.; Gu, H.; Fang, T.; Zhou, K.; Xu, J.; Yin, X. Hypoxia-induced let-7f-5p/TARBP2 feedback loop regulates osteosarcoma cell proliferation and invasion by inhibiting the Wnt signaling pathway. Aging 2020, 12, 6891–6903. [Google Scholar] [CrossRef]
- De Vito, C.; Riggi, N.; Cornaz, S.; Suvà, M.L.; Baumer, K.; Provero, P.; Stamenkovic, I. A TARBP2-dependent miRNA expression profile underlies cancer stem cell properties and provides candidate therapeutic reagents in Ewing sarcoma. Cancer Cell 2012, 21, 807–821. [Google Scholar] [CrossRef]
- Fujimura, H.A. The yeast G-protein homolog is involved in the mating pheromone signal transduction system. Mol. Cell. Biol. 1989, 9, 152–158. [Google Scholar] [PubMed]
- Shibata, K.; Tanaka, S.; Shiraishi, T.; Kitano, S.; Mori, M. G-protein gamma 7 is down-regulated in cancers and associated with p 27kip1-induced growth arrest. Cancer Res. 1999, 59, 1096–1101. [Google Scholar]
- Ohta, M.; Mimori, K.; Fukuyoshi, Y.; Kita, Y.; Motoyama, K.; Yamashita, K.; Ishii, H.; Inoue, H.; Mori, M. Clinical significance of the reduced expression of G protein gamma 7 (GNG7) in oesophageal cancer. Br. J. Cancer 2008, 98, 410–417. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Tian, H.; Yu, X.; Ren, P.; Yang, Q. G protein gamma 7 suppresses progression of lung adenocarcinoma by inhibiting E2F transcription factor 1. Int. J. Biol. Macromol. 2021, 182, 858–865. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Zhang, H.; Liu, T.; Chen, Y.; He, D.; Li, L. G Protein γ subunit 7 loss contributes to progression of clear cell renal cell carcinoma. J. Cell. Physiol. 2019, 234, 20002–20012. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, J.-Y.; Liu, J.-Y.; Tao, Y.; Chen, C.; Liu, S.-L. LINC01526 Promotes Proliferation and Metastasis of Gastric Cancer by Interacting with TARBP2 to Induce GNG7 mRNA Decay. Cancers 2022, 14, 4940. https://doi.org/10.3390/cancers14194940
Zhou J-Y, Liu J-Y, Tao Y, Chen C, Liu S-L. LINC01526 Promotes Proliferation and Metastasis of Gastric Cancer by Interacting with TARBP2 to Induce GNG7 mRNA Decay. Cancers. 2022; 14(19):4940. https://doi.org/10.3390/cancers14194940
Chicago/Turabian StyleZhou, Jin-Yong, Jin-Yan Liu, Yu Tao, Chen Chen, and Shen-Lin Liu. 2022. "LINC01526 Promotes Proliferation and Metastasis of Gastric Cancer by Interacting with TARBP2 to Induce GNG7 mRNA Decay" Cancers 14, no. 19: 4940. https://doi.org/10.3390/cancers14194940
APA StyleZhou, J.-Y., Liu, J.-Y., Tao, Y., Chen, C., & Liu, S.-L. (2022). LINC01526 Promotes Proliferation and Metastasis of Gastric Cancer by Interacting with TARBP2 to Induce GNG7 mRNA Decay. Cancers, 14(19), 4940. https://doi.org/10.3390/cancers14194940





