Development of a Clinically Applicable NanoString-Based Gene Expression Classifier for Muscle-Invasive Bladder Cancer Molecular Stratification
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Case Selection and Morphologic Review
2.2. Tissue Microarray Construction
2.3. Immunohistochemistry and Interpretation
2.4. RNA Extraction and mRNA Expression Analysis
2.5. Statistical Analysis
3. Results
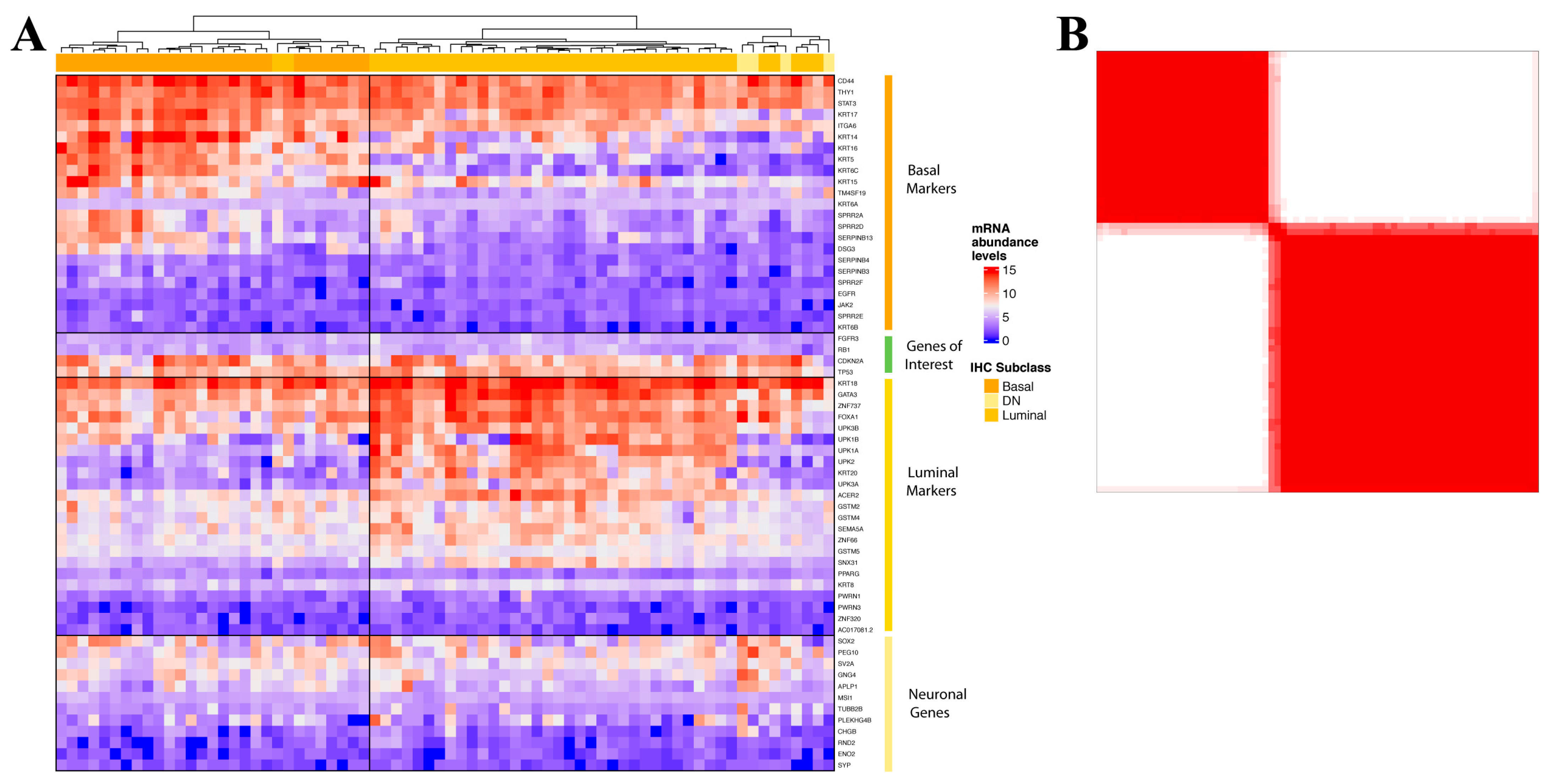
3.1. Development of NanoString nCounter Probe Set for Muscle-Invasive Bladder Cancer Molecular Stratification
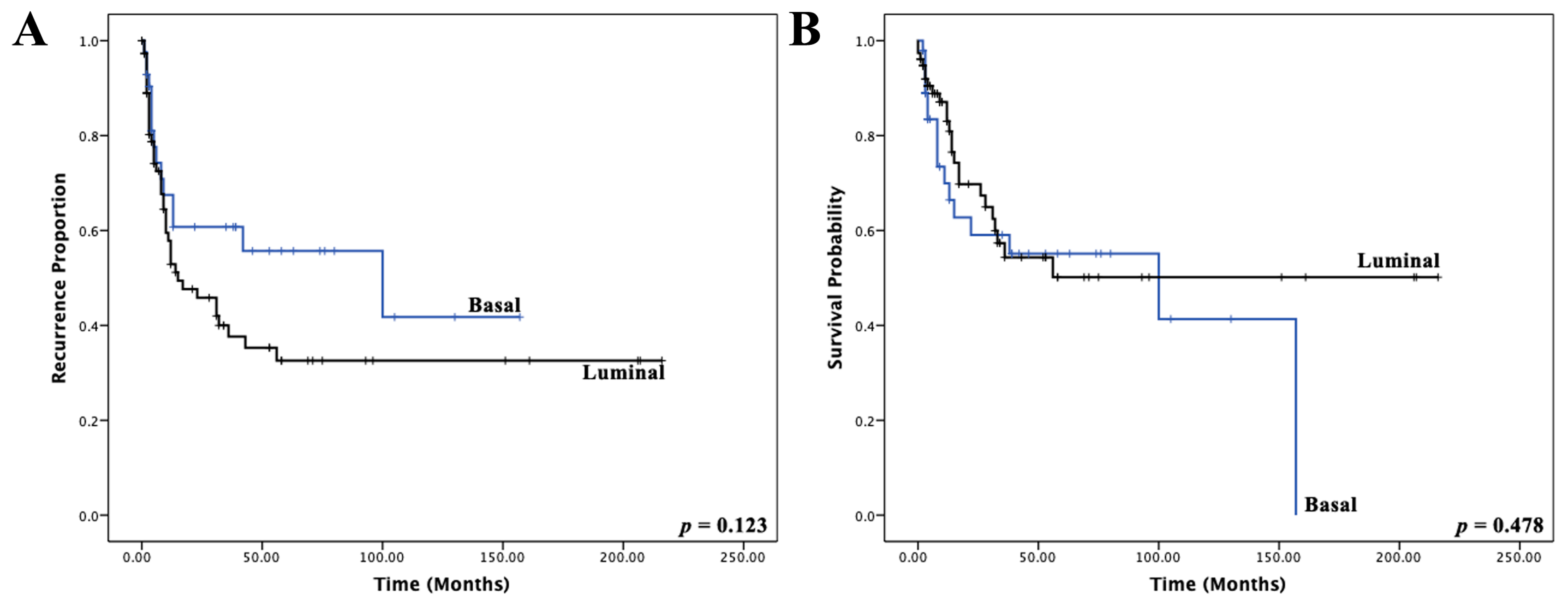
3.2. Validation of the Muscle-Invasive Bladder Cancer NanoString-Based Gene Expression Classifier
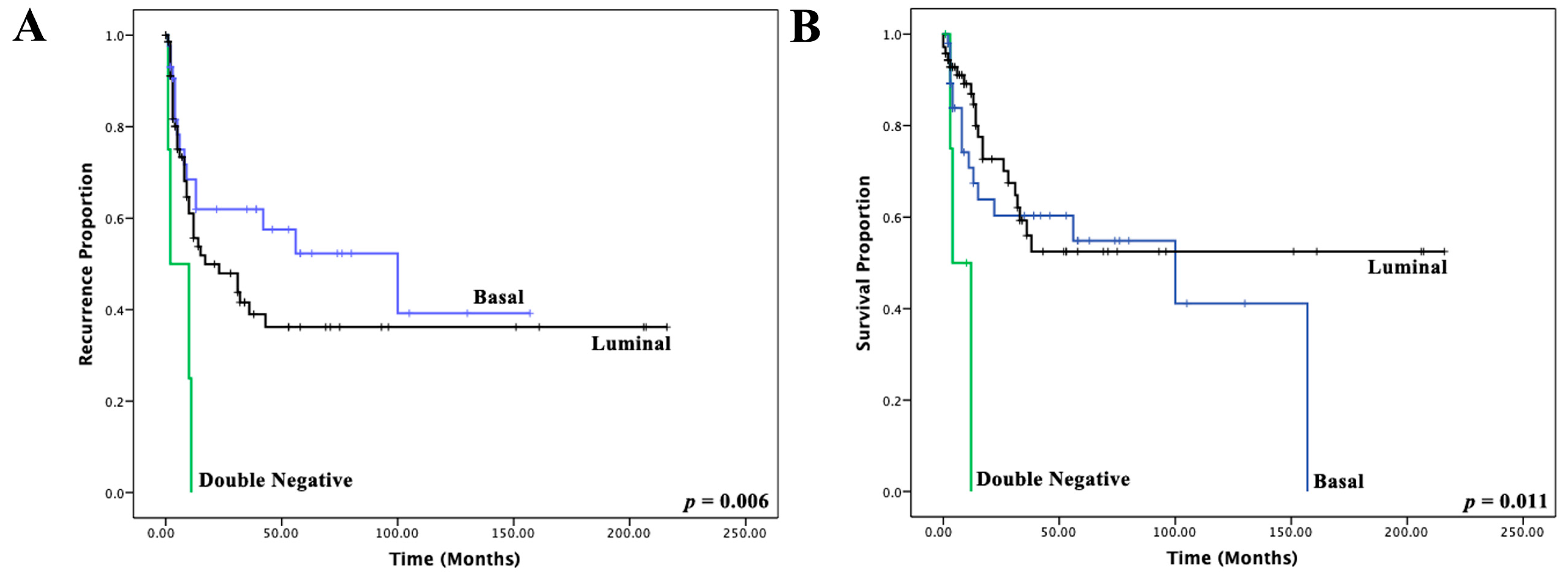
3.3. Immunohistochemical Muscle-Invasive Bladder Cancer Molecular Classifier
3.4. Development of a NanoString-Based MIBC Molecular Classifier
3.5. Technical Cost Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moch, H.; Humphrey, P.A.; Ulbright, T.M. WHO Classification of Tumours of the Urinary System and Male Genital Organs, 4th ed.; International Agency for Research on Cancer: Lyon, France, 2016; Volume 8, p. 356. ISBN 978-92-832-2437-2.
- Lenis, A.T.; Lec, P.M.; Chamie, K.; Mshs, M.D. Bladder Cancer: A Review. JAMA 2020, 324, 1980–1991. [Google Scholar] [CrossRef] [PubMed]
- Rink, M.; Lee, D.J.; Kent, M.; Xylinas, E.; Fritsche, H.M.; Babjuk, M.; Brisuda, A.; Hansen, J.; Green, D.A.; Aziz, A.; et al. Predictors of cancer-specific mortality after disease recurrence following radical cystectomy. BJU Int. 2013, 111, E30–E36. [Google Scholar] [CrossRef] [PubMed]
- Alfred Witjes, J.; Lebret, T.; Comperat, E.M.; Cowan, N.C.; De Santis, M.; Bruins, H.M.; Hernandez, V.; Espinos, E.L.; Dunn, J.; Rouanne, M.; et al. Updated 2016 EAU Guidelines on Muscle-invasive and Metastatic Bladder Cancer. Eur. Urol. 2017, 71, 462–475. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, R.W.; Barbie, D.A.; Flaherty, K.T. Mechanisms of resistance to immune checkpoint inhibitors. Br. J. Cancer 2018, 118, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Volkmer, J.P.; Sahoo, D.; Chin, R.K.; Ho, P.L.; Tang, C.; Kurtova, A.V.; Willingham, S.B.; Pazhanisamy, S.K.; Contreras-Trujillo, H.; Storm, T.A.; et al. Three differentiation states risk-stratify bladder cancer into distinct subtypes. Proc. Natl. Acad. Sci. USA 2012, 109, 2078–2083. [Google Scholar] [CrossRef] [PubMed]
- Sjodahl, G.; Lauss, M.; Lovgren, K.; Chebil, G.; Gudjonsson, S.; Veerla, S.; Patschan, O.; Aine, M.; Ferno, M.; Ringner, M.; et al. A molecular taxonomy for urothelial carcinoma. Clin. Cancer Res. 2012, 18, 3377–3386. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014, 507, 315–322. [Google Scholar] [CrossRef]
- Damrauer, J.S.; Hoadley, K.A.; Chism, D.D.; Fan, C.; Tiganelli, C.J.; Wobker, S.E.; Yeh, J.J.; Milowsky, M.I.; Iyer, G.; Parker, J.S.; et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc. Natl. Acad. Sci. USA 2014, 111, 3110–3115. [Google Scholar] [CrossRef]
- Choi, W.; Porten, S.; Kim, S.; Willis, D.; Plimack, E.R.; Hoffman-Censits, J.; Roth, B.; Cheng, T.; Tran, M.; Lee, I.L.; et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell 2014, 25, 152–165. [Google Scholar] [CrossRef]
- Rebouissou, S.; Bernard-Pierrot, I.; de Reynies, A.; Lepage, M.L.; Krucker, C.; Chapeaublanc, E.; Herault, A.; Kamoun, A.; Caillault, A.; Letouze, E.; et al. EGFR as a potential therapeutic target for a subset of muscle-invasive bladder cancers presenting a basal-like phenotype. Sci. Transl. Med. 2014, 6, 244ra291. [Google Scholar] [CrossRef]
- Dadhania, V.; Zhang, M.; Zhang, L.; Bondaruk, J.; Majewski, T.; Siefker-Radtke, A.; Guo, C.C.; Dinney, C.; Cogdell, D.E.; Zhang, S.; et al. Meta-Analysis of the Luminal and Basal Subtypes of Bladder Cancer and the Identification of Signature Immunohistochemical Markers for Clinical Use. EBioMedicine 2016, 12, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Sjodahl, G.; Eriksson, P.; Liedberg, F.; Hoglund, M. Molecular classification of urothelial carcinoma: Global mRNA classification versus tumour-cell phenotype classification. J. Pathol. 2017, 242, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.G.; Kim, J.; Al-Ahmadie, H.; Bellmunt, J.; Guo, G.; Cherniack, A.D.; Hinoue, T.; Laird, P.W.; Hoadley, K.A.; Akbani, R.; et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2017, 171, 540–556.e525. [Google Scholar] [CrossRef] [PubMed]
- Mo, Q.; Nikolos, F.; Chen, F.; Tramel, Z.; Lee, Y.C.; Hayashi, K.; Xiao, J.; Shen, J.; Chan, K.S. Prognostic Power of a Tumor Differentiation Gene Signature for Bladder Urothelial Carcinomas. J. Natl. Cancer Inst. 2018, 110, 448–459. [Google Scholar] [CrossRef] [PubMed]
- Marzouka, N.A.; Eriksson, P.; Rovira, C.; Liedberg, F.; Sjodahl, G.; Hoglund, M. A validation and extended description of the Lund taxonomy for urothelial carcinoma using the TCGA cohort. Sci. Rep. 2018, 8, 3737. [Google Scholar] [CrossRef] [PubMed]
- Seiler, R.; Ashab, H.A.D.; Erho, N.; van Rhijn, B.W.G.; Winters, B.; Douglas, J.; Van Kessel, K.E.; Fransen van de Putte, E.E.; Sommerlad, M.; Wang, N.Q.; et al. Impact of Molecular Subtypes in Muscle-invasive Bladder Cancer on Predicting Response and Survival after Neoadjuvant Chemotherapy. Eur. Urol. 2017, 72, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, J.E.; Hoffman-Censits, J.; Powles, T.; van der Heijden, M.S.; Balar, A.V.; Necchi, A.; Dawson, N.; O’Donnell, P.H.; Balmanoukian, A.; Loriot, Y.; et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: A single-arm, multicentre, phase 2 trial. Lancet 2016, 387, 1909–1920. [Google Scholar] [CrossRef]
- Sharma, P.; Retz, M.; Siefker-Radtke, A.; Baron, A.; Necchi, A.; Bedke, J.; Plimack, E.R.; Vaena, D.; Grimm, M.O.; Bracarda, S.; et al. Nivolumab in metastatic urothelial carcinoma after platinum therapy (CheckMate 275): A multicentre, single-arm, phase 2 trial. Lancet Oncol. 2017, 18, 312–322. [Google Scholar] [CrossRef]
- Kamoun, A.; de Reynies, A.; Allory, Y.; Sjodahl, G.; Robertson, A.G.; Seiler, R.; Hoadley, K.A.; Groeneveld, C.S.; Al-Ahmadie, H.; Choi, W.; et al. A Consensus Molecular Classification of Muscle-invasive Bladder Cancer. Eur. Urol. 2020, 77, 420–433. [Google Scholar] [CrossRef]
- Kim, J.; Kwiatkowski, D.; McConkey, D.J.; Meeks, J.J.; Freeman, S.S.; Bellmunt, J.; Getz, G.; Lerner, S.P. The Cancer Genome Atlas Expression Subtypes Stratify Response to Checkpoint Inhibition in Advanced Urothelial Cancer and Identify a Subset of Patients with High Survival Probability. Eur. Urol. 2019, 75, 961–964. [Google Scholar] [CrossRef]
- Guo, C.C.; Bondaruk, J.; Yao, H.; Wang, Z.; Zhang, L.; Lee, S.; Lee, J.G.; Cogdell, D.; Zhang, M.; Yang, G.; et al. Assessment of Luminal and Basal Phenotypes in Bladder Cancer. Sci. Rep. 2020, 10, 9743. [Google Scholar] [CrossRef]
- Weyerer, V.; Stoehr, R.; Bertz, S.; Lange, F.; Geppert, C.I.; Wach, S.; Taubert, H.; Sikic, D.; Wullich, B.; Hartmann, A.; et al. Prognostic impact of molecular muscle-invasive bladder cancer subtyping approaches and correlations with variant histology in a population-based mono-institutional cystectomy cohort. World J. Urol. 2021, 39, 4011–4019. [Google Scholar] [CrossRef]
- Bejrananda, T.; Kanjanapradit, K.; Saetang, J.; Sangkhathat, S. Impact of immunohistochemistry-based subtyping of GATA3, CK20, CK5/6, and CK14 expression on survival after radical cystectomy for muscle-invasive bladder cancer. Sci. Rep. 2021, 11, 21186. [Google Scholar] [CrossRef]
- Olkhov-Mitsel, E.; Hodgson, A.; Liu, S.K.; Vesprini, D.; Xu, B.; Downes, M.R. Three-antibody classifier for muscle invasive urothelial carcinoma and its correlation with p53 expression. J. Clin. Pathol. 2021. [Google Scholar] [CrossRef]
- Geiss, G.K.; Bumgarner, R.E.; Birditt, B.; Dahl, T.; Dowidar, N.; Dunaway, D.L.; Fell, H.P.; Ferree, S.; George, R.D.; Grogan, T.; et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 2008, 26, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Reis, P.P.; Waldron, L.; Goswami, R.S.; Xu, W.; Xuan, Y.; Perez-Ordonez, B.; Gullane, P.; Irish, J.; Jurisica, I.; Kamel-Reid, S. mRNA transcript quantification in archival samples using multiplexed, color-coded probes. BMC Biotechnol. 2011, 11, 46. [Google Scholar] [CrossRef]
- Lopez-Beltran, A.; Blanca, A.; Cimadamore, A.; Gogna, R.; Montironi, R.; Cheng, L. Molecular Classification of Bladder Urothelial Carcinoma Using NanoString-Based Gene Expression Analysis. Cancers 2021, 13, 5500. [Google Scholar] [CrossRef]
- Kardos, J.; Rose, T.L.; Manocha, U.; Wobker, S.E.; Damrauer, J.S.; Bivalaqua, T.J.; Kates, M.; Moore, K.J.; Parker, J.S.; Kim, W.Y. Development and validation of a NanoString BASE47 bladder cancer gene classifier. PLoS ONE 2020, 15, e0243935. [Google Scholar] [CrossRef]
- Kardos, J.; Chai, S.; Mose, L.E.; Selitsky, S.R.; Krishnan, B.; Saito, R.; Iglesia, M.D.; Milowsky, M.I.; Parker, J.S.; Kim, W.Y.; et al. Claudin-low bladder tumors are immune infiltrated and actively immune suppressed. JCI Insight 2016, 1, e85902. [Google Scholar] [CrossRef] [PubMed]
- Aine, M.; Eriksson, P.; Liedberg, F.; Sjodahl, G.; Hoglund, M. Biological determinants of bladder cancer gene expression subtypes. Sci. Rep. 2015, 5, 10957. [Google Scholar] [CrossRef] [PubMed]
- Batista da Costa, J.; Gibb, E.A.; Bivalacqua, T.J.; Liu, Y.; Oo, H.Z.; Miyamoto, D.T.; Alshalalfa, M.; Davicioni, E.; Wright, J.; Dall’Era, M.A.; et al. Molecular Characterization of Neuroendocrine-like Bladder Cancer. Clin. Cancer Res. 2019, 25, 3908–3920. [Google Scholar] [CrossRef] [PubMed]
- Koll, F.J.; Schwarz, A.; Kollermann, J.; Banek, S.; Kluth, L.; Wittler, C.; Bankov, K.; Doring, C.; Becker, N.; Chun, F.K.H.; et al. CK5/6 and GATA3 Defined Phenotypes of Muscle-Invasive Bladder Cancer: Impact in Adjuvant Chemotherapy and Molecular Subtyping of Negative Cases. Front. Med. 2022, 9, 875142. [Google Scholar] [CrossRef] [PubMed]
- Serag Eldien, M.M.; Abdou, A.G.; Elghrabawy, G.R.A.; Alhanafy, A.M.; Mahmoud, S.F. Stratification of urothelial bladder carcinoma depending on immunohistochemical expression of GATA3 and CK5/6. J. Immunoassay Immunochem. 2021, 42, 662–678. [Google Scholar] [CrossRef]
- Queipo, F.J.; Unamunzaga, G.M.; Negro, B.F.; Fuertes, S.G.; Cortes, M.A.; Tejedor, E.C.; Manas, C.M.B.; Arino, A.B.; Sjodahl, G.; Beorlegui, C. Immunohistochemistry subtyping of urothelial carcinoma is feasible in the daily practice. Virchows Arch. 2022, 481, 191–200. [Google Scholar] [CrossRef]
- Kollberg, P.; Chebil, G.; Eriksson, P.; Sjodahl, G.; Liedberg, F. Molecular subtypes applied to a population-based modern cystectomy series do not predict cancer-specific survival. Urol. Oncol. 2019, 37, 791–799. [Google Scholar] [CrossRef]
- Satyal, U.; Sikder, R.K.; McConkey, D.; Plimack, E.R.; Abbosh, P.H. Clinical implications of molecular subtyping in bladder cancer. Curr. Opin. Urol. 2019, 29, 350–356. [Google Scholar] [CrossRef]


| Univariate | Multivariate | |||
|---|---|---|---|---|
| Variable | HR (95% CI) | p-Value | HR (95% CI) | p-Value |
| Age ≥ 74 (median) | 1.179 (0.637–2.183) | 0.600 | ||
| Stage pT4 | 2.111 (1.104–4.038) | 0.024 | 1.204 (0.548–2.644) | 0.644 |
| Positive surgical Margins | 2.228 (1.152–4.309) | 0.017 | 1.985 (0.881–4.474) | 0.098 |
| Lymph Node Involvement | 2.335 (1.232–4.425) | 0.009 | 2.251 (1.132–4.476) | 0.021 |
| Variant histologic subtype | 1.516 (0.781–2.945) | 0.219 | ||
| NanoString-based gene expression subtype | ||||
| Basal vs. Luminal | 1.125 (0.670–2.336) | 0.482 | ||
| Immunohistochemical subtype | 0.032 | |||
| Double-Negative vs. Luminal | 5.868 (1.607–21.427) | 0.007 | 5.326 (1.372–20.669) | 0.016 |
| Basal vs. luminal | 1.734 (0.864–3.480) | 0.122 | 1.714 (0.853–3.443) | 0.130 |
| Variables | Study Cohort | |||
|---|---|---|---|---|
| Total | Basal | Luminal | χ2 | |
| n = 138 | n = 52 | n = 86 | p-Value | |
| Sex | 0.939 | |||
| Female | 35 (25%) | 13 (37%) | 22 (63%) | |
| Male | 103 (75%) | 39 (38%) | 64 (62%) | |
| Age | 72.1 (33–90) | 73.4 (49–90) | 71.4 (33–88) | 0.277 |
| Histology | 0.144 | |||
| Urothelial carcinoma | 99 (72%) | 37 (37%) | 62 (63%) | |
| Squamous | 23 (17%) | 13 (57%) | 10 (43%) | |
| Sarcomatoid | 4 (3%) | 1 (25%) | 3 (75%) | |
| Nested | 3 (2%) | 0 | 3 (100%) | |
| Micropapillary | 4 (3%) | 0 | 4 (100%) | |
| Plasmacytoid | 4 (3%) | 1 (25%) | 3 (75%) | |
| Carcinoma in situ | 0.003 | |||
| Present | 56 (41%) | 13 (23%) | 43 (77%) | |
| Absent | 81 (59%) | 39 (48%) | 42 (52%) | |
| Stage | 0.859 | |||
| pT2 | 15 (11%) | 5 (33%) | 10 (67%) | |
| pT3 | 81 (59%) | 32 (40%) | 49 (60%) | |
| pT4 | 42 (30%) | 15 (36%) | 27 (64%) | |
| Node | 0.138 | |||
| N0 | 88 (64%) | 37 (42%) | 51 (58%) | |
| N1 | 45 (33%) | 13 (29%) | 32 (71%) | |
| N/A | 5 (4%) | 2 (40%) | 3 (60%) | |
| Margins | 0.377 | |||
| No | 103 (75%) | 41 (40%) | 62 (60%) | |
| Yes | 35 (25%) | 11 (31%) | 24 (69%) | |
| Lymphovascular invasion | 0.033 | |||
| No | 41 (30%) | 21 (51%) | 20 (49%) | |
| Yes | 97 (70%) | 31 (32%) | 66 (68%) | |
| Recurrence | 0.058 | |||
| No | 62 (45%) | 27 (44%) | 35 (56%) | |
| Yes | 56 (41%) | 15 (27%) | 41 (73%) | |
| N/A | 20 (14%) | 10 (50%) | 10 (50%) | |
| Death | 0.623 | |||
| No | 84 (61%) | 31 (37%) | 53 (61%) | |
| Yes | 41 (30%) | 17 (41%) | 24 (59%) | |
| N/A | 13 (9%) | 4 (31%) | 9 (69%) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olkhov-Mitsel, E.; Yu, Y.; Lajkosz, K.; Liu, S.K.; Vesprini, D.; Sherman, C.G.; Downes, M.R. Development of a Clinically Applicable NanoString-Based Gene Expression Classifier for Muscle-Invasive Bladder Cancer Molecular Stratification. Cancers 2022, 14, 4911. https://doi.org/10.3390/cancers14194911
Olkhov-Mitsel E, Yu Y, Lajkosz K, Liu SK, Vesprini D, Sherman CG, Downes MR. Development of a Clinically Applicable NanoString-Based Gene Expression Classifier for Muscle-Invasive Bladder Cancer Molecular Stratification. Cancers. 2022; 14(19):4911. https://doi.org/10.3390/cancers14194911
Chicago/Turabian StyleOlkhov-Mitsel, Ekaterina, Yanhong Yu, Katherine Lajkosz, Stanley K. Liu, Danny Vesprini, Christopher G. Sherman, and Michelle R. Downes. 2022. "Development of a Clinically Applicable NanoString-Based Gene Expression Classifier for Muscle-Invasive Bladder Cancer Molecular Stratification" Cancers 14, no. 19: 4911. https://doi.org/10.3390/cancers14194911
APA StyleOlkhov-Mitsel, E., Yu, Y., Lajkosz, K., Liu, S. K., Vesprini, D., Sherman, C. G., & Downes, M. R. (2022). Development of a Clinically Applicable NanoString-Based Gene Expression Classifier for Muscle-Invasive Bladder Cancer Molecular Stratification. Cancers, 14(19), 4911. https://doi.org/10.3390/cancers14194911







