Liquid Biopsy-Derived DNA Sources as Tools for Comprehensive Mutation Profiling in Multiple Myeloma: A Comparative Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
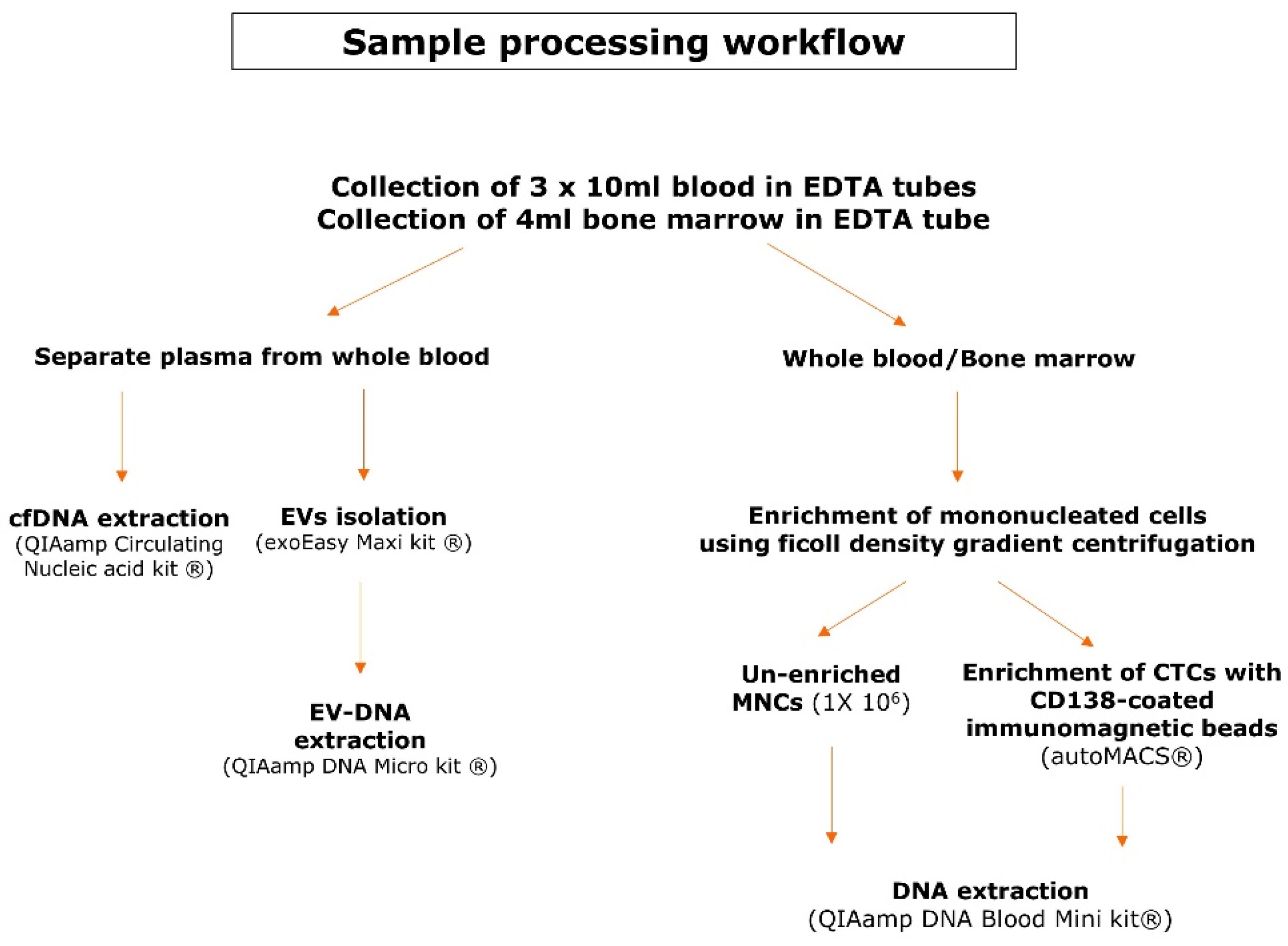
2.1. Patient Samples Collection and Pre-Analytical Sample Processing
2.2. Culturing of Human Myeloma Cell Lines (HMCLs)
2.3. Library Preparation and Targeted Gene Sequencing
2.4. Sequencing Data Processing, Variant Filtering and Interpretation
2.5. Monoclonal Immunoglobulin Detection with Next-Generation Sequencing
2.6. Statistics
3. Results
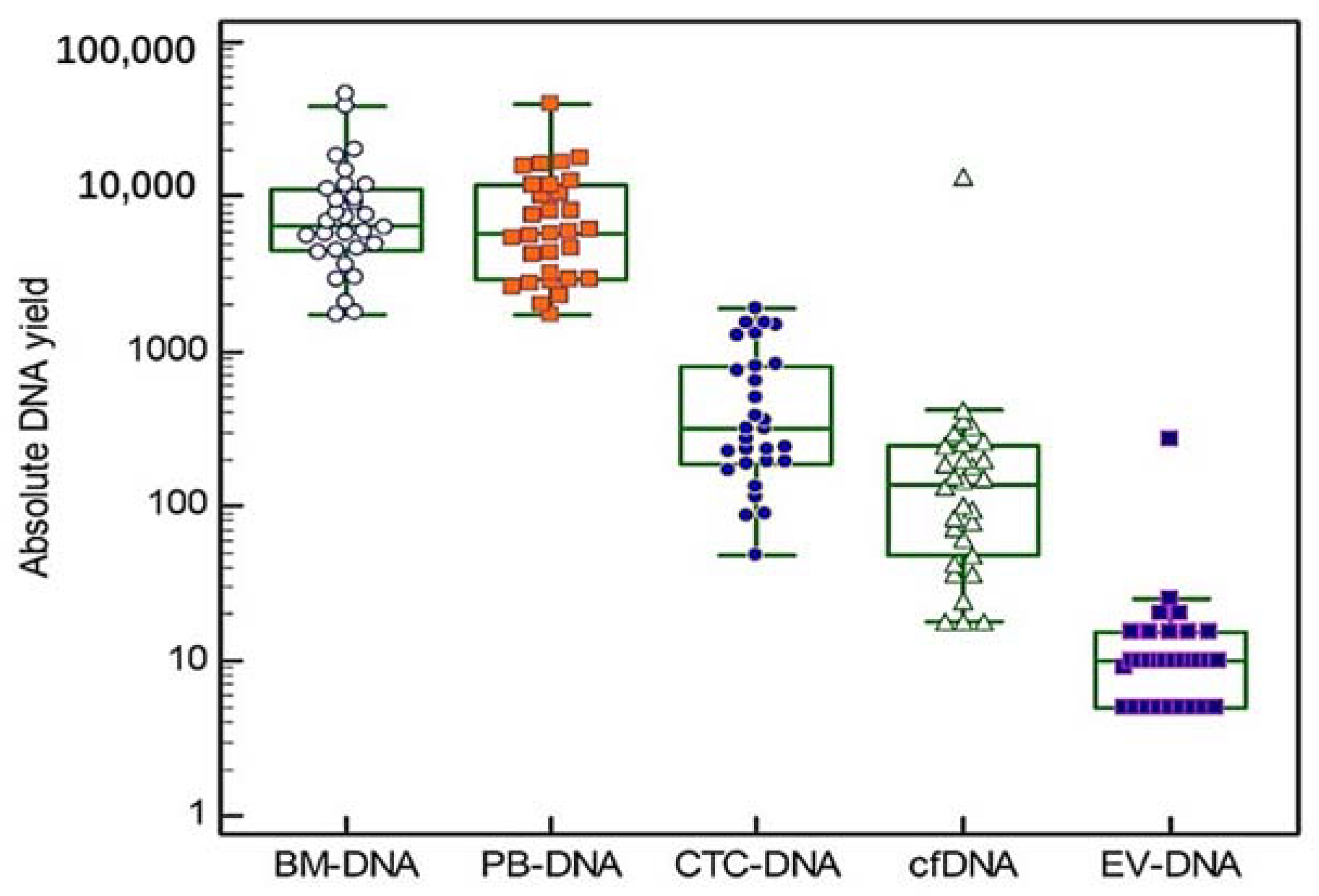
3.1. Patient Characteristics and Obtained DNA Yields
3.2. Detection of Previously Described Genetic Variants in HMCLs
3.3. Genetic Pathway Involvement and Biological Significance of Detected Variants in MM Patients
3.4. Differential Mutational Burden across MM Disease Stages and Correlation with Clinical Outcome
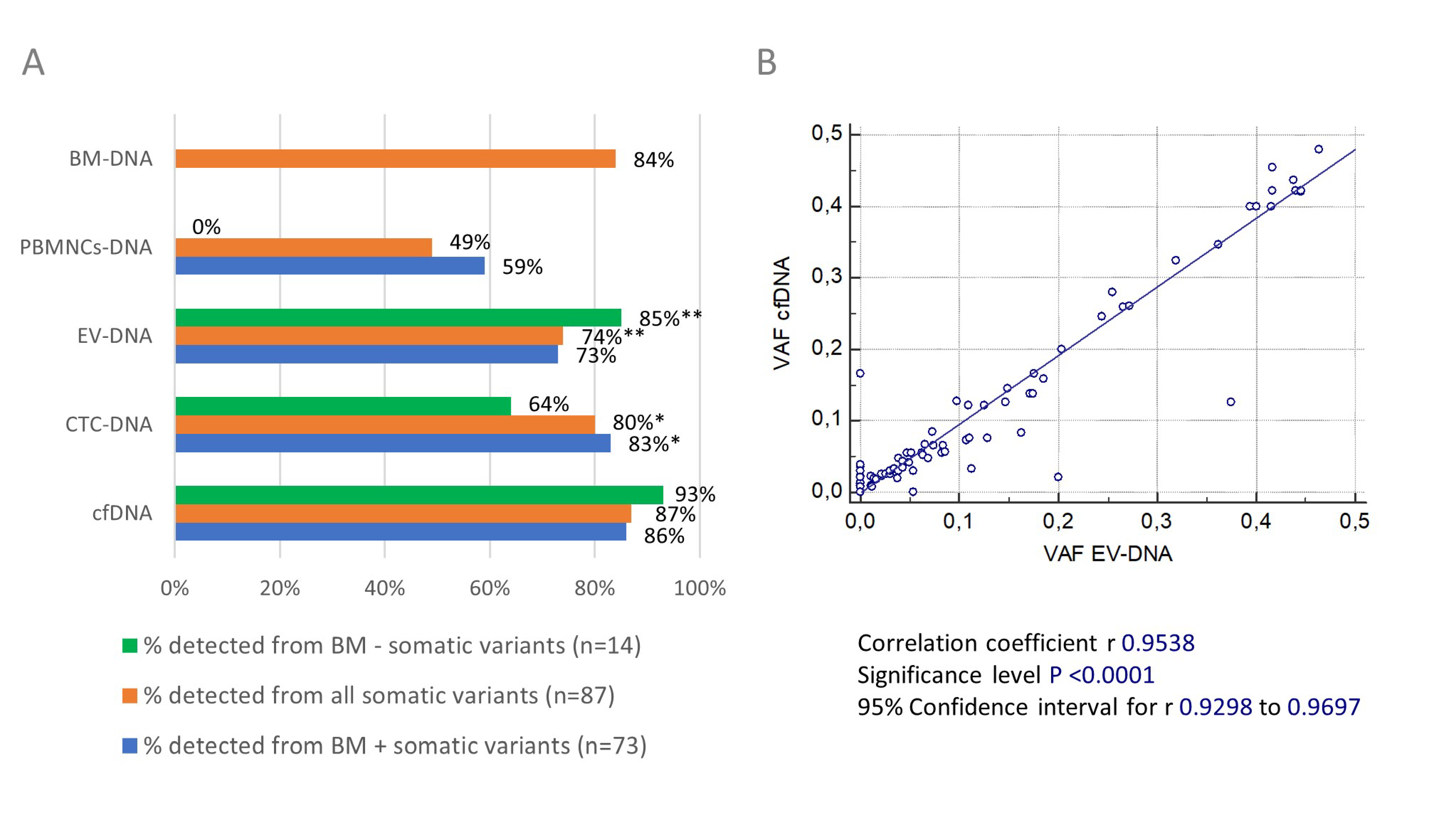
3.5. Detectability of Genetic Variants by Circulating Biomarkers and Concordance with Bone Marrow DNA
3.6. Discordant Results between Blood and Bone Marrow Compartment and Correlation with Monoclonal Immunoglobulin Percentages and Plasmacytosis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rollig, C.; Knop, S.; Bornhauser, M. Multiple myeloma. Lancet 2015, 385, 2197–2208. [Google Scholar] [CrossRef]
- Rasche, L.; Chavan, S.S.; Stephens, O.W.; Patel, P.H.; Tytarenko, R.; Ashby, C.; Bauer, M.; Stein, C.; Deshpande, S.; Wardell, C.; et al. Spatial genomic heterogeneity in multiple myeloma revealed by multi-region sequencing. Nat. Commun. 2017, 8, 268. [Google Scholar] [CrossRef] [PubMed]
- Manier, S.; Park, J.; Capelletti, M.; Bustoros, M.; Freeman, S.S.; Ha, G.; Rhoades, J.; Liu, C.J.; Huynh, D.; Reed, S.C.; et al. Whole-exome sequencing of cell-free DNA and circulating tumor cells in multiple myeloma. Nat. Commun. 2018, 9, 1691. [Google Scholar] [CrossRef] [PubMed]
- Bolli, N.; Avet-Loiseau, H.; Wedge, D.C.; Van Loo, P.; Alexandrov, L.B.; Martincorena, I.; Dawson, K.J.; Iorio, F.; Nik-Zainal, S.; Bignell, G.R.; et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat. Commun. 2014, 5, 2997. [Google Scholar] [CrossRef] [PubMed]
- Lohr, J.G.; Stojanov, P.; Carter, S.L.; Cruz-Gordillo, P.; Lawrence, M.S.; Auclair, D.; Sougnez, C.; Knoechel, B.; Gould, J.; Saksena, G.; et al. Widespread genetic heterogeneity in multiple myeloma: Implications for targeted therapy. Cancer Cell 2014, 25, 91–101. [Google Scholar] [CrossRef]
- John, L.; Poos, A.; Tirier, S.M.; Mallm, J.-P.; Prokoph, N.; Brobeil, A.; Lutz, R.; Schumacher, S.; Steiger, S.; Bauer, K.; et al. The Spatial Heterogeneity in Newly Diagnosed Multiple Myeloma Patients—From Sub-Clonal Architecture to the Immune Microenvironment. Blood 2021, 138, 729. [Google Scholar] [CrossRef]
- Mikulasova, A.; Wardell, C.P.; Murison, A.; Boyle, E.M.; Jackson, G.H.; Smetana, J.; Kufova, Z.; Pour, L.; Sandecka, V.; Almasi, M.; et al. The spectrum of somatic mutations in monoclonal gammopathy of undetermined significance indicates a less complex genomic landscape than that in multiple myeloma. Haematologica 2017, 102, 1617–1625. [Google Scholar] [CrossRef]
- Rossi, A.; Voigtlaender, M.; Janjetovic, S.; Thiele, B.; Alawi, M.; Marz, M.; Brandt, A.; Hansen, T.; Radloff, J.; Schon, G.; et al. Mutational landscape reflects the biological continuum of plasma cell dyscrasias. Blood Cancer J. 2017, 7, e537. [Google Scholar] [CrossRef]
- Lionetti, M.; Barbieri, M.; Manzoni, M.; Fabris, S.; Bandini, C.; Todoerti, K.; Nozza, F.; Rossi, D.; Musto, P.; Baldini, L.; et al. Molecular spectrum of TP53 mutations in plasma cell dyscrasias by next generation sequencing: An Italian cohort study and overview of the literature. Oncotarget 2016, 7, 21353–21361. [Google Scholar] [CrossRef]
- Mithraprabhu, S.; Khong, T.; Ramachandran, M.; Chow, A.; Klarica, D.; Mai, L.; Walsh, S.; Broemeling, D.; Marziali, A.; Wiggin, M.; et al. Circulating tumour DNA analysis demonstrates spatial mutational heterogeneity that coincides with disease relapse in myeloma. Leukemia 2017, 31, 1695–1705. [Google Scholar] [CrossRef]
- Weinhold, N.; Ashby, C.; Rasche, L.; Chavan, S.S.; Stein, C.; Stephens, O.W.; Tytarenko, R.; Bauer, M.A.; Meissner, T.; Deshpande, S.; et al. Clonal selection and double-hit events involving tumor suppressor genes underlie relapse in myeloma. Blood 2016, 128, 1735–1744. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.R.; Weinhold, N.; Ashby, C.; Walker, B.A.; Wardell, C.; Pawlyn, C.; Rasche, L.; Melchor, L.; Cairns, D.A.; Gregory, W.M.; et al. Clonal evolution in myeloma: The impact of maintenance lenalidomide and depth of response on the genetics and sub-clonal structure of relapsed disease in uniformly treated newly diagnosed patients. Haematologica 2019, 104, 1440. [Google Scholar] [CrossRef]
- Kortum, K.M.; Langer, C.; Monge, J.; Bruins, L.; Zhu, Y.X.; Shi, C.X.; Jedlowski, P.; Egan, J.B.; Ojha, J.; Bullinger, L.; et al. Longitudinal analysis of 25 sequential sample-pairs using a custom multiple myeloma mutation sequencing panel (M(3)P). Ann. Hematol 2015, 94, 1205–1211. [Google Scholar] [CrossRef]
- Kis, O.; Kaedbey, R.; Chow, S.; Danesh, A.; Dowar, M.; Li, T.; Li, Z.; Liu, J.; Mansour, M.; Masih-Khan, E.; et al. Circulating tumour DNA sequence analysis as an alternative to multiple myeloma bone marrow aspirates. Nat. Commun. 2017, 8, 15086. [Google Scholar] [CrossRef] [PubMed]
- Guo, G.; Raje, N.S.; Seifer, C.; Kloeber, J.; Isenhart, R.; Ha, G.; Yee, A.J.; O’Donnell, E.K.; Tai, Y.T.; Richardson, P.G.; et al. Genomic discovery and clonal tracking in multiple myeloma by cell-free DNA sequencing. Leukemia 2018, 32, 1838–1841. [Google Scholar] [CrossRef]
- Gerber, B.; Manzoni, M.; Spina, V.; Bruscaggin, A.; Lionetti, M.; Fabris, S.; Barbieri, M.; Ciceri, G.; Pompa, A.; Forestieri, G.; et al. Circulating tumor DNA as a liquid biopsy in plasma cell dyscrasias. Haematologica 2018, 103, e245–e248. [Google Scholar] [CrossRef] [PubMed]
- Lohr, J.G.; Kim, S.; Gould, J.; Knoechel, B.; Drier, Y.; Cotton, M.J.; Gray, D.; Birrer, N.; Wong, B.; Ha, G.; et al. Genetic interrogation of circulating multiple myeloma cells at single-cell resolution. Sci. Transl. Med. 2016, 8, 363ra147. [Google Scholar] [CrossRef] [PubMed]
- Mishima, Y.; Paiva, B.; Shi, J.; Park, J.; Manier, S.; Takagi, S.; Massoud, M.; Perilla-Glen, A.; Aljawai, Y.; Huynh, D.; et al. The Mutational Landscape of Circulating Tumor Cells in Multiple Myeloma. Cell Rep. 2017, 19, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Garcés, J.J.; Bretones, G.; Burgos, L.; Valdes-Mas, R.; Puig, N.; Cedena, M.T.; Alignani, D.; Rodriguez, I.; Puente, D.; Álvarez, M.G.; et al. Circulating tumor cells for comprehensive and multiregional non-invasive genetic characterization of multiple myeloma. Leukemia 2020, 34, 3007–3018. [Google Scholar] [CrossRef]
- Biancon, G.; Gimondi, S.; Vendramin, A.; Carniti, C.; Corradini, P. Noninvasive Molecular Monitoring in Multiple Myeloma Patients Using Cell-Free Tumor DNA: A Pilot Study. J. Mol. Diagn. 2018, 20, 859–870. [Google Scholar] [CrossRef]
- Mithraprabhu, S.; Hocking, J.; Ramachandran, M.; Choi, K.; Klarica, D.; Khong, T.; Reynolds, J.; Spencer, A. DNA-Repair Gene Mutations Are Highly Prevalent in Circulating Tumour DNA from Multiple Myeloma Patients. Cancers 2019, 11, 917. [Google Scholar] [CrossRef] [PubMed]
- Ciardiello, C.; Cavallini, L.; Spinelli, C.; Yang, J.; Reis-Sobreiro, M.; de Candia, P.; Minciacchi, V.R.; Di Vizio, D. Focus on Extracellular Vesicles: New Frontiers of Cell-to-Cell Communication in Cancer. Int. J. Mol. Sci. 2016, 17, 175. [Google Scholar] [CrossRef] [PubMed]
- Roma-Rodrigues, C.; Fernandes, A.R.; Baptista, P.V. Exosome in tumour microenvironment: Overview of the crosstalk between normal and cancer cells. Biomed. Res. Int. 2014, 2014, 179486. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R. The biology and function of exosomes in cancer. J. Clin. Investig. 2016, 126, 1208–1215. [Google Scholar] [CrossRef] [PubMed]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef] [PubMed]
- San Lucas, F.A.; Allenson, K.; Bernard, V.; Castillo, J.; Kim, D.U.; Ellis, K.; Ehli, E.A.; Davies, G.E.; Petersen, J.L.; Li, D.; et al. Minimally invasive genomic and transcriptomic profiling of visceral cancers by next-generation sequencing of circulating exosomes. Ann. Oncol. 2016, 27, 635–641. [Google Scholar] [CrossRef]
- Kahlert, C.; Melo, S.A.; Protopopov, A.; Tang, J.; Seth, S.; Koch, M.; Zhang, J.; Weitz, J.; Chin, L.; Futreal, A.; et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J. Biol. Chem. 2014, 289, 3869–3875. [Google Scholar] [CrossRef] [PubMed]
- Kahlert, C. Liquid Biopsy: Is There an Advantage to Analyzing Circulating Exosomal DNA Compared to cfDNA or Are They the Same? Cancer Res. 2019, 79, 2462–2465. [Google Scholar] [CrossRef] [PubMed]
- Vagner, T.; Spinelli, C.; Minciacchi, V.R.; Balaj, L.; Zandian, M.; Conley, A.; Zijlstra, A.; Freeman, M.R.; Demichelis, F.; De, S.; et al. Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J. Extracell. Vesicles 2018, 7, 1505403. [Google Scholar] [CrossRef]
- Mouliere, F.; El Messaoudi, S.; Gongora, C.; Guedj, A.S.; Robert, B.; Del Rio, M.; Molina, F.; Lamy, P.J.; Lopez-Crapez, E.; Mathonnet, M.; et al. Circulating Cell-Free DNA from Colorectal Cancer Patients May Reveal High KRAS or BRAF Mutation Load. Transl. Oncol. 2013, 6, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Parpart-Li, S.; Bartlett, B.; Popoli, M.; Adleff, V.; Tucker, L.; Steinberg, R.; Georgiadis, A.; Phallen, J.; Brahmer, J.; Azad, N.; et al. The Effect of Preservative and Temperature on the Analysis of Circulating Tumor DNA. Clin. Cancer Res. 2017, 23, 2471–2477. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Che, S.P.; Kurywchak, P.; Tavormina, J.L.; Gansmo, L.B.; Correa de Sampaio, P.; Tachezy, M.; Bockhorn, M.; Gebauer, F.; Haltom, A.R.; et al. Detection of mutant KRAS and TP53 DNA in circulating exosomes from healthy individuals and patients with pancreatic cancer. Cancer Biol. 2017, 18, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Allenson, K.; Castillo, J.; San Lucas, F.A.; Scelo, G.; Kim, D.U.; Bernard, V.; Davis, G.; Kumar, T.; Katz, M.; Overman, M.J.; et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann. Oncol. 2017, 28, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Bernard, V.; Kim, D.U.; San Lucas, F.A.; Castillo, J.; Allenson, K.; Mulu, F.C.; Stephens, B.M.; Huang, J.; Semaan, A.; Guerrero, P.A.; et al. Circulating Nucleic Acids Are Associated With Outcomes of Patients With Pancreatic Cancer. Gastroenterology 2019, 156, 108–118. [Google Scholar] [CrossRef]
- Hur, J.Y.; Kim, H.J.; Lee, J.S.; Choi, C.M.; Lee, J.C.; Jung, M.K.; Pack, C.G.; Lee, K.Y. Extracellular vesicle-derived DNA for performing EGFR genotyping of NSCLC patients. Mol. Cancer 2018, 17, 15. [Google Scholar] [CrossRef]
- Wan, Y.; Liu, B.; Lei, H.; Zhang, B.; Wang, Y.; Huang, H.; Chen, S.; Feng, Y.; Zhu, L.; Gu, Y.; et al. Nanoscale extracellular vesicle-derived DNA is superior to circulating cell-free DNA for mutation detection in early-stage non-small-cell lung cancer. Ann. Oncol. 2018, 29, 2379–2383. [Google Scholar] [CrossRef]
- Kontopoulou, E.; Strachan, S.; Reinhardt, K.; Kunz, F.; Walter, C.; Walkenfort, B.; Jastrow, H.; Hasenberg, M.; Giebel, B.; von Neuhoff, N.; et al. Evaluation of dsDNA from extracellular vesicles (EVs) in pediatric AML diagnostics. Ann. Hematol. 2020, 99, 459–475. [Google Scholar] [CrossRef]
- Wang, X.; Wilkinson, R.; Kildey, K.; Potriquet, J.; Mulvenna, J.; Lobb, R.J.; Möller, A.; Cloonan, N.; Mukhopadhyay, P.; Kassianos, A.J.; et al. Unique molecular profile of exosomes derived from primary human proximal tubular epithelial cells under diseased conditions. J. Extracell. Vesicles 2017, 6, 1314073. [Google Scholar] [CrossRef]
- Leich, E.; Weißbach, S.; Klein, H.U.; Grieb, T.; Pischimarov, J.; Stühmer, T.; Chatterjee, M.; Steinbrunn, T.; Langer, C.; Eilers, M.; et al. Multiple myeloma is affected by multiple and heterogeneous somatic mutations in adhesion- and receptor tyrosine kinase signaling molecules. Blood Cancer J. 2013, 3, e102. [Google Scholar] [CrossRef]
- Zhu, Y.X.; Shi, C.X.; Bruins, L.A.; Jedlowski, P.; Wang, X.; Kortum, K.M.; Luo, M.; Ahmann, J.M.; Braggio, E.; Stewart, A.K. Loss of FAM46C Promotes Cell Survival in Myeloma. Cancer Res. 2017, 77, 4317–4327. [Google Scholar] [CrossRef]
- Lionetti, M.; Barbieri, M.; Todoerti, K.; Agnelli, L.; Marzorati, S.; Fabris, S.; Ciceri, G.; Galletti, S.; Milesi, G.; Manzoni, M.; et al. Molecular spectrum of BRAF, NRAS and KRAS gene mutations in plasma cell dyscrasias: Implication for MEK-ERK pathway activation. Oncotarget 2015, 6, 24205–24217. [Google Scholar] [CrossRef] [PubMed]
- Moreaux, J.; Klein, B.; Bataille, R.; Descamps, G.; Maïga, S.; Hose, D.; Goldschmidt, H.; Jauch, A.; Rème, T.; Jourdan, M.; et al. A high-risk signature for patients with multiple myeloma established from the molecular classification of human myeloma cell lines. Haematologica 2011, 96, 574–582. [Google Scholar] [CrossRef]
- Froyen, G.; Le Mercier, M.; Lierman, E.; Vandepoele, K.; Nollet, F.; Boone, E.; Van der Meulen, J.; Jacobs, K.; Lambin, S.; Vander Borght, S.; et al. Standardization of Somatic Variant Classifications in Solid and Haematological Tumours by a Two-Level Approach of Biological and Clinical Classes: An Initiative of the Belgian ComPerMed Expert Panel. Cancers 2019, 11, 2030. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Wenger, A.M.; Zehir, A.; Mesirov, J.P. Variant Review with the Integrative Genomics Viewer. Cancer Res. 2017, 77, e31–e34. [Google Scholar] [CrossRef] [PubMed]
- Van der Straeten, J.; De Brouwer, W.; Kanjinga, E.K.; Dresse, M.F.; Fostier, K.; Schots, R.; Van Riet, I.; Bakkus, M. Validation of a PCR-based next-generation sequencing approach for the detection and quantification of minimal residual disease in acute lymphoblastic leukemia and multiple myeloma using gBlocks as calibrators. J. Mol. Diagn. 2021, 23, 599–611. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Sinkala, M.; Nkhoma, P.; Mulder, N.; Martin, D.P. Integrated molecular characterisation of the MAPK pathways in human cancers reveals pharmacologically vulnerable mutations and gene dependencies. Commun. Biol. 2021, 4, 9. [Google Scholar] [CrossRef] [PubMed]
- Wood, R.D.; Mitchell, M.; Lindahl, T. Human DNA repair genes, 2005. Mutat. Res. 2005, 577, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Coffey, D.G.; Wu, Q.V.; Towlerton, A.M.H.; Ornelas, S.; Morales, A.J.; Xu, Y.; Green, D.J.; Warren, E.H. Ultradeep, targeted sequencing reveals distinct mutations in blood compared to matched bone marrow among patients with multiple myeloma. Blood Cancer J. 2019, 9, 77. [Google Scholar] [CrossRef] [PubMed]
- Möhrmann, L.; Huang, H.J.; Hong, D.S.; Tsimberidou, A.M.; Fu, S.; Piha-Paul, S.A.; Subbiah, V.; Karp, D.D.; Naing, A.; Krug, A.; et al. Liquid Biopsies Using Plasma Exosomal Nucleic Acids and Plasma Cell-Free DNA Compared with Clinical Outcomes of Patients with Advanced Cancers. Clin. Cancer Res. 2018, 24, 181–188. [Google Scholar] [CrossRef]
- Kortum, K.M.; Langer, C.; Monge, J.; Bruins, L.; Egan, J.B.; Zhu, Y.X.; Shi, C.X.; Jedlowski, P.; Schmidt, J.; Ojha, J.; et al. Targeted sequencing using a 47 gene multiple myeloma mutation panel (M(3) P) in -17p high risk disease. Br. J. Haematol. 2015, 168, 507–510. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.A.; Wardell, C.P.; Melchor, L.; Hulkki, S.; Potter, N.E.; Johnson, D.C.; Fenwick, K.; Kozarewa, I.; Gonzalez, D.; Lord, C.J.; et al. Intraclonal heterogeneity and distinct molecular mechanisms characterize the development of t(4;14) and t(11;14) myeloma. Blood 2012, 120, 1077–1086. [Google Scholar] [CrossRef] [PubMed]
- Chapman, M.A.; Lawrence, M.S.; Keats, J.J.; Cibulskis, K.; Sougnez, C.; Schinzel, A.C.; Harview, C.L.; Brunet, J.P.; Ahmann, G.J.; Adli, M.; et al. Initial genome sequencing and analysis of multiple myeloma. Nature 2011, 471, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.A.; Mavrommatis, K.; Wardell, C.P.; Ashby, T.C.; Bauer, M.; Davies, F.E.; Rosenthal, A.; Wang, H.; Qu, P.; Hoering, A.; et al. Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood 2018, 132, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.A.; Boyle, E.M.; Wardell, C.P.; Murison, A.; Begum, D.B.; Dahir, N.M.; Proszek, P.Z.; Johnson, D.C.; Kaiser, M.F.; Melchor, L.; et al. Mutational Spectrum, Copy Number Changes, and Outcome: Results of a Sequencing Study of Patients With Newly Diagnosed Myeloma. J. Clin. Oncol. 2015, 33, 3911–3920. [Google Scholar] [CrossRef]
- Miller, A.; Asmann, Y.; Cattaneo, L.; Braggio, E.; Keats, J.; Auclair, D.; Lonial, S.; Russell, S.J.; Stewart, A.K. High somatic mutation and neoantigen burden are correlated with decreased progression-free survival in multiple myeloma. Blood Cancer J. 2017, 7, e612. [Google Scholar] [CrossRef]
- Wei, X.; Calvo-Vidal, M.N.; Chen, S.; Wu, G.; Revuelta, M.V.; Sun, J.; Zhang, J.; Walsh, M.F.; Nichols, K.E.; Joseph, V.; et al. Germline Lysine-Specific Demethylase 1 (LSD1/KDM1A) Mutations Confer Susceptibility to Multiple Myeloma. Cancer Res. 2018, 78, 2747–2759. [Google Scholar] [CrossRef]
- Pertesi, M.; Vallee, M.; Wei, X.; Revuelta, M.V.; Galia, P.; Demangel, D.; Oliver, J.; Foll, M.; Chen, S.; Perrial, E.; et al. Exome sequencing identifies germline variants in DIS3 in familial multiple myeloma. Leukemia 2019, 33, 2324–2330. [Google Scholar] [CrossRef] [PubMed]
- Campa, D.; Martino, A.; Macauda, A.; Dudzinski, M.; Suska, A.; Druzd-Sitek, A.; Raab, M.S.; Moreno, V.; Huhn, S.; Butrym, A.; et al. Genetic polymorphisms in genes of class switch recombination and multiple myeloma risk and survival: An IMMEnSE study. Leuk. Lymphoma 2019, 60, 1803–1811. [Google Scholar] [CrossRef]
- Merz, M.; Merz, A.M.A.; Wang, J.; Wei, L.; Hu, Q.; Hutson, N.; Rondeau, C.; Celotto, K.; Belal, A.; Alberico, R.; et al. Deciphering spatial genomic heterogeneity at a single cell resolution in multiple myeloma. Nat. Commun. 2022, 13, 807. [Google Scholar] [CrossRef]

| New Diagnosis (n = 10) | First Relapse (n = 10) | Second or Later Relapse (n = 10) | |
|---|---|---|---|
| Median age (range) | 72 (64–88) | 74 (68–83) | 73 (67–81) |
| Sex male (%) female (%) | |||
| 6 (60%) | 5 (50%) | 6 (60%) | |
| 4 (40%) | 5 (50%) | 4 (40%) | |
| Osteolytic bone lesions yes (%) no (%) unknown (%) | |||
6 (60%) | 5 (50%) | 8 (80%) | |
| 3 (30%) | 4 (40%) | 2 (20%) | |
| 1 (10%) | 1 (10%) | - | |
| Cytogenetics hyperdiploid (%) non-hyperdiploid (%) gain 1q (%) inconclusive (%) normal (%) | |||
| 3 (30%) | 3 (30%) | 3 (30%) | |
| 1 (10%) | 3 (30%) | 4 (40%) | |
| 2 (20%) | 3 (30%) | 4 (40%) | |
| 6 (60%) | 4 (40%) | 2 (20%) | |
| - | - | 1 (10%) |
| Cell Line | Gene | Position | gDNA Change | p. Change | VAF | Reference |
|---|---|---|---|---|---|---|
| OPM-2 | DIS3 | chr13:73355008 | g.73355008T>G | p.Tyr121Ser | 100% of 328 reads | Leich et al. [39] |
| OPM-2 | FAM46C | chr1:118166023 | g.118166023A>C | p.Glu178Ala | 100% of 135 reads | Zhu et al. [40] |
| U266 | BRAF | chr7:140453132 | g.140453132T>A | p.Lys601Asn | 65.4% of 665 reads | Lionetti et al. [41] |
| U266 | TP53 | chr17:7578449 | g.7578449C>T | p.Ala161Thr | 100% of 344 reads | Moreaux et al. [42] |
| RPMI-8226 | KRAS | chr12:25398284 | g.25398284C>G | p.Gly12Ala | 100% of 233 reads | Moreaux et al. [42] |
| Patient | Gene | cDNA Change | p. Change | Classification | % PCs BM | % Monoclonal Ig seq BM |
|---|---|---|---|---|---|---|
| 2019-008 | FAM46C | c.823_825del | p.Phe275del | VUS | 18% | 49.1% |
| 2019-015 | PIK3CA | c.3125_3128del | p.Gln1042Argfs*25 | VUS | 4.2% | 51.9% |
| 2019-015 | KRAS | c.183A>C | p.Gln61His | Pathogenic | ||
| 2020-024 | RB1 | c.610G>T | p.Glu204* | Likely pathogenic | 7.1% | 38.4% |
| 2020-025 | CDK12 | c.1132C>T | p.Arg378Cys | VUS | 3.8% | 59.3% |
| 2021-010 | KRAS | c.183A>C | p.Gln61His | Pathogenic | NA | NA |
| Patient | Gene | cDNA Change | p. Change | Classification | % PCs BM | % Monoclonal Ig seq BM |
|---|---|---|---|---|---|---|
| 2018-001 | RAD50 | c.3751G>A | p.Glu1251Lys | VUS | NA | 45.3% |
| 2018-005 † | NRAS | c.182A>G | p.Gln61Arg | Pathogenic | NA | 0.7% |
| 2018-005 † | FAM46C | c.416_417dup | p.Gln140Cysfs*4 | Likely pathogenic | ||
| 2018-005 † | CBL | c.2662G>A | p.Ala888Thr | VUS | ||
| 2018-011 | ATRX | c.3145A>G | p.Ile1049Val | VUS | NA | 50.5% |
| 2018-017 | MTOR | c.3286-1G>C | splice site | VUS | NA | 60.3% |
| 2019-007 † | KRAS | c.34G>T | p.Gly12Cys | Pathogenic | NA | NA |
| 2019-007 † | DIS3 | c.2339G>A | p.Arg780Lys | VUS | ||
| 2020-004 | STK11 | c.938A>G | p.His313Arg | VUS | NA | 16.0% |
| 2020-024 | NRAS | c.183A>T | p.Gln61His | Pathogenic | 7.1% | 38.4% |
| 2020-024 | NRAS | c.38G>T | p.Gly13Val | Pathogenic | ||
| 2020-024 | NRAS | c.38G>A | p.Gly13Asp | Pathogenic | ||
| 2020-024 | RB1 | c.2107-1G>A | splice site | Likely pathogenic | ||
| 2022-003 | TSC1 | c.2393C>T | p.Thr798Met | VUS | NA | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heestermans, R.; De Brouwer, W.; Maes, K.; Vande Broek, I.; Vaeyens, F.; Olsen, C.; Caljon, B.; De Becker, A.; Bakkus, M.; Schots, R.; et al. Liquid Biopsy-Derived DNA Sources as Tools for Comprehensive Mutation Profiling in Multiple Myeloma: A Comparative Study. Cancers 2022, 14, 4901. https://doi.org/10.3390/cancers14194901
Heestermans R, De Brouwer W, Maes K, Vande Broek I, Vaeyens F, Olsen C, Caljon B, De Becker A, Bakkus M, Schots R, et al. Liquid Biopsy-Derived DNA Sources as Tools for Comprehensive Mutation Profiling in Multiple Myeloma: A Comparative Study. Cancers. 2022; 14(19):4901. https://doi.org/10.3390/cancers14194901
Chicago/Turabian StyleHeestermans, Robbe, Wouter De Brouwer, Ken Maes, Isabelle Vande Broek, Freya Vaeyens, Catharina Olsen, Ben Caljon, Ann De Becker, Marleen Bakkus, Rik Schots, and et al. 2022. "Liquid Biopsy-Derived DNA Sources as Tools for Comprehensive Mutation Profiling in Multiple Myeloma: A Comparative Study" Cancers 14, no. 19: 4901. https://doi.org/10.3390/cancers14194901
APA StyleHeestermans, R., De Brouwer, W., Maes, K., Vande Broek, I., Vaeyens, F., Olsen, C., Caljon, B., De Becker, A., Bakkus, M., Schots, R., & Van Riet, I. (2022). Liquid Biopsy-Derived DNA Sources as Tools for Comprehensive Mutation Profiling in Multiple Myeloma: A Comparative Study. Cancers, 14(19), 4901. https://doi.org/10.3390/cancers14194901





