Regulation of Metastasis in Ewing Sarcoma
Abstract
Simple Summary
Abstract
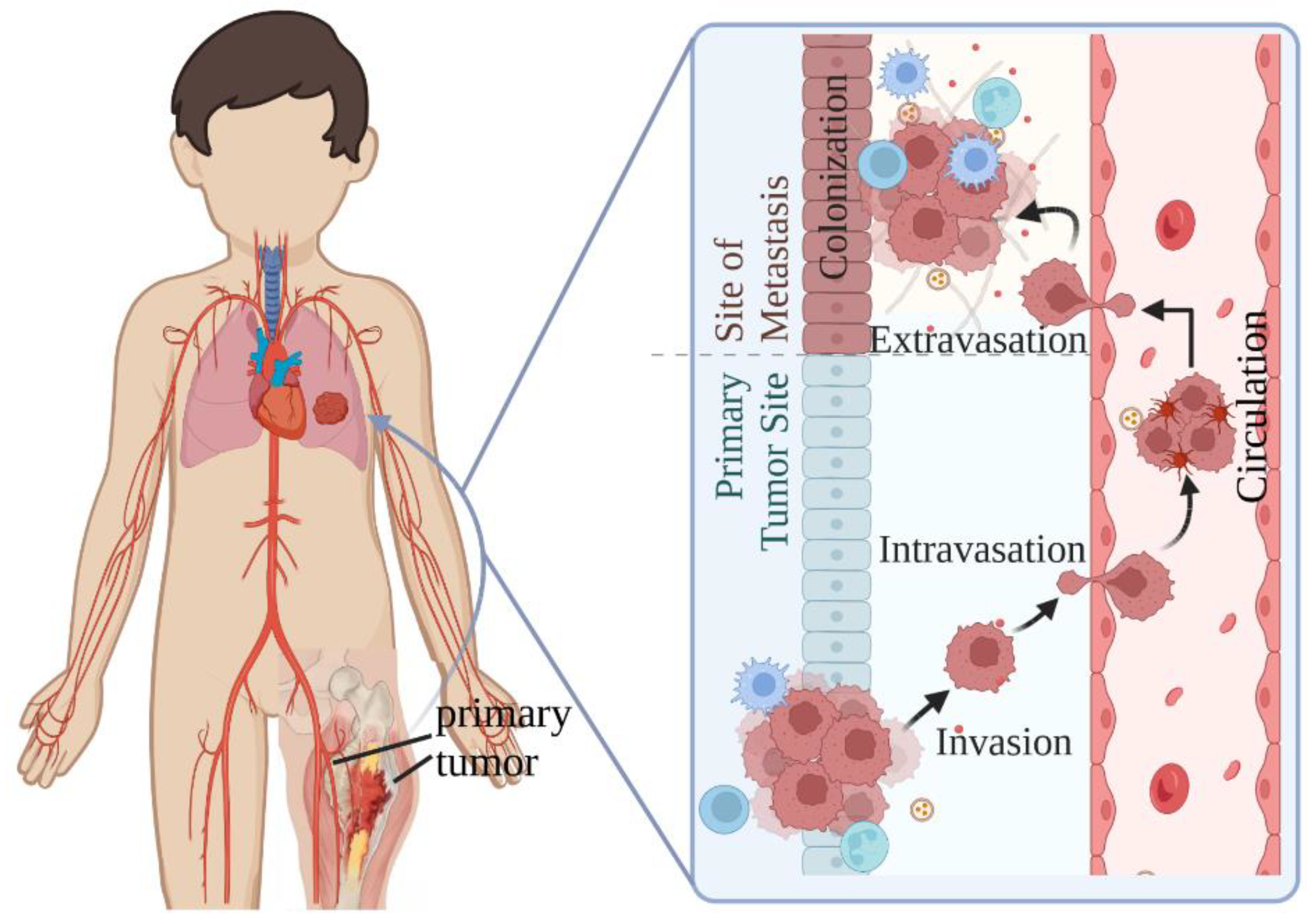
1. Introduction
2. Regulation of EwS Heterogeneity and Metastasis
2.1. Roles of EWSR1-FLI1 in Modulating Heterogeneity and Metastasis
2.1.1. Intra-Tumoral Heterogeneity Induced by the Fluctuation of EWSR1-FLI1 Expression
2.1.2. EWSR1-FLI1 Controls Metastatic Potential by Regulating Cytoskeleton Organization and Altering Gene Expression
2.2. Extracellular Signaling Contributing to EwS Metastasis
2.2.1. Hypoxia Stress-Induced Gene Expression Changes
2.2.2. Growth Factors and Immunosuppressive T Cells in the Bone Marrow
2.3. Intracellular Signaling Contributing to EwS Metastasis
2.3.1. Identification of the Gene Expression Signatures of Metastatic EwS at the Genome Level
2.3.2. Roles of Wnt/β-Catenin Signaling in Modulating EwS Metastatic Potential
2.3.3. Receptor Tyrosine Kinases (RTKs) in Modulating EwS Metastatic Potential
2.3.4. Roles of Hippo/YAP/TAZ/TEAD Signaling in Modulating EwS Metastatic Potential
2.3.5. Roles of Chromatin Modifiers in Modulating EwS Metastatic Potential
2.3.6. Transcription Factor ZEB2 Modulates EwS Metastasis
2.3.7. Other Genes Involved in EwS Metastasis
3. Discussion
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Grünewald, T.G.P.; Cidre-Aranaz, F.; Surdez, D.; Tomazou, E.M.; de Álava, E.; Kovar, H.; Sorensen, P.H.; Delattre, O.; Dirksen, U. Ewing sarcoma. Nat. Rev. Dis. Prim. 2018, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Lawlor, E.R.; Sorensen, P.H. Twenty Years on: What Do We Really Know about Ewing Sarcoma and What Is the Path Forward? Crit. Rev. Oncog. 2015, 20, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Riggi, N.; Suvà, M.L.; Stamenkovic, I. Ewing’s Sarcoma. N. Engl. J. Med. 2021, 384, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Zöllner, S.K.; Amatruda, J.F.; Bauer, S.; Collaud, S.; de Álava, E.; DuBois, S.G.; Hardes, J.; Hartmann, W.; Kovar, H.; Metzler, M.; et al. Ewing Sarcoma-Diagnosis, Treatment, Clinical Challenges and Future Perspectives. J. Clin. Med. 2021, 10, 1685. [Google Scholar] [CrossRef] [PubMed]
- Terrier, P.; Llombart-Bosch, A.; Contesso, G. Small round blue cell tumors in bone: Prognostic factors correlated to Ewing’s sarcoma and neuroectodermal tumors. Semin. Diagn. Pathol. 1996, 13, 250–257. [Google Scholar] [PubMed]
- Crompton, B.D.; Stewart, C.; Taylor-Weiner, A.; Alexe, G.; Kurek, K.C.; Calicchio, M.L.; Kiezun, A.; Carter, S.L.; Shukla, S.A.; Mehta, S.S.; et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov. 2014, 4, 1326–1341. [Google Scholar] [CrossRef]
- Delattre, O.; Zucman, J.; Plougastel, B.; Desmaze, C.; Melot, T.; Peter, M.; Kovar, H.; Joubert, I.; de Jong, P.; Rouleau, G.; et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 1992, 359, 162–165. [Google Scholar] [CrossRef]
- Toomey, E.C.; Schiffman, J.D.; Lessnick, S.L. Recent advances in the molecular pathogenesis of Ewing’s sarcoma. Oncogene 2010, 29, 4504–4516. [Google Scholar] [CrossRef]
- Herrero-Martin, D.; Fourtouna, A.; Niedan, S.; Riedmann, L.T.; Schwentner, R.; Aryee, D.N. Factors Affecting EWS-FLI1 Activity in Ewing’s Sarcoma. Sarcoma 2011, 2011, 352580. [Google Scholar] [CrossRef]
- Gangwal, K.; Close, D.; Enriquez, C.A.; Hill, C.P.; Lessnick, S.L. Emergent Properties of EWS/FLI Regulation via GGAA Microsatellites in Ewing’s Sarcoma. Genes Cancer 2010, 1, 177–187. [Google Scholar] [CrossRef]
- Monument, M.J.; Johnson, K.M.; Grossmann, A.H.; Schiffman, J.D.; Randall, R.L.; Lessnick, S.L. Microsatellites with macro-influence in ewing sarcoma. Genes 2012, 3, 444–460. [Google Scholar] [CrossRef]
- Johnson, K.M.; Mahler, N.R.; Saund, R.S.; Theisen, E.R.; Taslim, C.; Callender, N.W.; Crow, J.C.; Miller, K.R.; Lessnick, S.L. Role for the EWS domain of EWS/FLI in binding GGAA-microsatellites required for Ewing sarcoma anchorage independent growth. Proc. Natl. Acad. Sci. USA 2017, 114, 9870–9875. [Google Scholar] [CrossRef] [PubMed]
- Boulay, G.; Volorio, A.; Iyer, S.; Broye, L.C.; Stamenkovic, I.; Riggi, N.; Rivera, M.N. Epigenome editing of microsatellite repeats defines tumor-specific enhancer functions and dependencies. Genes Dev. 2018, 32, 1008–1019. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.M.; Taslim, C.; Saund, R.S.; Lessnick, S.L. Identification of two types of GGAA-microsatellites and their roles in EWS/FLI binding and gene regulation in Ewing sarcoma. PLoS ONE 2017, 12, e0186275. [Google Scholar] [CrossRef] [PubMed]
- Guillon, N.; Tirode, F.; Boeva, V.; Zynovyev, A.; Barillot, E.; Delattre, O. The oncogenic EWS-FLI1 protein binds in vivo GGAA microsatellite sequences with potential transcriptional activation function. PLoS ONE 2009, 4, e4932. [Google Scholar] [CrossRef] [PubMed]
- Riggi, N.; Knoechel, B.; Gillespie, S.M.; Rheinbay, E.; Boulay, G.; Suvà, M.L.; Rossetti, N.E.; Boonseng, W.E.; Oksuz, O.; Cook, E.B.; et al. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell 2014, 26, 668–681. [Google Scholar] [CrossRef]
- Boulay, G.; Sandoval, G.J.; Riggi, N.; Iyer, S.; Buisson, R.; Naigles, B.; Awad, M.E.; Rengarajan, S.; Volorio, A.; McBride, M.J.; et al. Cancer-Specific Retargeting of BAF Complexes by a Prion-like Domain. Cell 2017, 171, 163–178.e119. [Google Scholar] [CrossRef]
- Schwentner, R.; Papamarkou, T.; Kauer, M.O.; Stathopoulos, V.; Yang, F.; Bilke, S.; Meltzer, P.S.; Girolami, M.; Kovar, H. EWS-FLI1 employs an E2F switch to drive target gene expression. Nucleic Acids Res. 2015, 43, 2780–2789. [Google Scholar] [CrossRef]
- Tomazou, E.M.; Sheffield, N.C.; Schmidl, C.; Schuster, M.; Schönegger, A.; Datlinger, P.; Kubicek, S.; Bock, C.; Kovar, H. Epigenome mapping reveals distinct modes of gene regulation and widespread enhancer reprogramming by the oncogenic fusion protein EWS-FLI1. Cell Rep. 2015, 10, 1082–1095. [Google Scholar] [CrossRef]
- Sankar, S.; Theisen, E.R.; Bearss, J.; Mulvihill, T.; Hoffman, L.M.; Sorna, V.; Beckerle, M.C.; Sharma, S.; Lessnick, S.L. Reversible LSD1 inhibition interferes with global EWS/ETS transcriptional activity and impedes Ewing sarcoma tumor growth. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014, 20, 4584–4597. [Google Scholar] [CrossRef]
- Hernandez-Muñoz, I.; Figuerola, E.; Sanchez-Molina, S.; Rodriguez, E.; Fernández-Mariño, A.I.; Pardo-Pastor, C.; Bahamonde, M.I.; Fernández-Fernández, J.M.; García-Domínguez, D.J.; Hontecillas-Prieto, L.; et al. RING1B contributes to Ewing sarcoma development by repressing the NaV1.6 sodium channel and the NF-κB pathway, independently of the fusion oncoprotein. Oncotarget 2016, 7, 46283–46300. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Molina, S.; Figuerola-Bou, E.; Blanco, E.; Sánchez-Jiménez, M.; Táboas, P.; Gómez, S.; Ballaré, C.; García-Domínguez, D.J.; Prada, E.; Hontecillas-Prieto, L.; et al. RING1B recruits EWSR1-FLI1 and cooperates in the remodeling of chromatin necessary for Ewing sarcoma tumorigenesis. Sci. Adv. 2020, 6. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chen, C.W. Epigenetic and Transcriptional Signaling in Ewing Sarcoma-Disease Etiology and Therapeutic Opportunities. Biomedicines 2022, 10, 1325. [Google Scholar] [CrossRef] [PubMed]
- Dagogo-Jack, I.; Shaw, A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef]
- Dharia, N.V.; Kugener, G.; Guenther, L.M.; Malone, C.F.; Durbin, A.D.; Hong, A.L.; Howard, T.P.; Bandopadhayay, P.; Wechsler, C.S.; Fung, I.; et al. A first-generation pediatric cancer dependency map. Nat. Genet. 2021, 53, 529–538. [Google Scholar] [CrossRef]
- Shulman, D.S.; Whittle, S.B.; Surdez, D.; Bailey, K.M.; de Álava, E.; Yustein, J.T.; Shlien, A.; Hayashi, M.; Bishop, A.J.R.; Crompton, B.D.; et al. An international working group consensus report for the prioritization of molecular biomarkers for Ewing sarcoma. NPJ Precis. Oncol. 2022, 6, 65. [Google Scholar] [CrossRef]
- Mackintosh, C.; Ordóñez, J.L.; García-Domínguez, D.J.; Sevillano, V.; Llombart-Bosch, A.; Szuhai, K.; Scotlandi, K.; Alberghini, M.; Sciot, R.; Sinnaeve, F.; et al. 1q gain and CDT2 overexpression underlie an aggressive and highly proliferative form of Ewing sarcoma. Oncogene 2012, 31, 1287–1298. [Google Scholar] [CrossRef]
- Agelopoulos, K.; Richter, G.H.; Schmidt, E.; Dirksen, U.; von Heyking, K.; Moser, B.; Klein, H.U.; Kontny, U.; Dugas, M.; Poos, K.; et al. Deep Sequencing in Conjunction with Expression and Functional Analyses Reveals Activation of FGFR1 in Ewing Sarcoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2015, 21, 4935–4946. [Google Scholar] [CrossRef]
- Franzetti, G.A.; Laud-Duval, K.; van der Ent, W.; Brisac, A.; Irondelle, M.; Aubert, S.; Dirksen, U.; Bouvier, C.; de Pinieux, G.; Snaar-Jagalska, E.; et al. Cell-to-cell heterogeneity of EWSR1-FLI1 activity determines proliferation/migration choices in Ewing sarcoma cells. Oncogene 2017, 36, 3505–3514. [Google Scholar] [CrossRef]
- Sheffield, N.C.; Pierron, G.; Klughammer, J.; Datlinger, P.; Schönegger, A.; Schuster, M.; Hadler, J.; Surdez, D.; Guillemot, D.; Lapouble, E.; et al. DNA methylation heterogeneity defines a disease spectrum in Ewing sarcoma. Nat. Med. 2017, 23, 386–395. [Google Scholar] [CrossRef]
- Aynaud, M.M.; Mirabeau, O.; Gruel, N.; Grossetête, S.; Boeva, V.; Durand, S.; Surdez, D.; Saulnier, O.; Zaïdi, S.; Gribkova, S.; et al. Transcriptional Programs Define Intratumoral Heterogeneity of Ewing Sarcoma at Single-Cell Resolution. Cell Rep. 2020, 30, 1767–1779.e1766. [Google Scholar] [CrossRef] [PubMed]
- Khoogar, R.; Li, F.; Chen, Y.; Ignatius, M.; Lawlor, E.R.; Kitagawa, K.; Huang, T.H.; Phelps, D.A.; Houghton, P.J. Single-cell RNA profiling identifies diverse cellular responses to EWSR1/FLI1 downregulation in Ewing sarcoma cells. Cell. Oncol. 2022, 45, 19–40. [Google Scholar] [CrossRef] [PubMed]
- Steeg, P.S. Targeting metastasis. Nat. Rev. Cancer 2016, 16, 201–218. [Google Scholar] [CrossRef] [PubMed]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular principles of metastasis: A hallmark of cancer revisited. Signal Transduct. Target. Ther. 2020, 5, 28. [Google Scholar] [CrossRef]
- McCann, T.S.; Parrish, J.K.; Hsieh, J.; Sechler, M.; Sobral, L.M.; Self, C.; Jones, K.L.; Goodspeed, A.; Costello, J.C.; Jedlicka, P. KDM5A and PHF2 positively control expression of pro-metastatic genes repressed by EWS/Fli1, and promote growth and metastatic properties in Ewing sarcoma. Oncotarget 2020, 11, 3818–3831. [Google Scholar] [CrossRef]
- Pedersen, E.A.; Menon, R.; Bailey, K.M.; Thomas, D.G.; Van Noord, R.A.; Tran, J.; Wang, H.; Qu, P.P.; Hoering, A.; Fearon, E.R.; et al. Activation of Wnt/β-Catenin in Ewing Sarcoma Cells Antagonizes EWS/ETS Function and Promotes Phenotypic Transition to More Metastatic Cell States. Cancer Res. 2016, 76, 5040–5053. [Google Scholar] [CrossRef]
- Schaefer, K.L.; Eisenacher, M.; Braun, Y.; Brachwitz, K.; Wai, D.H.; Dirksen, U.; Lanvers-Kaminsky, C.; Juergens, H.; Herrero, D.; Stegmaier, S.; et al. Microarray analysis of Ewing’s sarcoma family of tumours reveals characteristic gene expression signatures associated with metastasis and resistance to chemotherapy. Eur. J. Cancer 2008, 44, 699–709. [Google Scholar] [CrossRef]
- Jin, Z.; Zhao, C.; Han, X.; Han, Y. Wnt5a promotes ewing sarcoma cell migration through upregulating CXCR4 expression. BMC Cancer 2012, 12, 480. [Google Scholar] [CrossRef]
- Katschnig, A.M.; Kauer, M.O.; Schwentner, R.; Tomazou, E.M.; Mutz, C.N.; Linder, M.; Sibilia, M.; Alonso, J.; Aryee, D.N.T.; Kovar, H. EWS-FLI1 perturbs MRTFB/YAP-1/TEAD target gene regulation inhibiting cytoskeletal autoregulatory feedback in Ewing sarcoma. Oncogene 2017, 36, 5995–6005. [Google Scholar] [CrossRef]
- Bierbaumer, L.; Katschnig, A.M.; Radic-Sarikas, B.; Kauer, M.O.; Petro, J.A.; Högler, S.; Gurnhofer, E.; Pedot, G.; Schäfer, B.W.; Schwentner, R.; et al. YAP/TAZ inhibition reduces metastatic potential of Ewing sarcoma cells. Oncogenesis 2021, 10, 2. [Google Scholar] [CrossRef]
- Mendoza-Naranjo, A.; El-Naggar, A.; Wai, D.H.; Mistry, P.; Lazic, N.; Ayala, F.R.; da Cunha, I.W.; Rodriguez-Viciana, P.; Cheng, H.; Tavares Guerreiro Fregnani, J.H.; et al. ERBB4 confers metastatic capacity in Ewing sarcoma. EMBO Mol. Med. 2013, 5, 1087–1102. [Google Scholar] [CrossRef] [PubMed]
- Potratz, J.; Tillmanns, A.; Berning, P.; Korsching, E.; Schaefer, C.; Lechtape, B.; Schleithoff, C.; Unland, R.; Schäfer, K.L.; Müller-Tidow, C.; et al. Receptor tyrosine kinase gene expression profiles of Ewing sarcomas reveal ROR1 as a potential therapeutic target in metastatic disease. Mol. Oncol. 2016, 10, 677–692. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, A.; Hoffman, L.M.; Welm, A.L.; Lessnick, S.L.; Beckerle, M.C. The EWS/FLI Oncogene Drives Changes in Cellular Morphology, Adhesion, and Migration in Ewing Sarcoma. Genes Cancer 2012, 3, 102–116. [Google Scholar] [CrossRef] [PubMed]
- Amsellem, V.; Kryszke, M.H.; Hervy, M.; Subra, F.; Athman, R.; Leh, H.; Brachet-Ducos, C.; Auclair, C. The actin cytoskeleton-associated protein zyxin acts as a tumor suppressor in Ewing tumor cells. Exp. Cell Res. 2005, 304, 443–456. [Google Scholar] [CrossRef]
- Chaturvedi, A.; Hoffman, L.M.; Jensen, C.C.; Lin, Y.C.; Grossmann, A.H.; Randall, R.L.; Lessnick, S.L.; Welm, A.L.; Beckerle, M.C. Molecular dissection of the mechanism by which EWS/FLI expression compromises actin cytoskeletal integrity and cell adhesion in Ewing sarcoma. Mol. Biol. Cell 2014, 25, 2695–2709. [Google Scholar] [CrossRef]
- Luo, W.; Xu, C.; Ayello, J.; Dela Cruz, F.; Rosenblum, J.M.; Lessnick, S.L.; Cairo, M.S. Protein phosphatase 1 regulatory subunit 1A in ewing sarcoma tumorigenesis and metastasis. Oncogene 2018, 37, 798–809. [Google Scholar] [CrossRef]
- von Heyking, K.; Calzada-Wack, J.; Göllner, S.; Neff, F.; Schmidt, O.; Hensel, T.; Schirmer, D.; Fasan, A.; Esposito, I.; Müller-Tidow, C.; et al. The endochondral bone protein CHM1 sustains an undifferentiated, invasive phenotype, promoting lung metastasis in Ewing sarcoma. Mol. Oncol. 2017, 11, 1288–1301. [Google Scholar] [CrossRef]
- Tirado, O.M.; Mateo-Lozano, S.; Villar, J.; Dettin, L.E.; Llort, A.; Gallego, S.; Ban, J.; Kovar, H.; Notario, V. Caveolin-1 (CAV1) is a target of EWS/FLI-1 and a key determinant of the oncogenic phenotype and tumorigenicity of Ewing’s sarcoma cells. Cancer Res. 2006, 66, 9937–9947. [Google Scholar] [CrossRef]
- Lagares-Tena, L.; García-Monclús, S.; López-Alemany, R.; Almacellas-Rabaiget, O.; Huertas-Martínez, J.; Sáinz-Jaspeado, M.; Mateo-Lozano, S.; Rodríguez-Galindo, C.; Rello-Varona, S.; Herrero-Martín, D.; et al. Caveolin-1 promotes Ewing sarcoma metastasis regulating MMP-9 expression through MAPK/ERK pathway. Oncotarget 2016, 7, 56889–56903. [Google Scholar] [CrossRef]
- Nicolaou, P.; Hajjar, R.J.; Kranias, E.G. Role of protein phosphatase-1 inhibitor-1 in cardiac physiology and pathophysiology. J. Mol. Cell. Cardiol. 2009, 47, 365–371. [Google Scholar] [CrossRef]
- Hauer, K.; Calzada-Wack, J.; Steiger, K.; Grunewald, T.G.; Baumhoer, D.; Plehm, S.; Buch, T.; Prazeres da Costa, O.; Esposito, I.; Burdach, S.; et al. DKK2 mediates osteolysis, invasiveness, and metastatic spread in Ewing sarcoma. Cancer Res. 2013, 73, 967–977. [Google Scholar] [CrossRef]
- Staege, M.S.; Hutter, C.; Neumann, I.; Foja, S.; Hattenhorst, U.E.; Hansen, G.; Afar, D.; Burdach, S.E. DNA microarrays reveal relationship of Ewing family tumors to both endothelial and fetal neural crest-derived cells and define novel targets. Cancer Res. 2004, 64, 8213–8221. [Google Scholar] [CrossRef] [PubMed]
- Joshi, B.; Strugnell, S.S.; Goetz, J.G.; Kojic, L.D.; Cox, M.E.; Griffith, O.L.; Chan, S.K.; Jones, S.J.; Leung, S.P.; Masoudi, H.; et al. Phosphorylated caveolin-1 regulates Rho/ROCK-dependent focal adhesion dynamics and tumor cell migration and invasion. Cancer Res. 2008, 68, 8210–8220. [Google Scholar] [CrossRef] [PubMed]
- Sáinz-Jaspeado, M.; Lagares-Tena, L.; Lasheras, J.; Navid, F.; Rodriguez-Galindo, C.; Mateo-Lozano, S.; Notario, V.; Sanjuan, X.; Garcia Del Muro, X.; Fabra, A.; et al. Caveolin-1 modulates the ability of Ewing’s sarcoma to metastasize. Mol. Cancer Res. 2010, 8, 1489–1500. [Google Scholar] [CrossRef] [PubMed]
- Muz, B.; de la Puente, P.; Azab, F.; Azab, A.K. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia 2015, 3, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Vaupel, P.; Mayer, A. Hypoxia in cancer: Significance and impact on clinical outcome. Cancer Metastasis Rev. 2007, 26, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Ratcliffe, P.J.; O’Rourke, J.F.; Maxwell, P.H.; Pugh, C.W. Oxygen sensing, hypoxia-inducible factor-1 and the regulation of mammalian gene expression. J. Exp. Biol. 1998, 201, 1153–1162. [Google Scholar] [CrossRef]
- Maxwell, P.H.; Dachs, G.U.; Gleadle, J.M.; Nicholls, L.G.; Harris, A.L.; Stratford, I.J.; Hankinson, O.; Pugh, C.W.; Ratcliffe, P.J. Hypoxia-inducible factor-1 modulates gene expression in solid tumors and influences both angiogenesis and tumor growth. Proc. Natl. Acad. Sci. USA 1997, 94, 8104–8109. [Google Scholar] [CrossRef]
- Rankin, E.B.; Giaccia, A.J. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 2008, 15, 678–685. [Google Scholar] [CrossRef]
- van der Schaft, D.W.; Hillen, F.; Pauwels, P.; Kirschmann, D.A.; Castermans, K.; Egbrink, M.G.; Tran, M.G.; Sciot, R.; Hauben, E.; Hogendoorn, P.C.; et al. Tumor cell plasticity in Ewing sarcoma, an alternative circulatory system stimulated by hypoxia. Cancer Res. 2005, 65, 11520–11528. [Google Scholar] [CrossRef]
- Knowles, H.J.; Schaefer, K.L.; Dirksen, U.; Athanasou, N.A. Hypoxia and hypoglycaemia in Ewing’s sarcoma and osteosarcoma: Regulation and phenotypic effects of Hypoxia-Inducible Factor. BMC Cancer 2010, 10, 372. [Google Scholar] [CrossRef] [PubMed]
- Aryee, D.N.; Niedan, S.; Kauer, M.; Schwentner, R.; Bennani-Baiti, I.M.; Ban, J.; Muehlbacher, K.; Kreppel, M.; Walker, R.L.; Meltzer, P.; et al. Hypoxia modulates EWS-FLI1 transcriptional signature and enhances the malignant properties of Ewing’s sarcoma cells in vitro. Cancer Res. 2010, 70, 4015–4023. [Google Scholar] [CrossRef] [PubMed]
- Bailey, K.M.; Airik, M.; Krook, M.A.; Pedersen, E.A.; Lawlor, E.R. Micro-Environmental Stress Induces Src-Dependent Activation of Invadopodia and Cell Migration in Ewing Sarcoma. Neoplasia 2016, 18, 480–488. [Google Scholar] [CrossRef] [PubMed]
- El-Naggar, A.M.; Veinotte, C.J.; Cheng, H.; Grunewald, T.G.; Negri, G.L.; Somasekharan, S.P.; Corkery, D.P.; Tirode, F.; Mathers, J.; Khan, D.; et al. Translational Activation of HIF1α by YB-1 Promotes Sarcoma Metastasis. Cancer Cell 2015, 27, 682–697. [Google Scholar] [CrossRef]
- El-Naggar, A.M.; Somasekharan, S.P.; Wang, Y.; Cheng, H.; Negri, G.L.; Pan, M.; Wang, X.Q.; Delaidelli, A.; Rafn, B.; Cran, J.; et al. Class I HDAC inhibitors enhance YB-1 acetylation and oxidative stress to block sarcoma metastasis. EMBO Rep. 2019, 20, e48375. [Google Scholar] [CrossRef]
- Somasekharan, S.P.; Stoynov, N.; Rotblat, B.; Leprivier, G.; Galpin, J.D.; Ahern, C.A.; Foster, L.J.; Sorensen, P.H. Identification and quantification of newly synthesized proteins translationally regulated by YB-1 using a novel Click-SILAC approach. J. Proteom. 2012, 77, e1–e10. [Google Scholar] [CrossRef]
- Evdokimova, V.; Tognon, C.; Ng, T.; Sorensen, P.H. Reduced proliferation and enhanced migration: Two sides of the same coin? Molecular mechanisms of metastatic progression by YB-1. Cell Cycle 2009, 8, 2901–2906. [Google Scholar] [CrossRef]
- Gluz, O.; Mengele, K.; Schmitt, M.; Kates, R.; Diallo-Danebrock, R.; Neff, F.; Royer, H.D.; Eckstein, N.; Mohrmann, S.; Ting, E.; et al. Y-box-binding protein YB-1 identifies high-risk patients with primary breast cancer benefiting from rapidly cycled tandem high-dose adjuvant chemotherapy. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2009, 27, 6144–6151. [Google Scholar] [CrossRef]
- Giménez-Bonafé, P.; Fedoruk, M.N.; Whitmore, T.G.; Akbari, M.; Ralph, J.L.; Ettinger, S.; Gleave, M.E.; Nelson, C.C. YB-1 is upregulated during prostate cancer tumor progression and increases P-glycoprotein activity. Prostate 2004, 59, 337–349. [Google Scholar] [CrossRef]
- Evdokimova, V.; Tognon, C.; Ng, T.; Ruzanov, P.; Melnyk, N.; Fink, D.; Sorokin, A.; Ovchinnikov, L.P.; Davicioni, E.; Triche, T.J.; et al. Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell 2009, 15, 402–415. [Google Scholar] [CrossRef]
- Krook, M.A.; Nicholls, L.A.; Scannell, C.A.; Chugh, R.; Thomas, D.G.; Lawlor, E.R. Stress-induced CXCR4 promotes migration and invasion of ewing sarcoma. Mol. Cancer Res. 2014, 12, 953–964. [Google Scholar] [CrossRef] [PubMed]
- Burger, J.A.; Kipps, T.J. CXCR4: A key receptor in the crosstalk between tumor cells and their microenvironment. Blood 2006, 107, 1761–1767. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Homey, B.; Soto, H.; Ge, N.; Catron, D.; Buchanan, M.E.; McClanahan, T.; Murphy, E.; Yuan, W.; Wagner, S.N.; et al. Involvement of chemokine receptors in breast cancer metastasis. Nature 2001, 410, 50–56. [Google Scholar] [CrossRef]
- Orimo, A.; Gupta, P.B.; Sgroi, D.C.; Arenzana-Seisdedos, F.; Delaunay, T.; Naeem, R.; Carey, V.J.; Richardson, A.L.; Weinberg, R.A. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 2005, 121, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, F.; Monti, P.; Leone, B.E.; Zerbi, A.; Vecchi, A.; Piemonti, L.; Mantovani, A.; Allavena, P. Increased survival, proliferation, and migration in metastatic human pancreatic tumor cells expressing functional CXCR4. Cancer Res. 2004, 64, 8420–8427. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.; Zhou, Z.; Jia, S.F.; Lee, T.H.; Morales-Arias, J.; Cao, Y.; Kleinerman, E.S. Stromal cell-derived factor-1 stimulates vasculogenesis and enhances Ewing’s sarcoma tumor growth in the absence of vascular endothelial growth factor. Int. J. Cancer 2008, 123, 831–837. [Google Scholar] [CrossRef] [PubMed]
- Bennani-Baiti, I.M.; Cooper, A.; Lawlor, E.R.; Kauer, M.; Ban, J.; Aryee, D.N.; Kovar, H. Intercohort gene expression co-analysis reveals chemokine receptors as prognostic indicators in Ewing’s sarcoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2010, 16, 3769–3778. [Google Scholar] [CrossRef]
- Berghuis, D.; Schilham, M.W.; Santos, S.J.; Savola, S.; Knowles, H.J.; Dirksen, U.; Schaefer, K.L.; Vakkila, J.; Hogendoorn, P.C.; Lankester, A.C. The CXCR4-CXCL12 axis in Ewing sarcoma: Promotion of tumor growth rather than metastatic disease. Clin. Sarcoma Res. 2012, 2, 24. [Google Scholar] [CrossRef]
- Kamura, S.; Matsumoto, Y.; Fukushi, J.I.; Fujiwara, T.; Iida, K.; Okada, Y.; Iwamoto, Y. Basic fibroblast growth factor in the bone microenvironment enhances cell motility and invasion of Ewing’s sarcoma family of tumours by activating the FGFR1-PI3K-Rac1 pathway. Br. J. Cancer 2010, 103, 370–381. [Google Scholar] [CrossRef]
- Brinkrolf, P.; Landmeier, S.; Altvater, B.; Chen, C.; Pscherer, S.; Rosemann, A.; Ranft, A.; Dirksen, U.; Juergens, H.; Rossig, C. A high proportion of bone marrow T cells with regulatory phenotype (CD4+CD25hiFoxP3+) in Ewing sarcoma patients is associated with metastatic disease. Int. J. Cancer 2009, 125, 879–886. [Google Scholar] [CrossRef]
- Ohali, A.; Avigad, S.; Zaizov, R.; Ophir, R.; Horn-Saban, S.; Cohen, I.J.; Meller, I.; Kollender, Y.; Issakov, J.; Yaniv, I. Prediction of high risk Ewing’s sarcoma by gene expression profiling. Oncogene 2004, 23, 8997–9006. [Google Scholar] [CrossRef] [PubMed]
- Zambelli, D.; Zuntini, M.; Nardi, F.; Manara, M.C.; Serra, M.; Landuzzi, L.; Lollini, P.L.; Ferrari, S.; Alberghini, M.; Llombart-Bosch, A.; et al. Biological indicators of prognosis in Ewing’s sarcoma: An emerging role for lectin galactoside-binding soluble 3 binding protein (LGALS3BP). Int. J. Cancer 2010, 126, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Iacobelli, S.; Arnò, E.; D’Orazio, A.; Coletti, G. Detection of antigens recognized by a novel monoclonal antibody in tissue and serum from patients with breast cancer. Cancer Res. 1986, 46, 3005–3010. [Google Scholar] [PubMed]
- Koths, K.; Taylor, E.; Halenbeck, R.; Casipit, C.; Wang, A. Cloning and characterization of a human Mac-2-binding protein, a new member of the superfamily defined by the macrophage scavenger receptor cysteine-rich domain. J. Biol. Chem. 1993, 268, 14245–14249. [Google Scholar] [CrossRef]
- Iacobelli, S.; Sismondi, P.; Giai, M.; D’Egidio, M.; Tinari, N.; Amatetti, C.; Di Stefano, P.; Natoli, C. Prognostic value of a novel circulating serum 90K antigen in breast cancer. Br. J. Cancer 1994, 69, 172–176. [Google Scholar] [CrossRef]
- Fornarini, B.; D’Ambrosio, C.; Natoli, C.; Tinari, N.; Silingardi, V.; Iacobelli, S. Adhesion to 90K (Mac-2 BP) as a mechanism for lymphoma drug resistance in vivo. Blood 2000, 96, 3282–3285. [Google Scholar] [CrossRef]
- Iacovazzi, P.A.; Guerra, V.; Elba, S.; Sportelli, F.; Manghisi, O.G.; Correale, M. Are 90K/MAC-2BP serum levels correlated with poor prognosis in HCC patients? Preliminary results. Int. J. Biol. Markers 2003, 18, 222–226. [Google Scholar] [CrossRef]
- Strizzi, L.; Muraro, R.; Vianale, G.; Natoli, C.; Talone, L.; Catalano, A.; Mutti, L.; Tassi, G.; Procopio, A. Expression of glycoprotein 90K in human malignant pleural mesothelioma: Correlation with patient survival. J. Pathol. 2002, 197, 218–223. [Google Scholar] [CrossRef]
- Gentiloni, N.; Caradonna, P.; Costamagna, G.; D’Ostilio, N.; Perri, V.; Mutignani, M.; Febbraro, S.; Tinari, N.; Iacobelli, S.; Natoli, C. Pancreatic juice 90K and serum CA 19-9 combined determination can discriminate between pancreatic cancer and chronic pancreatitis. Am. J. Gastroenterol. 1995, 90, 1069–1072. [Google Scholar]
- Marchetti, A.; Tinari, N.; Buttitta, F.; Chella, A.; Angeletti, C.A.; Sacco, R.; Mucilli, F.; Ullrich, A.; Iacobelli, S. Expression of 90K (Mac-2 BP) correlates with distant metastasis and predicts survival in stage I non-small cell lung cancer patients. Cancer Res. 2002, 62, 2535–2539. [Google Scholar]
- Volchenboum, S.L.; Andrade, J.; Huang, L.; Barkauskas, D.A.; Krailo, M.; Womer, R.B.; Ranft, A.; Potratz, J.; Dirksen, U.; Triche, T.J.; et al. Gene Expression Profiling of Ewing Sarcoma Tumors Reveals the Prognostic Importance of Tumor-Stromal Interactions: A Report from the Children’s Oncology Group. J. Pathol. Clin. Res. 2015, 1, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H.; Nusse, R. Wnt/β-catenin signaling and disease. Cell 2012, 149, 1192–1205. [Google Scholar] [CrossRef] [PubMed]
- Nusse, R.; Clevers, H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017, 169, 985–999. [Google Scholar] [CrossRef] [PubMed]
- Scannell, C.A.; Pedersen, E.A.; Mosher, J.T.; Krook, M.A.; Nicholls, L.A.; Wilky, B.A.; Loeb, D.M.; Lawlor, E.R. LGR5 is Expressed by Ewing Sarcoma and Potentiates Wnt/β-Catenin Signaling. Front. Oncol. 2013, 3, 81. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Yamazaki, Y.; Kanno, Y.; Igarashi, K.; Aisaki, K.; Kanno, J.; Nakamura, T. Ewing’s sarcoma precursors are highly enriched in embryonic osteochondrogenic progenitors. J. Clin. Investig. 2014, 124, 3061–3074. [Google Scholar] [CrossRef]
- Yamamoto, H.; Oue, N.; Sato, A.; Hasegawa, Y.; Yamamoto, H.; Matsubara, A.; Yasui, W.; Kikuchi, A. Wnt5a signaling is involved in the aggressiveness of prostate cancer and expression of metalloproteinase. Oncogene 2010, 29, 2036–2046. [Google Scholar] [CrossRef]
- Lemmon, M.A.; Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 2010, 141, 1117–1134. [Google Scholar] [CrossRef]
- Du, Z.; Lovly, C.M. Mechanisms of receptor tyrosine kinase activation in cancer. Mol. Cancer 2018, 17, 58. [Google Scholar] [CrossRef]
- Steinestel, K.; Trautmann, M.; Jansen, E.P.; Dirksen, U.; Rehkämper, J.; Mikesch, J.H.; Gerke, J.S.; Orth, M.F.; Sannino, G.; Arteaga, M.F.; et al. Focal adhesion kinase confers pro-migratory and antiapoptotic properties and is a potential therapeutic target in Ewing sarcoma. Mol. Oncol. 2020, 14, 248–260. [Google Scholar] [CrossRef]
- Johnson, R.; Halder, G. The two faces of Hippo: Targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat. Rev. Drug Discov. 2014, 13, 63–79. [Google Scholar] [CrossRef]
- Chan, S.W.; Lim, C.J.; Guo, K.; Ng, C.P.; Lee, I.; Hunziker, W.; Zeng, Q.; Hong, W. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res. 2008, 68, 2592–2598. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Teng, L. YAP/TAZ for cancer therapy: Opportunities and challenges (review). Int. J. Oncol. 2015, 46, 1444–1452. [Google Scholar] [CrossRef]
- Sechler, M.; Parrish, J.K.; Birks, D.K.; Jedlicka, P. The histone demethylase KDM3A, and its downstream target MCAM, promote Ewing Sarcoma cell migration and metastasis. Oncogene 2017, 36, 4150–4160. [Google Scholar] [CrossRef] [PubMed]
- Richter, G.H.; Plehm, S.; Fasan, A.; Rössler, S.; Unland, R.; Bennani-Baiti, I.M.; Hotfilder, M.; Löwel, D.; von Luettichau, I.; Mossbrugger, I.; et al. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc. Natl. Acad. Sci. USA 2009, 106, 5324–5329. [Google Scholar] [CrossRef] [PubMed]
- Wiles, E.T.; Bell, R.; Thomas, D.; Beckerle, M.; Lessnick, S.L. ZEB2 Represses the Epithelial Phenotype and Facilitates Metastasis in Ewing Sarcoma. Genes Cancer 2013, 4, 486–500. [Google Scholar] [CrossRef]
- Choo, S.; Wang, P.; Newbury, R.; Roberts, W.; Yang, J. Reactivation of TWIST1 contributes to Ewing sarcoma metastasis. Pediatr. Blood Cancer 2018, 65. [Google Scholar] [CrossRef]
- Sand, L.G.; Berghuis, D.; Szuhai, K.; Hogendoorn, P.C. Expression of CCL21 in Ewing sarcoma shows an inverse correlation with metastases and is a candidate target for immunotherapy. Cancer Immunol. Immunother. CII 2016, 65, 995–1002. [Google Scholar] [CrossRef]
- Meynet, O.; Scotlandi, K.; Pradelli, E.; Manara, M.C.; Colombo, M.P.; Schmid-Antomarchi, H.; Picci, P.; Bernard, A.; Bernard, G. Xg expression in Ewing’s sarcoma is of prognostic value and contributes to tumor invasiveness. Cancer Res. 2010, 70, 3730–3738. [Google Scholar] [CrossRef]
- Rocchi, A.; Manara, M.C.; Sciandra, M.; Zambelli, D.; Nardi, F.; Nicoletti, G.; Garofalo, C.; Meschini, S.; Astolfi, A.; Colombo, M.P.; et al. CD99 inhibits neural differentiation of human Ewing sarcoma cells and thereby contributes to oncogenesis. J. Clin. Investig. 2010, 120, 668–680. [Google Scholar] [CrossRef]
- Lo, J.C.; Chin, R.K.; Lee, Y.; Kang, H.S.; Wang, Y.; Weinstock, J.V.; Banks, T.; Ware, C.F.; Franzoso, G.; Fu, Y.X. Differential regulation of CCL21 in lymphoid/nonlymphoid tissues for effectively attracting T cells to peripheral tissues. J. Clin. Investig. 2003, 112, 1495–1505. [Google Scholar] [CrossRef]
- Sharma, S.; Yang, S.C.; Hillinger, S.; Zhu, L.X.; Huang, M.; Batra, R.K.; Lin, J.F.; Burdick, M.D.; Strieter, R.M.; Dubinett, S.M. SLC/CCL21-mediated anti-tumor responses require IFNgamma, MIG/CXCL9 and IP-10/CXCL10. Mol. Cancer 2003, 2, 22. [Google Scholar] [CrossRef] [PubMed]
- Meacham, C.E.; Morrison, S.J. Tumour heterogeneity and cancer cell plasticity. Nature 2013, 501, 328–337. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Chen, C. Regulation of Metastasis in Ewing Sarcoma. Cancers 2022, 14, 4902. https://doi.org/10.3390/cancers14194902
Li M, Chen C. Regulation of Metastasis in Ewing Sarcoma. Cancers. 2022; 14(19):4902. https://doi.org/10.3390/cancers14194902
Chicago/Turabian StyleLi, Mingli, and Chunwei Chen. 2022. "Regulation of Metastasis in Ewing Sarcoma" Cancers 14, no. 19: 4902. https://doi.org/10.3390/cancers14194902
APA StyleLi, M., & Chen, C. (2022). Regulation of Metastasis in Ewing Sarcoma. Cancers, 14(19), 4902. https://doi.org/10.3390/cancers14194902







