Circulating Cell-Free DNA-Based Methylation Pattern in Saliva for Early Diagnosis of Head and Neck Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. cfDNA and ctDNA Biology
Pre-Analytical and Analytical Phase Examination
3. cfDNA Methylation Biomarkers for the Early Detection of HNCs
3.1. Targeted Gene Analyses
3.2. Genome-Wide Methylation Analyses
4. ctDNA Methylation-Based Sequencing Techniques
4.1. Next-Generation Sequencing
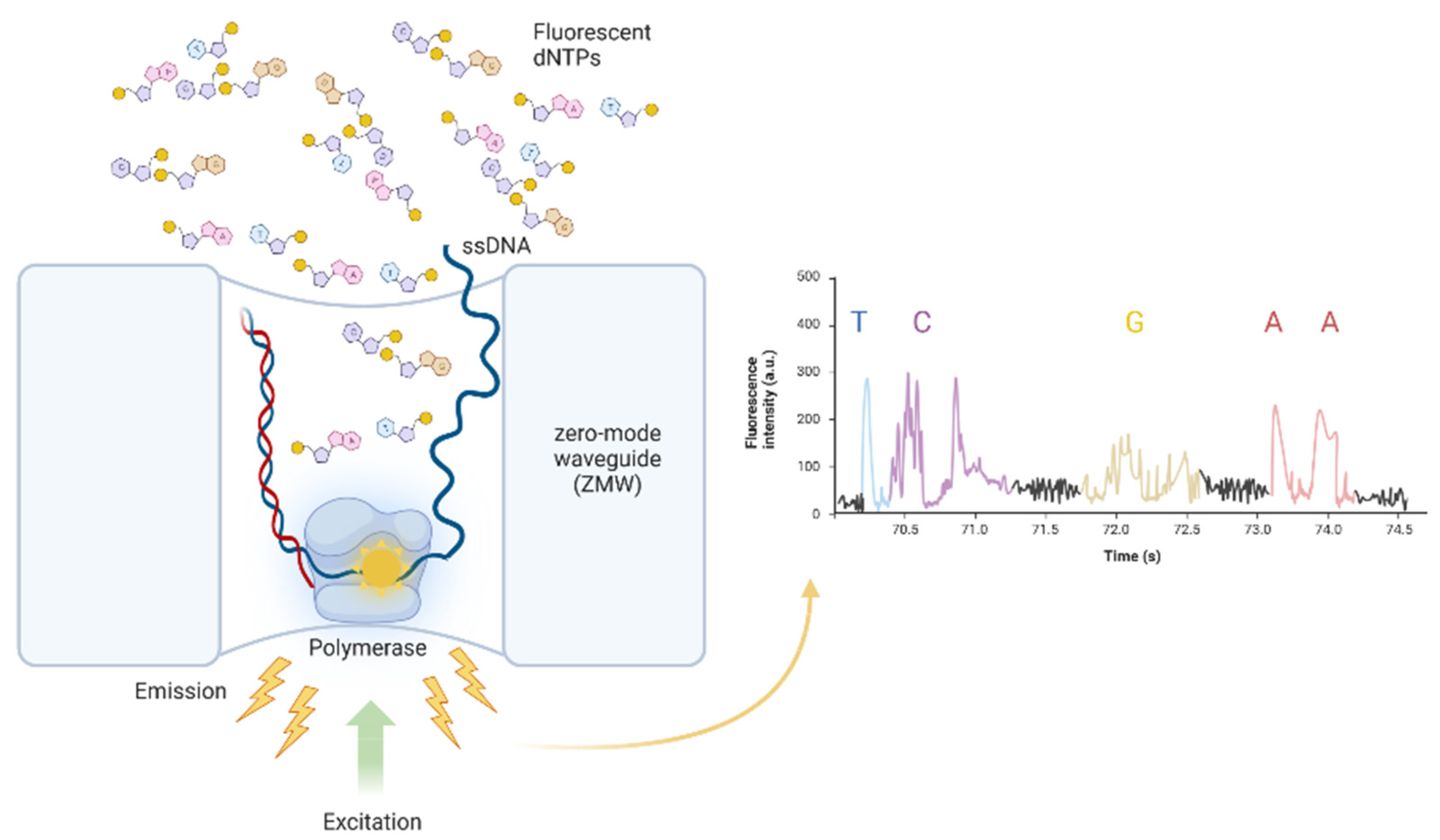
4.2. Third-Generation Sequencing: PacBio SMRT Sequencing
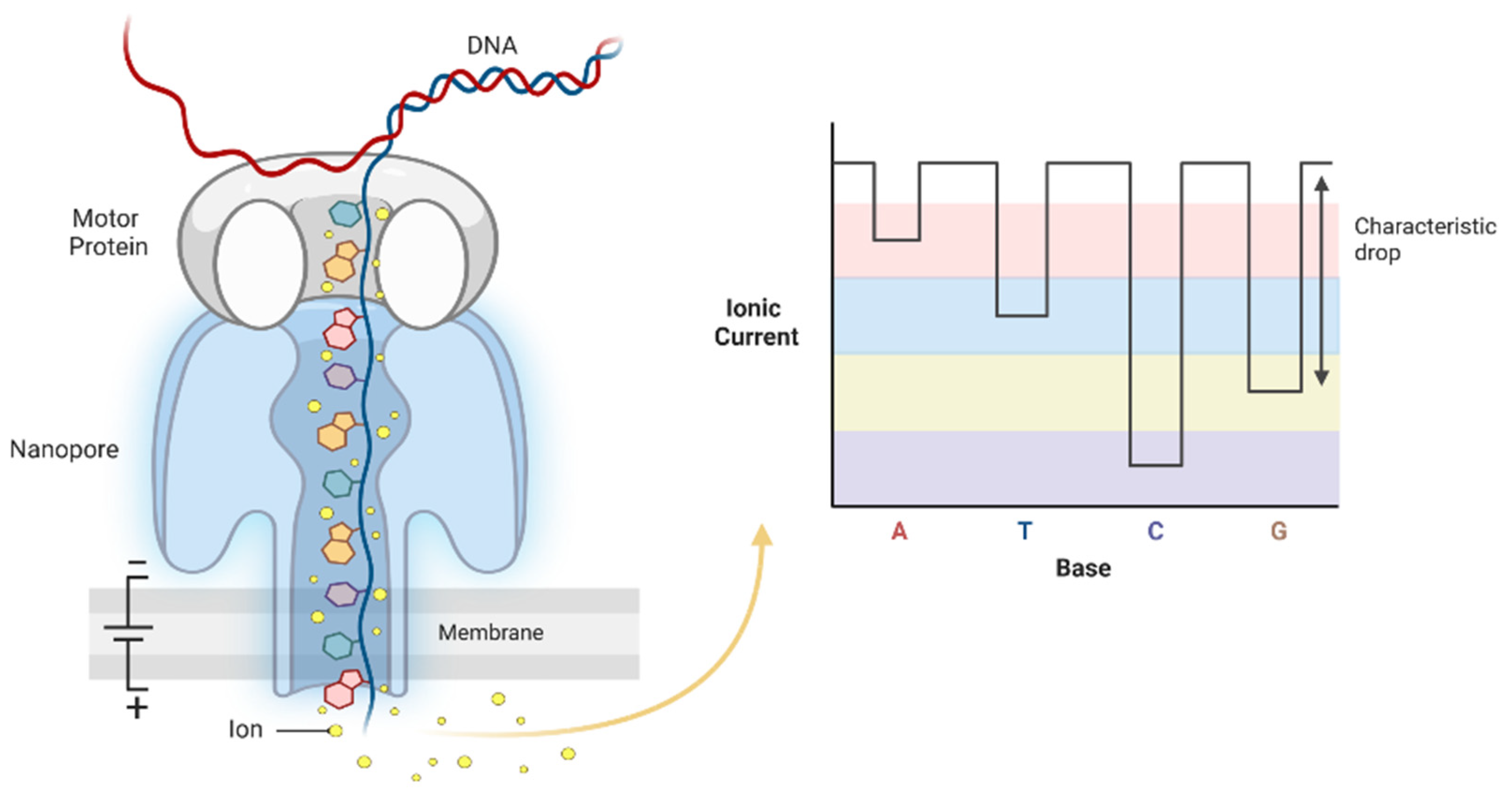
4.3. Third-Generation Sequencing: Oxford Nanopore Technology
5. Current Challenges and Future Perspectives
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- International Agency for Research on Cancer. Global Cancer Observatory. Available online: https://gco.iarc.fr/today/home (accessed on 24 April 2022).
- Guo, K.; Xiao, W.; Chen, X.; Zhao, Z.; Lin, Y.; Chen, G. Epidemiological trends of head and neck cancer: A population-based study. BioMed Res. Int. 2021, 2021, 1738932. [Google Scholar] [CrossRef] [PubMed]
- Schiff, B.A. MSD Manual for the Professional. Available on: Overview of Head and Neck Tumors—Ear, Nose, and Throat Disorders—MSD Manual Professional Edition. Available online: https://www.msdmanuals.com/professional/ear,-nose,-and-throat-disorders/tumors-of-the-head-and-neck/overview-of-head-and-neck-tumors (accessed on 13 April 2022).
- Schutte, H.W.; Heutink, F.; Wellenstein, D.J.; van den Broek, G.B.; van den Hoogen, F.J.A.; Marres, H.A.M.; van Herpen, C.M.L.; Kaanders, J.H.A.M.; Merkx, T.M.A.W.; Takes, R.P. Impact of time to diagnosis and treatment in head and neck cancer: A systematic review. Otolaryngol. Head Neck Surg. 2020, 162, 446–457. [Google Scholar] [CrossRef]
- Lo Nigro, C.; Denaro, N.; Merlotti, A.; Merlano, M. Head and neck cancer: Improving outcomes with a multidisciplinary approach. Cancer Manag. Res. 2017, 9, 363–371. [Google Scholar] [CrossRef]
- Birknerová, N.; Kovaříková, H.; Baranová, I.; Přikrylová, A.; Laco, J.; Vošmiková, H.; Gajdošová, B.; Hodek, M.; Vošmik, M.; Palička, V.; et al. DNA hypermethylation of CADM1, PAX5, WT1, RARβ, and PAX6 genes in oropharyngeal cancer associated with human papillomavirus. Epigenetics 2022. [Google Scholar] [CrossRef]
- Kuhlin, B.; Kramer, B.; Nefas, V.; Rotter, N.; Aderhold, C. Indicators for secondary carcinoma in head and neck cancer patients following curative therapy: A retrospective clinical study. Mol. Clin. Oncol. 2020, 12, 403–410. [Google Scholar] [CrossRef]
- Wang, S.; Liu, Y.; Feng, Y.; Zhang, J.; Swinnen, J.; Li, Y.; Ni, Y. A review on curability of cancers: More efforts for novel therapeutic options are needed. Cancers 2019, 11, 1782. [Google Scholar] [CrossRef]
- Fernández-Lázaro, D.; García Hernández, J.L.; García, A.C.; Córdova Martínez, A.; Mielgo-Ayuso, J.; Cruz-Hernández, J.J. Liquid biopsy as novel tool in precision medicine: Origins, properties, identification and clinical perspective of cancer’s biomarkers. Diagnostics 2020, 10, 215. [Google Scholar] [CrossRef]
- Mazouji, O.; Ouhajjou, A.; Incitti, R.; Mansour, H. Updates on clinical use of liquid biopsy in colorectal cancer screening, diagnosis, follow-up, and treatment guidance. Front. Cell Dev. Biol. 2021, 9, 660924. [Google Scholar] [CrossRef] [PubMed]
- Chiappin, S.; Antonelli, G.; Gatti, R.; De Palo, E.F. Saliva specimen: A new laboratory tool for diagnostic and basic investigation. Clin. Chim. Acta 2007, 383, 30–40. [Google Scholar] [CrossRef]
- Vila, T.; Rizk, A.M.; Sultan, A.S.; Jabra-Rizk, M.A. The power of saliva: Antimicrobial and beyond. PLoS Pathog. 2019, 15, e1008058. [Google Scholar] [CrossRef] [PubMed]
- Kaczor-Urbanowicz, K.E.; Martin Carreras-Presas, C.; Aro, K.; Tu, M.; Garcia-Godoy, F.; Wong, D.T. Saliva diagnostics—Current views and directions. Exp. Biol. Med. 2017, 242, 459–472. [Google Scholar] [CrossRef]
- Patel, A.; Patel, S.; Patel, P.; Tanavde, V. Saliva based liquid biopsies in head and neck cancer: How far are we from the clinic? Front. Oncol. 2022, 12, 828434. [Google Scholar] [CrossRef]
- Michela, B. Liquid biopsy: A family of possible diagnostic tools. Diagnostics 2021, 11, 1391. [Google Scholar] [CrossRef]
- Martignano, F. Cell-free DNA: An overview of sample types and isolation procedures. In Cell-free DNA as Diagnostic Markers; Casadio, V., Salvi, S., Eds.; Methods in Molecular Biology; Springer New York: New York, NY, USA, 2019; Volume 1909, pp. 13–27. ISBN 978-1-4939-8972-0. [Google Scholar]
- Mandel, P.; Metais, P. Nuclear acids in human blood plasma. C R Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar]
- Rostami, A.; Lambie, M.; Yu, C.W.; Stambolic, V.; Waldron, J.N.; Bratman, S.V. Senescence, necrosis, and apoptosis govern circulating cell-free DNA release kinetics. Cell Rep. 2020, 31, 107830. [Google Scholar] [CrossRef]
- Snyder, M.W.; Kircher, M.; Hill, A.J.; Daza, R.M.; Shendure, J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 2016, 164, 57–68. [Google Scholar] [CrossRef]
- Thierry, A.R.; El Messaoudi, S.; Gahan, P.B.; Anker, P.; Stroun, M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016, 35, 347–376. [Google Scholar] [CrossRef]
- Aucamp, J.; Bronkhorst, A.J.; Badenhorst, C.P.S.; Pretorius, P.J. The diverse origins of circulating cell-free DNA in the human body: A critical re-evaluation of the literature. Biol. Rev. 2018, 93, 1649–1683. [Google Scholar] [CrossRef] [PubMed]
- Heitzer, E.; Auinger, L.; Speicher, M.R. Cell-free DNA and apoptosis: How dead cells inform about the living. Trends Mol. Med. 2020, 26, 519–528. [Google Scholar] [CrossRef]
- Luo, H.; Wei, W.; Ye, Z.; Zheng, J.; Xu, R. Liquid biopsy of methylation biomarkers in cell-free DNA. Trends Mol. Med. 2021, 27, 482–500. [Google Scholar] [CrossRef]
- Pös, Z.; Pös, O.; Styk, J.; Mocova, A.; Strieskova, L.; Budis, J.; Kadasi, L.; Radvanszky, J.; Szemes, T. Technical and methodological aspects of cell-free nucleic acids analyzes. Int. J. Mol. Sci. 2020, 21, 8634. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef]
- Mulki, S.; Shetty, P.; Pai, P. Oral rinse-based cytology and conventional exfoliative cytology: A comparative study. J. Cancer Res. Ther. 2015, 11, 129. [Google Scholar] [CrossRef]
- Pereira, T.; Kesarkar, K.; Tamgadge, A.; Bhalerao, S.; Shetty, S. Comparative analysis of oral rinse-based cytology and conventional exfoliative cytology: A pilot study. J. Cancer Res. Ther. 2018, 14, 921. [Google Scholar] [CrossRef]
- Wang, Z.; Li, F.; Rufo, J.; Chen, C.; Yang, S.; Li, L.; Zhang, J.; Cheng, J.; Kim, Y.; Wu, M.; et al. Acoustofluidic salivary exosome isolation. J. Mol. Diagn. 2020, 22, 50–59. [Google Scholar] [CrossRef]
- Tivey, A.; Church, M.; Rothwell, D.; Dive, C.; Cook, N. Circulating tumour DNA—Looking beyond the blood. Nat. Rev. Clin. Oncol. 2022, 19, 600–612. [Google Scholar] [CrossRef]
- Lim, Y.; Punyadeera, C. A pilot study to investigate the feasibility of transporting saliva samples at room temperature with MAWI cell stabilization buffer. Cogent Biol. 2018, 4, 1470895. [Google Scholar] [CrossRef]
- Green, S.F. The cost of poor blood specimen quality and errors in preanalytical processes. Clin. Biochem. 2013, 46, 1175–1179. [Google Scholar] [CrossRef]
- Aro, K.; Wei, F.; Wong, D.T.; Tu, M. Saliva liquid biopsy for point-of-care applications. Front. Public Health 2017, 5, 77. [Google Scholar] [CrossRef]
- Rosas, S.L.; Koch, W.; da Costa Carvalho, M.G.; Wu, L.; Califano, J.; Westra, W.; Jen, J.; Sidransky, D. Promoter hypermethylation patterns of P16, O6-methylguanine-DNA-methyltransferase, and death-associated protein kinase in tumors and saliva of head and neck cancer patients. Cancer Res. 2001, 61, 939–942. [Google Scholar]
- Righini, C.A.; de Fraipont, F.; Timsit, J.-F.; Faure, C.; Brambilla, E.; Reyt, E.; Favrot, M.-C. Tumor-specific methylation in saliva: A promising biomarker for early detection of head and neck cancer recurrence. Clin. Cancer Res. 2007, 13, 1179–1185. [Google Scholar] [CrossRef]
- Franzmann, E.J.; Reategui, E.P.; Pedroso, F.; Pernas, F.G.; Karakullukcu, B.M.; Carraway, K.L.; Hamilton, K.; Singal, R.; Goodwin, W.J. Soluble CD44 is a potential marker for the early detection of head and neck cancer. Cancer Epidemiol. Biomark. Prev. 2007, 16, 1348–1355. [Google Scholar] [CrossRef]
- Viet, C.T.; Jordan, R.C.K.; Schmidt, B.L. DNA Promoter hypermethylation in saliva for the early diagnosis of oral cancer. J. Calif. Dent. Assoc. 2007, 35, 844–849. [Google Scholar]
- Viet, C.T.; Schmidt, B.L. Methylation array analysis of preoperative and postoperative saliva DNA in oral cancer patients. Cancer Epidemiol. Biomark. Prev. 2008, 17, 3603–3611. [Google Scholar] [CrossRef]
- Carvalho, A.L.; Jeronimo, C.; Kim, M.M.; Henrique, R.; Zhang, Z.; Hoque, M.O.; Chang, S.; Brait, M.; Nayak, C.S.; Jiang, W.-W.; et al. Evaluation of promoter hypermethylation detection in body fluids as a screening/diagnosis tool for head and neck squamous cell carcinoma. Clin. Cancer Res. 2008, 14, 97–107. [Google Scholar] [CrossRef]
- Pattani, K.M.; Zhang, Z.; Demokan, S.; Glazer, C.; Loyo, M.; Goodman, S.; Sidransky, D.; Bermudez, F.; Jean-Charles, G.; McCaffrey, T.; et al. Endothelin Receptor Type B gene promoter hypermethylation in salivary rinses is independently associated with risk of oral cavity cancer and premalignancy. Cancer Prev. Res. 2010, 3, 1093–1103. [Google Scholar] [CrossRef]
- Demokan, S.; Chang, X.; Chuang, A.; Mydlarz, W.K.; Kaur, J.; Huang, P.; Khan, Z.; Khan, T.; Ostrow, K.L.; Brait, M.; et al. KIF1A and EDNRB are differentially methylated in primary HNSCC and salivary rinses. Int. J. Cancer 2010, 127, 2351–2359. [Google Scholar] [CrossRef]
- Guerrero-Preston, R.; Soudry, E.; Acero, J.; Orera, M.; Moreno-López, L.; Macía-Colón, G.; Jaffe, A.; Berdasco, M.; Ili-Gangas, C.; Brebi-Mieville, P.; et al. NID2 and HOXA9 promoter hypermethylation as biomarkers for prevention and early detection in oral cavity squamous cell carcinoma tissues and saliva. Cancer Prev. Res. 2011, 4, 1061–1072. [Google Scholar] [CrossRef]
- Nagata, S.; Hamada, T.; Yamada, N.; Yokoyama, S.; Kitamoto, S.; Kanmura, Y.; Nomura, M.; Kamikawa, Y.; Yonezawa, S.; Sugihara, K. Aberrant DNA methylation of tumor-related genes in oral rinse: A noninvasive method for detection of oral squamous cell carcinoma. Cancer 2012, 118, 4298–4308. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, Z.-T.; He, Q.-B.; Jiang, W.-W. DAPK promoter hypermethylation in tissues and body fluids of oral precancer patients. Med. Oncol. 2012, 29, 729–733. [Google Scholar] [CrossRef]
- Kusumoto, T.; Hamada, T.; Yamada, N.; Nagata, S.; Kanmura, Y.; Houjou, I.; Kamikawa, Y.; Yonezawa, S.; Sugihara, K. Comprehensive epigenetic analysis using oral rinse samples: A pilot study. J. Oral Maxillofac. Surg. 2012, 70, 1486–1494. [Google Scholar] [CrossRef]
- Ovchinnikov, D.A.; Cooper, M.A.; Pandit, P.; Coman, W.B.; Cooper-White, J.J.; Keith, P.; Wolvetang, E.J.; Slowey, P.D.; Punyadeera, C. Tumor-suppressor gene promoter hypermethylation in saliva of head and neck cancer patients. Transl. Oncol. 2012, 5, 321–326. [Google Scholar] [CrossRef]
- Rettori, M.M.; de Carvalho, A.C.; Bomfim Longo, A.L.; de Oliveira, C.Z.; Kowalski, L.P.; Carvalho, A.L.; Vettore, A.L. Prognostic significance of TIMP3 hypermethylation in post-treatment salivary rinse from head and neck squamous cell carcinoma patients. Carcinogenesis 2013, 34, 20–27. [Google Scholar] [CrossRef]
- Puttipanyalears, C.; Subbalekha, K.; Mutirangura, A.; Kitkumthorn, N. Alu hypomethylation in smoke-exposed epithelia and oral squamous carcinoma. Asian Pac. J. Cancer Prev. 2013, 14, 5495–5501. [Google Scholar] [CrossRef][Green Version]
- Schussel, J.; Zhou, X.C.; Zhang, Z.; Pattani, K.; Bermudez, F.; Jean-Charles, G.; McCaffrey, T.; Padhya, T.; Phelan, J.; Spivakovsky, S.; et al. EDNRB and DCC salivary rinse hypermethylation has a similar performance as expert clinical examination in discrimination of oral cancer/dysplasia versus benign lesions. Clin. Cancer Res. 2013, 19, 3268–3275. [Google Scholar] [CrossRef]
- Ovchinnikov, D.A.; Wan, Y.; Coman, W.B.; Pandit, P.; Cooper-White, J.J.; Herman, J.G.; Punyadeera, C. DNA methylation at the novel CpG sites in the promoter of MED15/PCQAP gene as a biomarker for head and neck cancers. Biomark. Insights 2014, 9, 53–60. [Google Scholar] [CrossRef]
- Gaykalova, D.A.; Vatapalli, R.; Wei, Y.; Tsai, H.-L.; Wang, H.; Zhang, C.; Hennessey, P.T.; Guo, T.; Tan, M.; Li, R.; et al. Outlier analysis defines zinc finger gene family DNA methylation in tumors and saliva of head and neck cancer patients. PLoS ONE 2015, 10, e0142148. [Google Scholar] [CrossRef]
- Lim, Y.; Wan, Y.; Vagenas, D.; Ovchinnikov, D.A.; Perry, C.F.L.; Davis, M.J.; Punyadeera, C. Salivary DNA methylation panel to diagnose HPV-positive and HPV-negative head and neck cancers. BMC Cancer 2016, 16, 749. [Google Scholar] [CrossRef]
- Ferlazzo, N.; Currò, M.; Zinellu, A.; Caccamo, D.; Isola, G.; Ventura, V.; Carru, C.; Matarese, G.; Ientile, R. Influence of MTHFR genetic background on P16 and MGMT methylation in oral squamous cell cancer. Int. J. Mol. Sci. 2017, 18, 724. [Google Scholar] [CrossRef]
- Cheng, S.-J.; Chang, C.-F.; Ko, H.-H.; Lee, J.-J.; Chen, H.-M.; Wang, H.-J.; Lin, H.-S.; Chiang, C.-P. Hypermethylated ZNF582 and PAX1 genes in mouth rinse samples as biomarkers for oral dysplasia and oral cancer detection. Head Neck 2018, 40, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Puttipanyalears, C.; Arayataweegool, A.; Chalertpet, K.; Rattanachayoto, P.; Mahattanasakul, P.; Tangjaturonsasme, N.; Kerekhanjanarong, V.; Mutirangura, A.; Kitkumthorn, N. TRH site-specific methylation in oral and oropharyngeal squamous cell carcinoma. BMC Cancer 2018, 18, 786. [Google Scholar] [CrossRef] [PubMed]
- Liyanage, C.; Wathupola, A.; Muraleetharan, S.; Perera, K.; Punyadeera, C.; Udagama, P. Promoter hypermethylation of tumor-suppressor genes P16INK4a, RASSF1A, TIMP3, and PCQAP/MED15 in salivary DNA as a quadruple biomarker panel for early detection of oral and oropharyngeal cancers. Biomolecules 2019, 9, 148. [Google Scholar] [CrossRef] [PubMed]
- Srisuttee, R.; Arayataweegool, A.; Mahattanasakul, P.; Tangjaturonrasme, N.; Kerekhanjanarong, V.; Keelawat, S.; Mutirangura, A.; Kitkumthorn, N. Evaluation of NID2 promoter methylation for screening of oral squamous cell carcinoma. BMC Cancer 2020, 20, 218. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Saito, Y.; Ren, S.; Liu, C.; Guo, T.; Qualliotine, J.; Khan, Z.; Sadat, S.; Califano, J.A. Targeting viral DNA and promoter hypermethylation in salivary rinses for recurrent HPV-positive oropharyngeal cancer. Otolaryngol. Head Neck Surg. 2020, 162, 512–519. [Google Scholar] [CrossRef]
- Rapado-González, Ó.; Martínez-Reglero, C.; Salgado-Barreira, Á.; Muinelo-Romay, L.; Muinelo-Lorenzo, J.; López-López, R.; Díaz-Lagares, Á.; Suárez-Cunqueiro, M.M. Salivary DNA methylation as an epigenetic biomarker for head and neck cancer. part I: A diagnostic accuracy meta-analysis. J. Pers. Med. 2021, 11, 568. [Google Scholar] [CrossRef] [PubMed]
- Henson, B.S.; Wong, D.T. Collection, storage, and processing of saliva samples for downstream molecular applications. In Oral Biology; Seymour, G.J., Cullinan, M.P., Heng, N.C.K., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2010; Volume 666, pp. 21–30. ISBN 978-1-60761-819-5. [Google Scholar]
- Chiang, S.H.; Thomas, G.A.; Liao, W.; Grogan, T.; Buck, R.L.; Fuentes, L.; Yakob, M.; Laughlin, M.J.; Schafer, C.; Nazmul-Hossain, A.; et al. RNAPro•SAL: A device for rapid and standardized collection of saliva RNA and proteins. Biotechniques 2015, 58, 69–76. [Google Scholar] [CrossRef]
- Abbosh, C.; Birkbak, N.J.; Wilson, G.A.; Jamal-Hanjani, M.; Constantin, T.; Salari, R.; Le Quesne, J.; Moore, D.A.; Veeriah, S.; Rosenthal, R.; et al. Phylogenetic CtDNA analysis depicts early-stage lung cancer evolution. Nature 2017, 545, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Ye, M.; Ni, S.; Li, Q.; Ye, D.; Li, J.; Shen, Z.; Deng, H. DNA methylation biomarkers for head and neck squamous cell carcinoma. Epigenetics 2018, 13, 398–409. [Google Scholar] [CrossRef]
- Xu, T.; Gao, H. Hydroxymethylation and tumors: Can 5-hydroxymethylation be used as a marker for tumor diagnosis and treatment? Hum. Genom. 2020, 14, 15. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; de Medeiros, M.C.; Fernandez, E.M.; Zarins, K.R.; Cavalcante, R.G.; Qin, T.; Wolf, G.T.; Figueroa, M.E.; D’Silva, N.J.; Rozek, L.S.; et al. 5-hydroxymethylation highlights the heterogeneity in keratinization and cell junctions in head and neck cancers. Clin. Epigenetics 2020, 12, 175. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, G.P.; Xiong, W.; Hahn, M.A.; Jin, S.-G. The role of 5-hydroxymethylcytosine in human cancer. Cell Tissue Res. 2014, 356, 631–641. [Google Scholar] [CrossRef]
- Li, W.; Zhang, X.; Lu, X.; You, L.; Song, Y.; Luo, Z.; Zhang, J.; Nie, J.; Zheng, W.; Xu, D.; et al. 5-hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res. 2017, 27, 1243–1257. [Google Scholar] [CrossRef]
- Köhler, F.; Rodríguez-Paredes, M. DNA methylation in epidermal differentiation, aging, and cancer. J. Investig. Dermatol. 2020, 140, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Dor, Y.; Cedar, H. Principles of DNA methylation and their implications for biology and medicine. Lancet 2018, 392, 777–786. [Google Scholar] [CrossRef]
- Nishiyama, A.; Nakanishi, M. Navigating the DNA methylation landscape of cancer. Trends Genet. 2021, 37, 1012–1027. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Gole, J.; Gore, A.; He, Q.; Lu, M.; Min, J.; Yuan, Z.; Yang, X.; Jiang, Y.; Zhang, T.; et al. Non-invasive early detection of cancer four years before conventional diagnosis using a blood test. Nat. Commun. 2020, 11, 3475. [Google Scholar] [CrossRef]
- Wang, Y.; Springer, S.; Mulvey, C.L.; Silliman, N.; Schaefer, J.; Sausen, M.; James, N.; Rettig, E.M.; Guo, T.; Pickering, C.R.; et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci. Transl. Med. 2015, 7, 293ra104. [Google Scholar] [CrossRef]
- Davenport, A.P.; Maguire, J.J. Endothelin. In The Vascular Endothelium I.; Moncada, S., Higgs, A., Eds.; Handbook of Experimental Pharmacology; Springer Berlin Heidelberg: Berlin/Heidelberg, Germany, 2006; Volume 176/I, pp. 295–329. ISBN 978-3-540-32966-4. [Google Scholar]
- Mousavi Ardehaie, R.; Hashemzadeh, S.; Behrouz Sharif, S.; Ghojazadeh, M.; Teimoori-Toolabi, L.; Sakhinia, E. Aberrant methylated EDNRB can act as a potential diagnostic biomarker in sporadic colorectal cancer while KISS1 is controversial. Bioengineered 2017, 8, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Dang, D.; Ye, Y.; Aouizerat, B.E.; Patel, Y.K.; Viet, D.T.; Chan, K.C.; Ono, K.; Doan, C.; Figueroa, J.D.; Yu, G.; et al. Targeting the endothelin axis as a therapeutic strategy for oral cancer metastasis and pain. Sci. Rep. 2020, 10, 20832. [Google Scholar] [CrossRef] [PubMed]
- Meng, R.-W.; Li, Y.-C.; Chen, X.; Huang, Y.-X.; Shi, H.; Du, D.-D.; Niu, X.; Lu, C.; Lu, M.-X. Aberrant methylation of RASSF1A closely associated with HNSCC, a meta-analysis. Sci. Rep. 2016, 6, 20756. [Google Scholar] [CrossRef]
- Wang, H. Cell-free DNA methylation profiling analysis—Technologies and bioinformatics. Cancers 2019, 11, 1741. [Google Scholar] [CrossRef]
- Wen, L.; Li, J.; Guo, H.; Liu, X.; Zheng, S.; Zhang, D.; Zhu, W.; Qu, J.; Guo, L.; Du, D.; et al. Genome-scale detection of hypermethylated CpG islands in circulating cell-free DNA of hepatocellular carcinoma patients. Cell Res. 2015, 25, 1250–1264. [Google Scholar] [CrossRef]
- Aberg, K.A.; Chan, R.F.; Shabalin, A.A.; Zhao, M.; Turecki, G.; Staunstrup, N.H.; Starnawska, A.; Mors, O.; Xie, L.Y.; van den Oord, E.J. A MBD-seq protocol for large-scale methylome-wide studies with (very) low amounts of DNA. Epigenetics 2017, 12, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.-T.; Whyte, J.J.; Hopkins, G.M.; Kirk, M.D.; Prather, R.S. Methylated DNA immunoprecipitation and high-throughput sequencing (MeDIP-seq) using low amounts of genomic DNA. Cell. Reprogramming Former. Cloning Stem Cells 2014, 16, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.Y.; Singhania, R.; Fehringer, G.; Chakravarthy, A.; Roehrl, M.H.A.; Chadwick, D.; Zuzarte, P.C.; Borgida, A.; Wang, T.T.; Li, T.; et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 2018, 563, 579–583. [Google Scholar] [CrossRef]
- Maunakea, A.K.; Nagarajan, R.P.; Bilenky, M.; Ballinger, T.J.; D’Souza, C.; Fouse, S.D.; Johnson, B.E.; Hong, C.; Nielsen, C.; Zhao, Y.; et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 2010, 466, 253–257. [Google Scholar] [CrossRef]
- Marcozzi, A.; Jager, M.; Elferink, M.; Straver, R.; van Ginkel, J.H.; Peltenburg, B.; Chen, L.-T.; Renkens, I.; van Kuik, J.; Terhaard, C.; et al. Accurate detection of circulating tumor DNA using nanopore consensus sequencing. NPJ Genom. Med. 2021, 6, 106. [Google Scholar] [CrossRef] [PubMed]
- Genereux, D.P.; Johnson, W.C.; Burden, A.F.; Stoger, R.; Laird, C.D. Errors in the bisulfite conversion of DNA: Modulating inappropriate- and failed-conversion frequencies. Nucleic Acids Res. 2008, 36, e150. [Google Scholar] [CrossRef] [PubMed]
- Katsman, E.; Orlanski, S.; Martignano, F.; Eden, A.; Petrini, I.; Conticello, S.G.; Berman, B.P. Detecting cell-of-origin and cancer-specific features of cell-free dna with nanopore sequencing. Genome Biol. 2022, 23, 158. [Google Scholar] [CrossRef]
- Korlach, J.; Bjornson, K.P.; Chaudhuri, B.P.; Cicero, R.L.; Flusberg, B.A.; Gray, J.J.; Holden, D.; Saxena, R.; Wegener, J.; Turner, S.W. Real-time DNA sequencing from single polymerase molecules. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2010; Volume 472, pp. 431–455. ISBN 978-0-12-374954-3. [Google Scholar]
- Feng, Z.; Fang, G.; Korlach, J.; Clark, T.; Luong, K.; Zhang, X.; Wong, W.; Schadt, E. Detecting DNA modifications from SMRT sequencing data by modeling sequence context dependence of polymerase kinetic. PLoS Comput. Biol. 2013, 9, e1002935. [Google Scholar] [CrossRef] [PubMed]
- Flusberg, B.A.; Webster, D.R.; Lee, J.H.; Travers, K.J.; Olivares, E.C.; Clark, T.A.; Korlach, J.; Turner, S.W. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods 2010, 7, 461–465. [Google Scholar] [CrossRef]
- Eid, J.; Fehr, A.; Gray, J.; Luong, K.; Lyle, J.; Otto, G.; Peluso, P.; Rank, D.; Baybayan, P.; Bettman, B.; et al. Real-time DNA sequencing from single polymerase molecules. Science 2009, 323, 133–138. [Google Scholar] [CrossRef]
- Ardui, S.; Ameur, A.; Vermeesch, J.R.; Hestand, M.S. Single molecule real-time (SMRT) sequencing comes of age: Applications and utilities for medical diagnostics. Nucleic Acids Res. 2018, 46, 2159–2168. [Google Scholar] [CrossRef]
- Tse, O.Y.O.; Jiang, P.; Cheng, S.H.; Peng, W.; Shang, H.; Wong, J.; Chan, S.L.; Poon, L.C.Y.; Leung, T.Y.; Chan, K.C.A.; et al. Genome-wide detection of cytosine methylation by single molecule real-time sequencing. Proc. Natl. Acad. Sci. USA. 2021, 118, e2019768118. [Google Scholar] [CrossRef]
- Clark, T.A.; Lu, X.; Luong, K.; Dai, Q.; Boitano, M.; Turner, S.W.; He, C.; Korlach, J. Enhanced 5-methylcytosine detection in single-molecule, real-time sequencing via Tet1 oxidation. BMC Biol. 2013, 11, 4. [Google Scholar] [CrossRef]
- Choy, L.Y.L.; Peng, W.; Jiang, P.; Cheng, S.H.; Yu, S.C.Y.; Shang, H.; Olivia Tse, O.Y.; Wong, J.; Wong, V.W.S.; Wong, G.L.H.; et al. Single-molecule sequencing enables long cell-free DNA detection and direct methylation analysis for cancer patients. Clin. Chem. 2022, 68, 1151–1163. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, Y.; Ying, C.; Wang, D.; Du, C. Nanopore-based fourth-generation DNA sequencing technology. Genom. Proteom. Bioinform. 2015, 13, 4–16. [Google Scholar] [CrossRef]
- Deamer, D.; Akeson, M.; Branton, D. Three decades of nanopore sequencing. Nat. Biotechnol. 2016, 34, 518–524. [Google Scholar] [CrossRef]
- Martignano, F.; Munagala, U.; Crucitta, S.; Mingrino, A.; Semeraro, R.; Del Re, M.; Petrini, I.; Magi, A.; Conticello, S.G. Nanopore sequencing from liquid biopsy: Analysis of copy number variations from cell-free DNA of lung cancer patients. Mol. Cancer 2021, 20, 32. [Google Scholar] [CrossRef]
- Kuschel, L.P.; Hench, J.; Frank, S.; Hench, I.B.; Girard, E.; Blanluet, M.; Masliah-Planchon, J.; Misch, M.; Onken, J.; Czabanka, M.; et al. Robust Methylation-based classification of brain tumors using nanopore sequencing. medRxiv 2021. [Google Scholar] [CrossRef]
- Djirackor, L.; Halldorsson, S.; Niehusmann, P.; Leske, H.; Capper, D.; Kuschel, L.P.; Pahnke, J.; Due-Tønnessen, B.J.; Langmoen, I.A.; Sandberg, C.J.; et al. Intraoperative DNA methylation classification of brain tumors impacts neurosurgical strategy. Neurooncol. Adv. 2021, 3, vdab149. [Google Scholar] [CrossRef] [PubMed]
- Rang, F.J.; Kloosterman, W.P.; de Ridder, J. From squiggle to basepair: Computational approaches for improving nanopore sequencing read accuracy. Genome Biol. 2018, 19, 90. [Google Scholar] [CrossRef] [PubMed]
- Tyler, A.D.; Mataseje, L.; Urfano, C.J.; Schmidt, L.; Antonation, K.S.; Mulvey, M.R.; Corbett, C.R. Evaluation of Oxford Nanopore’s MinION sequencing device for microbial whole genome sequencing applications. Sci. Rep. 2018, 8, 10931. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, Y.; Bollas, A.; Wang, Y.; Au, K.F. Nanopore sequencing technology, bioinformatics and applications. Nat. Biotechnol. 2021, 39, 1348–1365. [Google Scholar] [CrossRef] [PubMed]
- Oxford Nanopore Releases Short Fragment Mode: A New Tool For Real-Time Sequencing Of Short Fragments of DNA. Available online: https://nanoporetech.com/about-us/news/oxford-nanopore-releases-short-fragment-mode-new-tool-real-time-sequencing-short (accessed on 20 June 2022). Oxford Nanopore Technology News.
- Oxford Nanopore Integrates “Remora”: A Tool To Enable Real-Time, High-Accuracy Epigenetic Insights with Nanopore Sequencing Software MinKNOW. Available online: https://nanoporetech.com/about-us/news/oxford-nanopore-integrates-remora-tool-enable-real-time-high-accuracy-epigenetic (accessed on 20 June 2022). Oxford Nanopore Technology News.
- Brown, C.G. Oxford Nanopore Technology News. Oxford Nanopore Technology Update: CTO Clive G Brown Unveils Latest Sequencing Chemistry with Highest Performance to Date, Short Fragment Mode and Latest Methylation Performance Evaluations. Available online: https://nanoporetech.com/about-us/news/oxford-nanopore-technology-update-cto-clive-g-brown-unveils-latest-sequencing (accessed on 20 June 2022).
- Klutstein, M.; Nejman, D.; Greenfield, R.; Cedar, H. DNA methylation in cancer and aging. Cancer Res. 2016, 76, 3446–3450. [Google Scholar] [CrossRef]
- Glezeva, N.; Moran, B.; Collier, P.; Moravec, C.S.; Phelan, D.; Donnellan, E.; Russell-Hallinan, A.; O’Connor, D.P.; Gallagher, W.M.; Gallagher, J.; et al. Targeted DNA methylation profiling of human cardiac tissue reveals novel epigenetic traits and gene deregulation across different heart failure patient subtypes. Circ. Heart Fail. 2019, 12, e005765. [Google Scholar] [CrossRef]
- Imperiale, T.F.; Ransohoff, D.F.; Itzkowitz, S.H.; Levin, T.R.; Lavin, P.; Lidgard, G.P.; Ahlquist, D.A.; Berger, B.M. Multitarget stool DNA testing for colorectal-cancer screening. N. Engl. J. Med. 2014, 370, 1287–1297. [Google Scholar] [CrossRef]
- Lin, J.S.; Piper, M.A.; Perdue, L.A.; Rutter, C.M.; Webber, E.M.; O’Connor, E.; Smith, N.; Whitlock, E.P. Screening for colorectal cancer: Updated evidence report and systematic review for the US preventive services task force. JAMA 2016, 315, 2576. [Google Scholar] [CrossRef]
- Potter, N.T.; Hurban, P.; White, M.N.; Whitlock, K.D.; Lofton-Day, C.E.; Tetzner, R.; Koenig, T.; Quigley, N.B.; Weiss, G. Validation of a real-time PCR—Based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin. Chem. 2014, 60, 1183–1191. [Google Scholar] [CrossRef]
- Oh, T.; Kim, N.; Moon, Y.; Kim, M.S.; Hoehn, B.D.; Park, C.H.; Kim, T.S.; Kim, N.K.; Chung, H.C.; An, S. Genome-wide identification and validation of a novel methylation biomarker, SDC2, for blood-based detection of colorectal cancer. J. Mol. Diagn. 2013, 15, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.D.; Oh, T.J.; Chung, T.-H.; Jang, H.W.; Kim, Y.N.; An, S.; Kim, N.K. Early detection of colorectal cancer based on presence of methylated syndecan-2 (SDC2) in stool DNA. Clin. Epigenetics 2019, 11, 51. [Google Scholar] [CrossRef]
- Aubele, M.; Schmitt, M.; Napieralski, R.; Paepke, S.; Ettl, J.; Absmaier, M.; Magdolen, V.; Martens, J.; Foekens, J.A.; Wilhelm, O.G.; et al. The predictive value of PITX2 DNA methylation for high-risk breast cancer therapy: Current guidelines, medical needs, and challenges. Dis. Markers 2017, 2017, 4934608. [Google Scholar] [CrossRef]
- Schricker, G.; Napieralski, R.; Noske, A.; Piednoir, E.; Manner, O.; Schüren, E.; Lauber, J.; Perkins, J.; Magdolen, V.; Schmitt, M.; et al. Clinical performance of an analytically validated assay in comparison to microarray technology to assess PITX2 DNA-methylation in breast cancer. Sci. Rep. 2018, 8, 16861. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, D. Performance evaluation of the DNA methylation biomarker SHOX2 for the Aid in Diagnosis of Lung Cancer Based on the Analysis of Bronchial Aspirates. Int. J. Oncol. 2012, 40, 825–832. [Google Scholar] [CrossRef] [PubMed]
- Weiss, G.; Schlegel, A.; Kottwitz, D.; König, T.; Tetzner, R. Validation of the SHOX2/PTGER4 DNA methylation marker panel for plasma-based discrimination between patients with malignant and nonmalignant lung disease. J. Thorac. Oncol. 2017, 12, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, B.; Liebenberg, V.; Dietrich, D.; Schlegel, T.; Kneip, C.; Seegebarth, A.; Flemming, N.; Seemann, S.; Distler, J.; Lewin, J.; et al. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer based on bronchial aspirates. BMC Cancer 2010, 10, 600. [Google Scholar] [CrossRef]
- Beltrán-García, J.; Osca-Verdegal, R.; Mena-Mollá, S.; García-Giménez, J.L. Epigenetic IVD tests for personalized precision medicine in cancer. Front. Genet. 2019, 10, 621. [Google Scholar] [CrossRef]
- Srivastava, S. Cancer biomarker discovery and development in gastrointestinal cancers: Early detection research network—A collaborative approach. Gastrointest. Cancer Res. GCR 2007, 1, S60–S63. [Google Scholar]

| Biomarker Name | Biological Function | Tumor Site | Sample Type | Sample Size | DNA Processing | Technical Approach (Methods) | Methylation Status | Application | Sensitivity (%) | Specificity (%) | AUC (95% CI) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p16 | Cell cycle regulation | HNC (SCC) | Oral rinse (NaCl) | 30 tumors and 30 saliva controls | Volume: - Kit: Phenol-chloroform extraction/Bisulfite treatment (Sigma, Burlington, MA, USA) | MSP | Hypermethylation | Diagnosis | NA | NA | NA | Rosas 2001 [34] |
| MGMT | DNA repair | NA | NA | NA | ||||||||
| DAPK | Cell death regulation | NA | NA | NA | ||||||||
| TIMP3 | Cell cycle regulation | HNC (SCC) | Oral rinse (25 mL NaCl 0.9%) | 60 patients | Volume: - Kit: QIAmp tissue kit (Qiagen, Hilden, Germany)/CpGenome DNA modification kit (MP Biomedicals, Irvine, CA, USA) | MSP | Hypermethylation | Diagnosis/ disease monitoring | 28 | NA | NA | Righini 2007 [35] |
| ECAD | Cell adhesion | 20 | NA | NA | ||||||||
| p16 | Cell cycle regulation | 27 | NA | NA | ||||||||
| MGMT | DNA repair | 22 | NA | NA | ||||||||
| DAPK | Cell death regulation | 15 | NA | NA | ||||||||
| RASSF1A | Cell cycle regulation | 17 | NA | NA | ||||||||
| soluble CD44 | Cell adhesion | HNC (SCC) | Oral rinse (5 mL NaCl, 5 s) | 102 patients and 69 controls | Volume: - Kit: QIAmp DNA mini kit (Qiagen, Hilden, Germany)/bisulfite solution | MSP | Hypermethylation | Diagnosis | 62–70 | 75–88 | NA | Franzmann 2007 [36] |
| p16 | Cell cycle regulation | OC (SCC) | Unstimulated saliva (7.5 mL) | 14 patients and 5 controls | Volume: 1 mL Kit: QIAamp Blood (Qiagen, Hilden, Germany)/EpiTect Bisulfite (Qiagen, Hilden, Germany) | Methylight | Hypermethylation | Diagnosis | 35 | NA | NA | Viet 2007 [37] |
| MGMT | DNA repair | 29 | NA | NA | ||||||||
| p15 | Cell growth regulation, cell death regulation | 29 | NA | NA | ||||||||
| APC | Cell growth regulation | 14 | NA | NA | ||||||||
| ECAD | Cell adhesion | 7 | NA | NA | ||||||||
| GABRB3 + IL11 + INSR + NOTCH3 + NTRK3 + PXN | Cell signaling, cell differentiation, cell adhesion | OC (SCC) | Unstimulated saliva (7.5 mL) | 13 patients and 10 controls | Volume: 1 mL Kit:iPrep Chargeswitch Buccal Cell kit (Invitrogen, Waltham, MA, USA)/EZ DNA Methylation kit (Zymo Research, Irvine, CA, USA) | GoldenGate Methylation Array (Illumina, San Diego, CA, USA) | Hypermethylation | Diagnosis | 77 | 87 | NA | Viet 2008 [38] |
| CCNA1 | Cell cycle regulation | HNC (SCC) | Oral rinse (20 mL NaCl) | 175 patients and 444 controls | Volume: - Kit: Phenol-chloroform extraction/Bisulfite solution | qMSP | Hypermethylation | Diagnosis | 20 | 97 | >0.60 | Carvalho 2008 [39] |
| DAPK | Cell death regulation | 176 patients and 451 controls | 16 | 96 | >0.60 | |||||||
| DCC | Cell cycle regulation | 176 patients and 462 controls | 12 | 99 | >0.60 | |||||||
| MGMT | DNA repair | 149 patients and 239 controls | 13 | 95 | >0.60 | |||||||
| TIMP3 | Cell cycle regulation | 176 patients and 450 controls | 11 | 93 | >0.60 | |||||||
| MINT31 | Calcium channel regulator | 175 patients and 492 controls | 5 | 100 | >0.60 | |||||||
| p16 | Cell cycle regulation | 177 patients and 500 controls | 5 | 100 | >0.60 | |||||||
| PGP9.5 | Cell cycle regulation | 34 patients and 112 controls | 82 | 30 | >0.60 | |||||||
| AIM1 | Cell signaling | 23 patients and 73 controls | 4 | 99 | >0.60 | |||||||
| ESR | Cell cycle regulation, cell signaling | 33 patients and 119 controls | 3 | 98 | >0.60 | |||||||
| CCND2 | Cell cycle regulation | 136 patients and 97 controls | 7 | 90 | >0.60 | |||||||
| MINT1 | Cell adhesion | 131 patients and 296 controls | 35 | 66 | >0.60 | |||||||
| CDH1 | Cell adhesion | 66 patients and 116 controls | 30 | 38 | >0.60 | |||||||
| EDNRB | Cell signaling | OC (SCC) | Oral rinse (25 mL NaCl, 15 s) | 161 patients | Volume: - Kit: Phenol-chloroform extraction/EpiTect Bisulfite kit (Qiagen, Hilden, Germany) | qMSP | Hypermethylation | Diagnosis | 65 | 51 | 0.61 | Pattani 2010 [40] |
| KIF1A | Cell signaling, extracellular transport | HNC (SCC) | Oral rinse (20 mL NaCl) | 71 patients and 61 controls | Volume: - Kit: Phenol-chloroform extraction/EpiTect Bisulfite kit (Qiagen, Hilden, Germany) | qMSP | Hypermethylation | Diagnosis | 37 | 98 | NA | Demokan 2010 [41] |
| EDNRB | Cell signaling | 68 | 93 | NA | ||||||||
| KIF1A + EDNRB | - | 77 | 93 | NA | ||||||||
| HOXA9 | Homeodomain control, cell differentiation | OC (SCC) | Oral rinse (20 mL NaCl) | 16 OC patients, 16 OPC patients and 19 controls | Volume: - Kit: Phenol-chloroform extraction/EpiTect Bisulfite kit (Qiagen, Hilden, Germany) | Discovery: Illumina Infinium HumanMethylation27 BeadChips. Validation: qMSP | Hypermethylation | Diagnosis | 63 | 53 | 0.65 | Guerrero-Preston 2011 [42] |
| NID2 | Cell adhesion, cell differentiation | 72 | 21 | 0.57 | ||||||||
| ECAD | Cell adhesion | OC (SCC) | Oral rinse (20 mL NaCl, 30–60 s) | 34 patients and 24 controls | Volume: 5 mL Kit: DNeasy Blood and Tissue (Qiagen, Hilden, Germany)/EpiTect Bisulfite (Qiagen, Hilden, Germany) | MSP | Hypermethylation | Diagnosis | 94 | 80 | 0.91 | Nagata 2012 [43] |
| TMEFF2 | Cell cycle regulation, cell differentiation, cell signaling | 85 | 87 | 0.90 | ||||||||
| RARß | Cell signaling, cell cycle regulation, cell differentiation | 82 | 92 | 0.88 | ||||||||
| MGMT | DNA repair | 77 | 80 | 0.81 | ||||||||
| FHIT | Cell death regulation | 80 | 67 | 0.75 | ||||||||
| WIF-1 | Cell cycle regulation | 71 | 79 | 0.69 | ||||||||
| DAPK | Cell death regulation | 56 | 75 | 0.66 | ||||||||
| p16 | Cell cycle regulation | 38 | 92 | 0.66 | ||||||||
| HIN-1 | Cell cycle regulation, cell death regulation, cell growth regulation | 29 | 92 | 0.61 | ||||||||
| TIMP3 | Cell cycle regulation | 24 | 96 | 0.60 | ||||||||
| p15 | Cell growth regulation, cell death regulation | 65 | 63 | 0.58 | ||||||||
| APC | Cell growth regulation | 63 | 63 | 0.56 | ||||||||
| SPARC | Cell adhesion, cell differentiation | 41 | 67 | 0.51 | ||||||||
| ECAD + TMEFF2 + RARB + MGMT | - | 100 | 88 | NA | ||||||||
| ECAD + TMEFF2 + MGMT | - | 97 | 92 | NA | ||||||||
| ECAD + TMEFF2 + RARB | - | 94 | 96 | NA | ||||||||
| ECAD + RARB + MGMT | - | 91 | 92 | NA | ||||||||
| DAPK | Cell death regulation | OC (SCC) | Oral rinse (4 mL NaCl) | 77 oral precancer patients and 32 OC(SCC) samples | Volume: - Kit: Phenol-chloroform extraction/Bisulfite solution | qMSP | Hypermethylation | Diagnosis | 3 | NA | NA | Liu 2012 [44] |
| p16 | Cell cycle regulation | OC (SCC) | Oral rinse (16 mL NaCl, 30 s) | 10 patients and 3 controls | Volume: 3 mL Kit: Methylamp Whole Cell Bisulfite Modification (Epigentek, Farmingdale, NY, USA) | MSP | Hypermethylation | Diagnosis | 40 | 100 | NA | Kusumoto 2012 [45] |
| p16INK4a | Cell cycle regulation | HNC (SCC) | Unstimulated saliva (DNA-SAL Salivary DNA Collection Device) | 143 patients and 46 controls | Volume: - Kit: EpiTect Plus Kit (Qiagen, Hilden, Germany) | Nested MSP | Hypermethylation | Diagnosis | 58 | 91 | NA | Ovchinnikov 2012 [46] |
| RASSF1A | Cell cycle regulation | 55 | 80 | NA | ||||||||
| DAPK1 | Cell death regulation | 13 | 98 | NA | ||||||||
| p16INK4a + RASSF1A + DAPK1 | - | 80 | 87 | NA | ||||||||
| DCC | Cell cycle regulation | HNC (SCC) | Oral rinse (10 mL NaCl 0.9%) | 146 pretreated patients and 60 controls | Volume: - Kit: Phenol-chloroform extraction/Bisulfite solution | qMSP | Hypermethylation | Diagnosis | 52 | 90 | NA | Rettori 2013 [47] |
| CCNA1 | Cell cycle regulation | 11 | 97 | NA | ||||||||
| DAPK | Cell death regulation | 8 | 98 | NA | ||||||||
| MGMT | DNA repair | 8 | 97 | NA | ||||||||
| TIMP3 | Cell cycle regulation | 5 | 98 | NA | ||||||||
| MINT31 | Calcium channel regulator | 4 | 100 | NA | ||||||||
| AIM1 | Cell signaling | 3 | 100 | NA | ||||||||
| SFRP1 | Cell growth regulation, cell differentiation | 3 | 100 | NA | ||||||||
| APC | Cell growth regulation | 3 | 100 | NA | ||||||||
| p16 | Cell cycle regulation | 1 | 100 | NA | ||||||||
| HIN-1 | Cell cycle regulation, cell death regulation, cell growth regulation | 12 | 81 | NA | ||||||||
| CCNA1 + DAPK + DCC + MGMT + TIMP3 | - | 55 | 76 | NA | ||||||||
| CCNA1 + DAPK + MGMT + TIMP3 | - | 20 | 82 | NA | ||||||||
| CCNA1 + DAPK + MGMT | - | 18 | 85 | NA | ||||||||
| CCNA1 + MGMT + TIMP3 | - | 18 | 85 | NA | ||||||||
| CCNA1 + DAPK + TIMP3 | - | 16 | 92 | NA | ||||||||
| DAPK + MGMT + TIMP3 | - | 16 | 85 | NA | ||||||||
| CCNA1 + MGMT | - | 16 | 88 | NA | ||||||||
| CCNA1 + DAPK | - | 15 | 95 | NA | ||||||||
| CCNA1 + TIMP3 | - | 14 | 93 | NA | ||||||||
| Alu | Cell cycle regulation, cell signaling | OC (SCC) | Oral rinse (10 mL NaCl 0.9%, 15 s) | 43 patients and 108 controls | Volume: - Kit: Phenol-chloroform extraction/Bisulfite solution | COBRA | Hypomethylation | Diagnosis | 87 | 57 | NA | Puttipanyalears 2013 [48] |
| EDNRB | Cell signaling | HNC (SCC) | Oral rinse (20 mL NaCl) | 191 patients | Volume: - Kit: Phenol-chloroform extraction/EpiTect Bisulfite kit (Qiagen, Hilden, Germany) | qMSP | Hypermethylation | Diagnosis | 73 | 51 | 0.65 | Schussel 2013 [49] |
| DCC | Cell cycle regulation | 69 | 59 | 0.65 | ||||||||
| EDNRB + DCC | - | 75 | 48 | 0.67 | ||||||||
| MED15/PCQAP3′ | Cell cycle regulation | HNC (SCC) | Unstimulated saliva | 44 patients and 45 controls | Volume: - Kit: EpiTect Plus Kit (Qiagen, Hilden, Germany) | MSP | Hypermethylation | Diagnosis | 68 | 58 | 0.63 | Ovchinnikov 2014 [50] |
| MED15/PCQAP5′ | 46 patients and 49 controls | 70 | 63 | 0.70 | ||||||||
| ZNF14 | Cell cycle regulation | HNC (SCC) | Oral rinse | 59 patients and 35 controls | Volume: 250 μL Kit: Phenol-chloroform extraction/EpiTect Bisulfite kit (Qiagen, Hilden, Germany) | Discovery: Illumina Infinium HumanMethylation27 BeadChips. Validation: qMSP | Hypermethylation | Diagnosis | 8 | 100 | NA | Gaykalova 2015 [51] |
| ZNF160 | Cell cycle regulation | 17 | 100 | NA | ||||||||
| ZNF420 | Cell cycle regulation, cell death regulation | 14 | 100 | NA | ||||||||
| RASSF1α + p16INK4a + TIMP3 + PCQAP5′ + PCQAP3′ | - | HNC (SCC) | Unstimulated saliva | 88 HPV- patients and 122 controls | Volume: - Kit: The Epitect Plus DNA Bisulfite Kit (Qiagen, Hilden, Germany) | MSP | Hypermethylation | Diagnosis | 71 | 80 | 0.86 | Lim 2016 [52] |
| 45 HPV+ patients and 122 controls | 80 | 74 | 0.80 | |||||||||
| p16 | Cell cycle regulation | OC (SCC) | Saliva (Oragene® DNA Self-Collection kit) | 58 patients and 90 controls | Volume: - Kit: Oragene® DNA/Bisulfite treatment (Sigma, Burlington, MA, USA) | MSP | Hypermethylation | Diagnosis | 17 | 94 | NA | Ferlazzo 2017 [53] |
| MGMT | DNA repair | 28 | 92 | NA | ||||||||
| p16 + MGMT | - | 21 | NA | NA | ||||||||
| ZNF582 | Cell cycle regulation | OC (SCC) | Oral rinse (20 mL mouth rinse solution containing 0.12% chlorhexidine, 20 s) | 94 patients and 65 controls | Volume: 0.4 mL Kit: Epigene Nucleic Acid Extraction (iStat Biomedical, Taipei City, Taiwan)/Bisulfite conversion (iStat Biomedical, Taipei City, Taiwan) | qMSP | Hypermethylation | Diagnosis | 65 | 75 | NA | Cheng 2018 [54] |
| PAX1 | Cell differentiation | 64 | 82 | NA | ||||||||
| TRH | Cell cycle regulation, thyroid hormone regulation | OC (SCC) | Oral rinse (10 mL 0.9% NaCl, 15 s) | 42 patients and 54 controls | Volume: - Kit: QIAamp DNA FFPE Tissue (Qiagen, Hilden, Germany)/EZ DNA Methylation-Gold (Zymo Research, Irvine, CA, USA) | qMSP | Hypermethylation | Diagnosis | 88 | 93 | 0.93 | Puttipanyalears 2018 [55] |
| OPC (SCC) | 24 patients and 54 controls | 83 | 93 | 0.88 | ||||||||
| p16 + RASSF1α + TIMP3 + PCQAP/MED15 | - | OC (SCC) | Unstimulated saliva (2 mL) | 54 OC patients, 34 OPC patients and 60 controls | Volume: - Kit: DNeasy Blood and Tissue (Qiagen, Hilden, Germany)/EpiTect Plus DNA Bisulfite (Qiagen, Hilden, Germany) | MSP | Hypermethylation | Diagnosis | 92 | 92 | 0.92 | Liyanage 2019 [56] |
| OPC (SCC) | 100 | 92 | 0.97 | |||||||||
| NID2 | Cell adhesion, cell differentiation | OC (SCC) | Oral rinse (0.9% NaCl, 15 s) | 43 patients and 90 controls | Volume: - Kit: Phenol-chloroform extraction/EZ DNA Methylation (Zymo Research, Irvine, CA, USA) | qMSP | Hypermethylation | Diagnosis | 79 | 100 | NA | Srisuttee 2020 [57] |
| EDNRB | Cell signaling | OPC (SCC) | Oral rinse (15 mL NaCl) | 21 patients and 40 controls | Volume: - Kit: EpiTect Plus DNA Bisulfite (Qiagen, Hilden, Germany) | qMSP | Hypermethylation | Diagnosis/recurence detection | 72 | 95 | 0.83 | Shen 2020 [58] |
| PAX5 | Cell differentiation, cell cycle regulation | 70 | 91 | 0.78 | ||||||||
| p16INK4a | Cell cycle regulation | 17 | 100 | 0.59 | ||||||||
| p16 | Cell cycle regulation | OC (SCC) | Unstimulated saliva (5 mL) | 43 patients and 40 controls | Volume: 200 μL Kit: QIAamp DNA blood mini kit (Qiagen, Hilden, Germany)/Epitect Plus DNA Bisulfite Kit (Qiagen, Hilden, Germany) | MSP | Hypermethylation | Diagnosis | 44 | 90 | NA | Rapado-González 2021 [59] |
| RASSF1A | Cell cycle regulation | 23 | 95 | NA | ||||||||
| p16 + RASSF1A | - | 54 | 88 | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Birknerova, N.; Mancikova, V.; Paul, E.D.; Matyasovsky, J.; Cekan, P.; Palicka, V.; Parova, H. Circulating Cell-Free DNA-Based Methylation Pattern in Saliva for Early Diagnosis of Head and Neck Cancer. Cancers 2022, 14, 4882. https://doi.org/10.3390/cancers14194882
Birknerova N, Mancikova V, Paul ED, Matyasovsky J, Cekan P, Palicka V, Parova H. Circulating Cell-Free DNA-Based Methylation Pattern in Saliva for Early Diagnosis of Head and Neck Cancer. Cancers. 2022; 14(19):4882. https://doi.org/10.3390/cancers14194882
Chicago/Turabian StyleBirknerova, Natalia, Veronika Mancikova, Evan David Paul, Jan Matyasovsky, Pavol Cekan, Vladimir Palicka, and Helena Parova. 2022. "Circulating Cell-Free DNA-Based Methylation Pattern in Saliva for Early Diagnosis of Head and Neck Cancer" Cancers 14, no. 19: 4882. https://doi.org/10.3390/cancers14194882
APA StyleBirknerova, N., Mancikova, V., Paul, E. D., Matyasovsky, J., Cekan, P., Palicka, V., & Parova, H. (2022). Circulating Cell-Free DNA-Based Methylation Pattern in Saliva for Early Diagnosis of Head and Neck Cancer. Cancers, 14(19), 4882. https://doi.org/10.3390/cancers14194882





