Integrative Analysis of N6-Methyladenosine-Related Enhancer RNAs Identifies Distinct Prognosis and Tumor Immune Micro-Environment Patterns in Head and Neck Squamous Cell Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. HNSCC Samples and Cell Culture
2.2. Data Collection, Processing, and Correlation Analysis
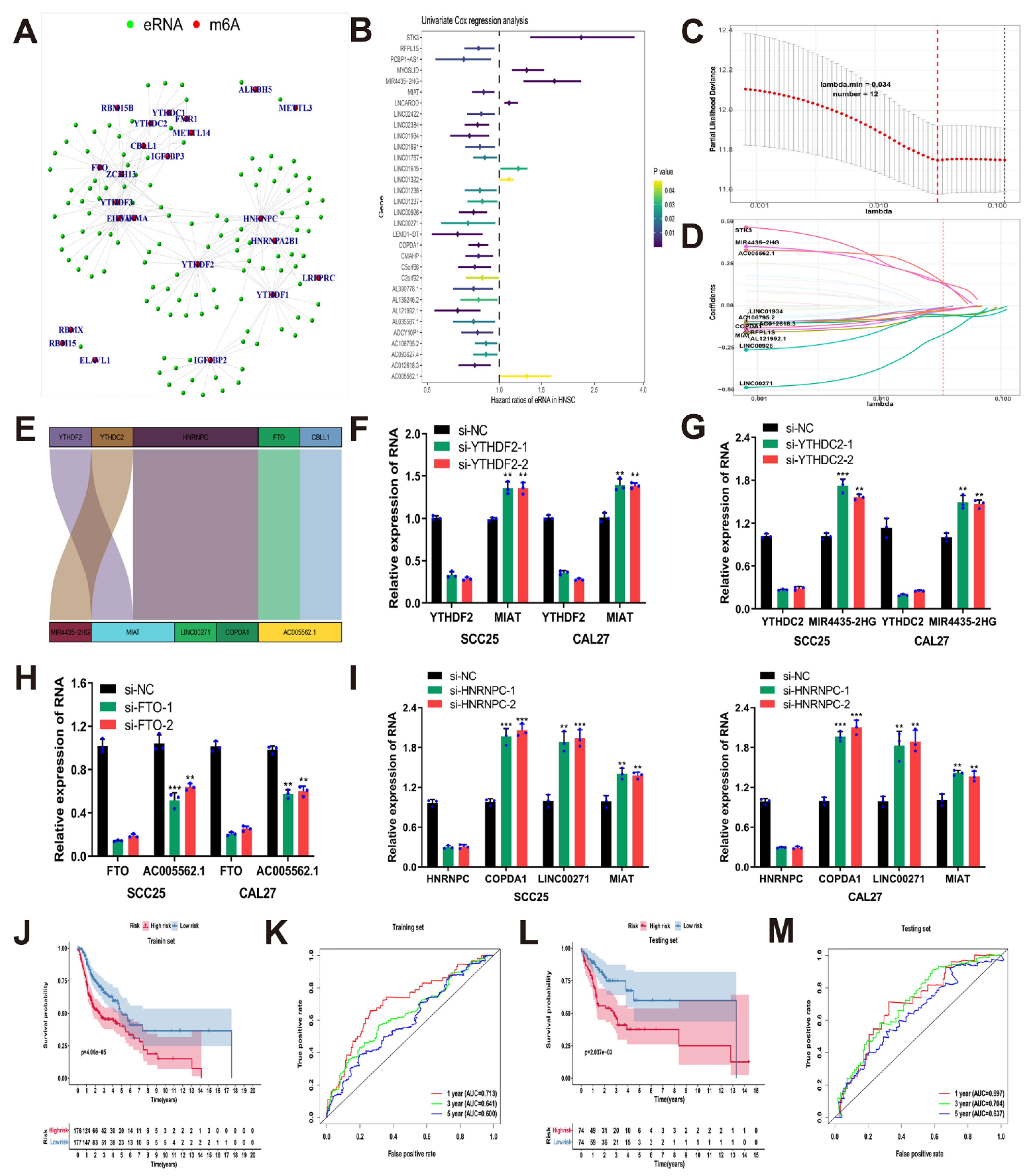
2.3. Identification of Prognostic m6A-Related eRNAs
2.4. Construction and Validation of the Risk Model Based on Prognostic m6A-Related eRNAs
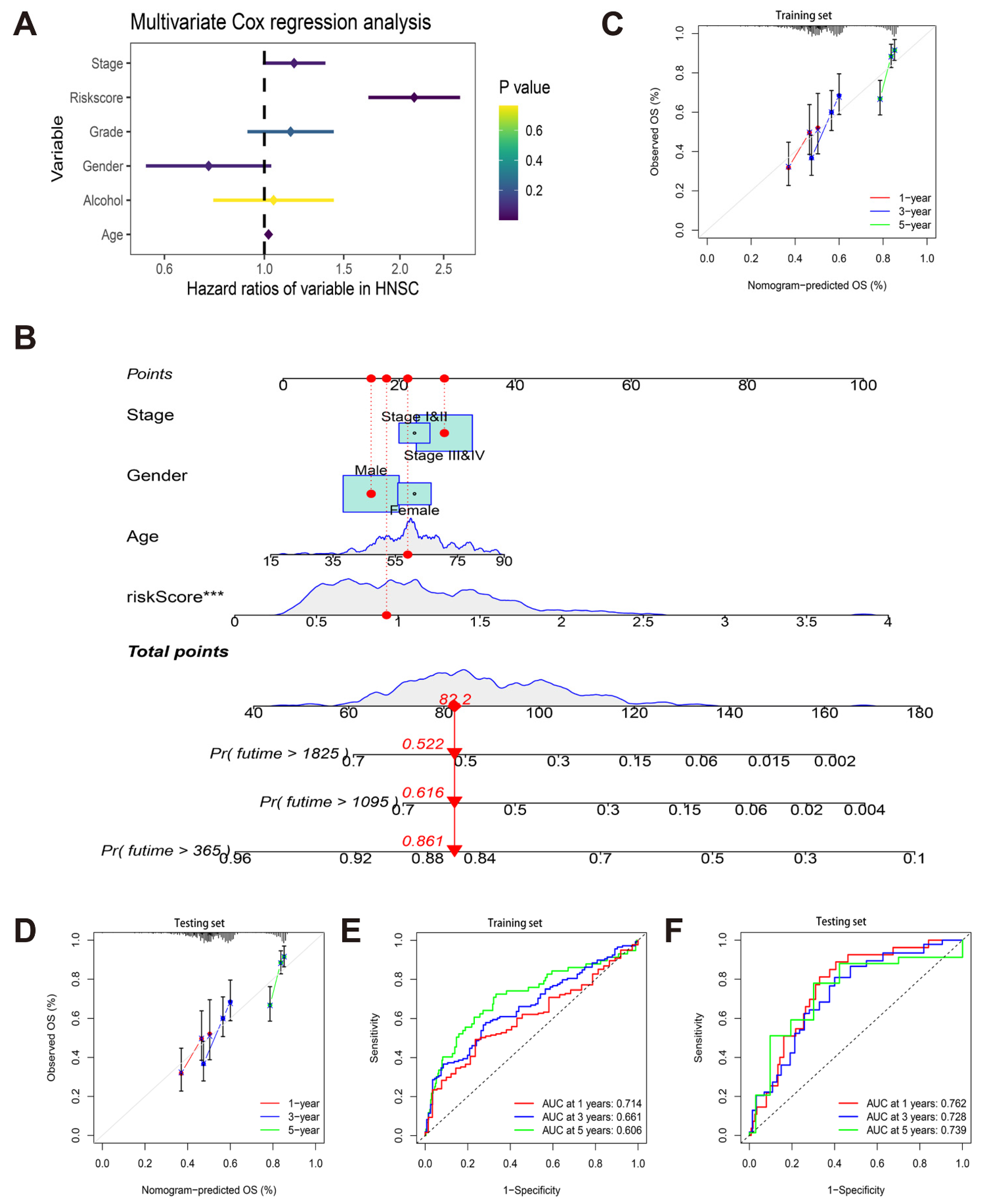
2.5. Construction and Validation of Nomogram
2.6. Gene Set Enrichment Analysis
2.7. Weighted Gene Coexpression Network Analysis
2.8. Correlation between Risk Model and Immune Cell Infiltration Patterns
2.9. Immune-Related Gene Analysis of Risk Model and Prediction of Immunotherapy Efficacy
2.10. Small Interfering RNA Transfection
2.11. Quantitative Real-Time PCR (qRT-PCR)
2.12. Wound-Healing, Migration, and Invasion Assays
2.13. Statistical Analysis
3. Results
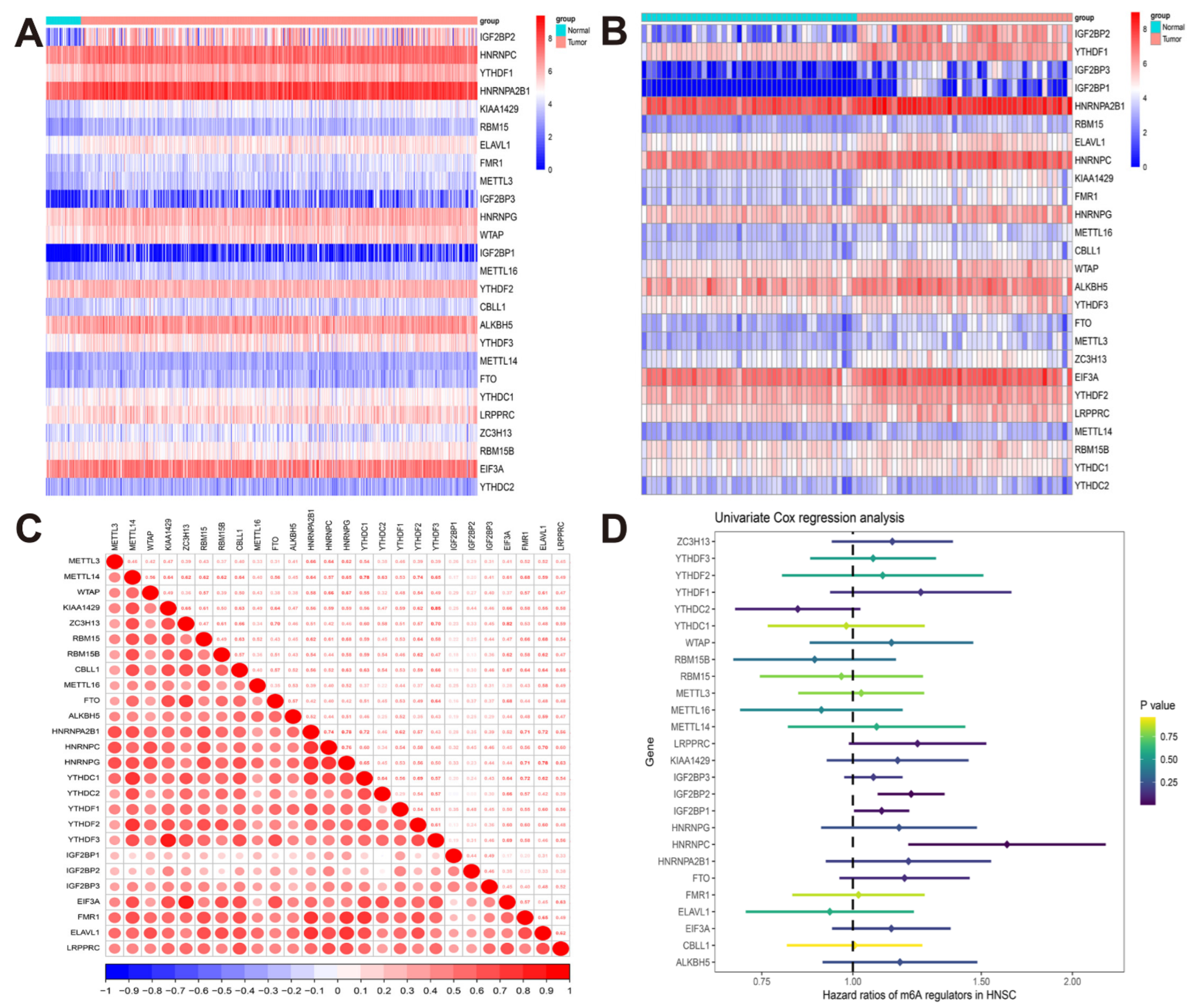
3.1. The Landscape of m6A Regulators in TCGA-HNSCC Cohort
3.2. Construction and Validation of the Risk Model Based on Prognostic m6A-Related eRNAs
3.3. Construction and Verification of a Nomogram
3.4. Identification of the Risk Model-Associated Enriched Biological Functions and Pathways
3.5. Correlation between Risk Model and Immune Cell Infiltration Patterns in TCGA-HNSCC Cohort
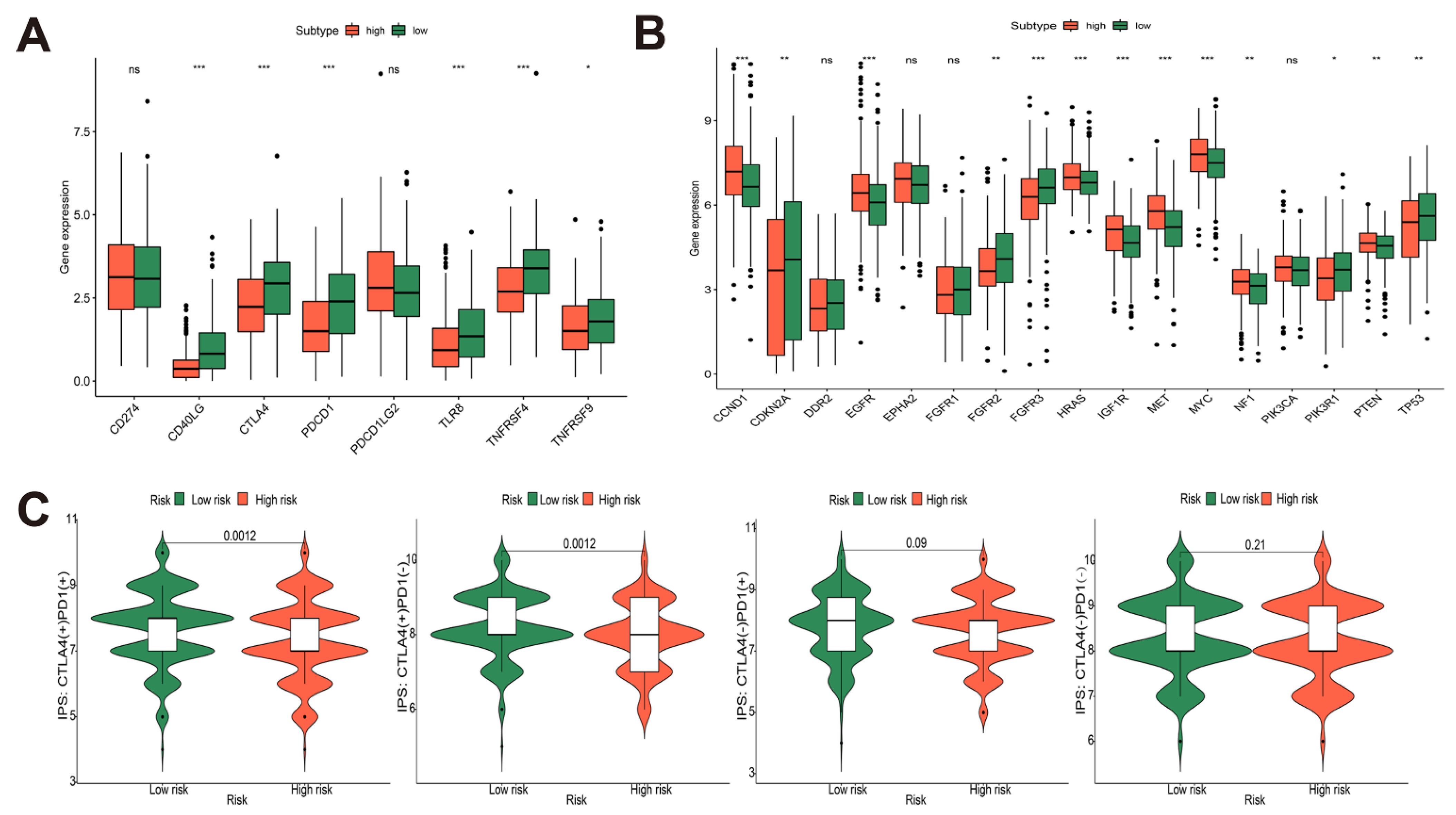
3.6. Immune-Related Gene Analysis of Risk Model and Prediction of Immunotherapy Efficacy in TCGA-HNSCC Cohort
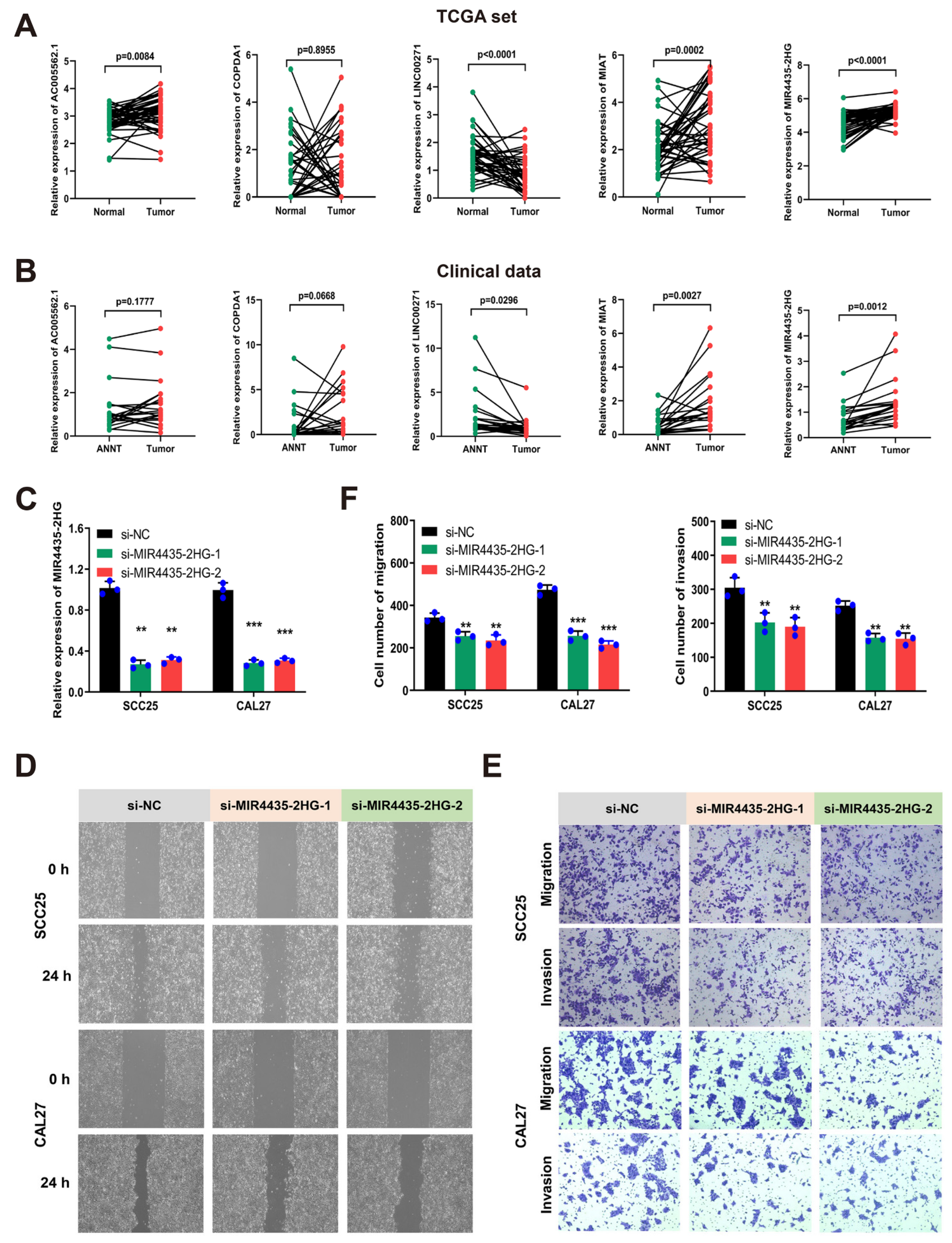
3.7. MIR4435-2HG Was Highly Expressed in HNSCC Tissues and Promoted Cell Migration and Invasion
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| m6A | N6-methyladenosine |
| m6A | N6-methyladenosine |
| HNSCC | head and neck squamous cell carcinoma |
| TCGA | The Cancer Genome Atlas |
| LASSO | least absolute shrinkage and selection operator |
| ROC | receiver operating characteristic |
| C-index | concordance index |
| GSEA | gene set enrichment analysis |
| WGCNA | weighted gene co-expression network analysis |
| SsGSEA | single-sample gene set enrichment analysis |
| OS | overall survival |
| TCIA | the Cancer Immunome Atlas |
| AUC | area under the curve |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| IPS | immunophenoscore |
| qRT-PCR | quantitative real-time PCR |
| HR | hazard ratio |
| CI | confidence interval |
| EMT | epithelial–mesenchymal transition |
References
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Prim. 2020, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Zhang, S.; Zeng, H.; Wang, S.; Sun, K.; Chen, R.; Li, L.; Wei, W.; He, J. Cancer incidence and mortality in China, 2016. J. Natl. Cancer Cent. 2022, 2, 1–9. [Google Scholar] [CrossRef]
- Chow, L.Q.M. Head and Neck Cancer. N. Engl. J. Med. 2020, 382, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, W.V.; Bell, T.A.; Schaening, C. Messenger RNA modifications: Form, distribution, and function. Science 2016, 352, 1408–1412. [Google Scholar] [CrossRef] [PubMed]
- Erson-Bensan, A.E.; Begik, O. m6A Modification and Implications for microRNAs. MicroRNA 2017, 6, 97–101. [Google Scholar] [CrossRef]
- Meng, S.; Zhou, H.; Feng, Z.; Xu, Z.; Tang, Y.; Wu, M. Epigenetics in Neurodevelopment: Emerging Role of Circular RNA. Front. Cell. Neurosci. 2019, 13, 327. [Google Scholar] [CrossRef]
- He, P.C.; He, C. m6A RNA methylation: From mechanisms to therapeutic potential. EMBO J. 2021, 40, e105977. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Z.; Yue, Y.N.; Han, D.L.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.F.; Yu, M.; Lu, Z.K.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N-6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Ping, X.L.; Sun, B.F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Feng, J.; Xue, Y.; Guan, Z.; Zhang, D.; Liu, Z.; Gong, Z.; Wang, Q.; Huang, J.; Tang, C.; et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 2016, 534, 575–578. [Google Scholar] [CrossRef]
- Patil, D.P.; Chen, C.K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef]
- Ruzicka, K.; Zhang, M.; Campilho, A.; Bodi, Z.; Kashif, M.; Saleh, M.; Eeckhout, D.; El-Showk, S.; Li, H.; Zhong, S.; et al. Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 2017, 215, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Pendleton, K.E.; Chen, B.; Liu, K.; Hunter, O.V.; Xie, Y.; Tu, B.P.; Conrad, N.K. The U6 snRNA m6A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 2017, 169, 824–835.e814. [Google Scholar] [CrossRef]
- Wen, J.; Lv, R.; Ma, H.; Shen, H.; He, C.; Wang, J.; Jiao, F.; Liu, H.; Yang, P.; Tan, L.; et al. Zc3h13 Regulates Nuclear RNA m6A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cell 2018, 69, 1028–1038.e1026. [Google Scholar] [CrossRef]
- Yue, Y.; Liu, J.; Cui, X.; Cao, J.; Luo, G.; Zhang, Z.; Cheng, T.; Gao, M.; Shu, X.; Ma, H.; et al. VIRMA mediates preferential m6A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yan, J.L.; Li, Q.; Li, J.F.; Gong, S.Z.; Zhou, H.; Gan, J.H.; Jiang, H.L.; Jia, G.F.; Luo, C.; et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015, 43, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef]
- Xu, C.; Wang, X.; Liu, K.; Roundtree, I.A.; Tempel, W.; Li, Y.; Lu, Z.; He, C.; Min, J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 2014, 10, 927–929. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Wang, X.; Lu, Z.; Zhao, B.S.; Ma, H.; Hsu, P.J.; Liu, C.; He, C. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017, 27, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Roundtree, I.A.; Wang, P.; Wang, X.; Wang, L.; Sun, C.; Tian, Y.; Li, J.; He, C.; Xu, Y. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res. 2014, 24, 1493–1496. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.J.; Zhu, Y.; Ma, H.; Guo, Y.; Shi, X.; Liu, Y.; Qi, M.; Lu, Z.; Shi, H.; Wang, J.; et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017, 27, 1115–1127. [Google Scholar] [CrossRef] [PubMed]
- Alarcon, C.R.; Goodarzi, H.; Lee, H.; Liu, X.; Tavazoie, S.; Tavazoie, S.F. HNRNPA2B1 is a Mediator of m6A-Dependent Nuclear RNA Processing Events. Cell 2015, 162, 1299–1308. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.B.; Jaffrey, S.R. 5′ UTR m6A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L.; et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018, 20, 285–295. [Google Scholar] [CrossRef]
- Liu, N.; Zhou, K.I.; Parisien, M.; Dai, Q.; Diatchenko, L.; Pan, T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017, 45, 6051–6063. [Google Scholar] [CrossRef]
- Song, N.; Cui, K.; Zhang, K.; Yang, J.; Liu, J.; Miao, Z.; Zhao, F.; Meng, H.; Chen, L.; Chen, C.; et al. The Role of m6A RNA Methylation in Cancer: Implication for Nature Products Anti-Cancer Research. Front. Pharmacol. 2022, 13, 933332. [Google Scholar] [CrossRef]
- Ling, Z.; Chen, L.; Zhao, J. m6A-dependent up-regulation of DRG1 by METTL3 and ELAVL1 promotes growth, migration, and colony formation in osteosarcoma. Biosci. Rep. 2020, 40, BSR20200282. [Google Scholar] [CrossRef]
- Wu, R.; Li, A.; Sun, B.; Sun, J.G.; Zhang, J.; Zhang, T.; Chen, Y.; Xiao, Y.; Gao, Y.; Zhang, Q.; et al. A novel m6A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 2018, 29, 23–41. [Google Scholar] [CrossRef]
- Liang, Y.; Zhan, G.; Chang, K.J.; Yang, Y.P.; Wang, L.; Lin, J.; Hsu, C.H. The roles of m6A RNA modifiers in human cancer. J. Chin. Med Assoc. 2020, 83, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Notani, D.; Rosenfeld, M.G. Enhancers as non-coding RNA transcription units: Recent insights and future perspectives. Nat. Rev. Genet. 2016, 17, 207–223. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.K.; Hemberg, M.; Gray, J.M.; Costa, A.M.; Bear, D.M.; Wu, J.; Harmin, D.A.; Laptewicz, M.; Barbara-Haley, K.; Kuersten, S.; et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 2010, 465, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Xiong, F.; Li, W. Enhancer RNAs in cancer: Regulation, mechanisms and therapeutic potential. RNA Biol. 2020, 17, 1550–1559. [Google Scholar] [CrossRef]
- McCleland, M.L.; Mesh, K.; Lorenzana, E.; Chopra, V.S.; Segal, E.; Watanabe, C.; Haley, B.; Mayba, O.; Yaylaoglu, M.; Gnad, F.; et al. CCAT1 is an enhancer-templated RNA that predicts BET sensitivity in colorectal cancer. J. Clin. Investig. 2016, 126, 639–652. [Google Scholar] [CrossRef]
- Zhang, Z.; Luo, M.; Li, Q.; Liu, Y.; Lussier, C.; Zhang, J.; Ye, Y.; Guo, A.Y.; Han, L. Genetic, Pharmacogenomic, and Immune Landscapes of Enhancer RNAs Across Human Cancers. Cancer Res. 2022, 82, 785–790. [Google Scholar] [CrossRef]
- Lee, J.H.; Wang, R.; Xiong, F.; Krakowiak, J.; Liao, Z.; Nguyen, P.T.; Moroz-Omori, E.V.; Shao, J.; Zhu, X.; Bolt, M.J.; et al. Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation. Mol. Cell 2021, 81, 3368–3385.e3369. [Google Scholar] [CrossRef]
- Simon, N.; Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Cox’s Proportional Hazards Model via Coordinate Descent. J. Stat. Softw. 2011, 39, 1–13. [Google Scholar] [CrossRef]
- Hou, C.; Cai, H.; Zhu, Y.; Huang, S.; Song, F.; Hou, J. Development and Validation of Autophagy-Related Gene Signature and Nomogram for Predicting Survival in Oral Squamous Cell Carcinoma. Front. Oncol. 2020, 10, 558596. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Khodadoust, M.S.; Liu, C.L.; Newman, A.M.; Alizadeh, A.A. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol. Biol. 2018, 1711, 243–259. [Google Scholar] [CrossRef]
- Hanzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Yoshihara, K.; Kim, H.; Verhaak, R.G. Estimate: Estimate of Stromal and Immune Cells in Malignant Tumor Tissues from Expression Data. Available online: https://R-Forge.R-project.org/projects/estimate/ (accessed on 26 November 2021).
- Cramer, J.D.; Burtness, B.; Le, Q.T.; Ferris, R.L. The changing therapeutic landscape of head and neck cancer. Nat. Rev. Clin. Oncol. 2019, 16, 669–683. [Google Scholar] [CrossRef] [PubMed]
- Van Allen, E.M.; Miao, D.; Schilling, B.; Shukla, S.A.; Blank, C.; Zimmer, L.; Sucker, A.; Hillen, U.; Foppen, M.H.G.; Goldinger, S.M.; et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015, 350, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Hugo, W.; Zaretsky, J.M.; Sun, L.; Song, C.; Moreno, B.H.; Hu-Lieskovan, S.; Berent-Maoz, B.; Pang, J.; Chmielowski, B.; Cherry, G.; et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell 2016, 165, 35–44. [Google Scholar] [CrossRef]
- Cai, H.; Li, J.; Zhang, Y.; Liao, Y.; Zhu, Y.; Wang, C.; Hou, J. LDHA Promotes Oral Squamous Cell Carcinoma Progression through Facilitating Glycolysis and Epithelial-Mesenchymal Transition. Front. Oncol. 2019, 9, 1446. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Li, H.; Cai, H.; Liu, W.; Pan, E.; Yu, D.; He, S. Upregulation of IGF2BP2 Promotes Oral Squamous Cell Carcinoma Progression That Is Related to Cell Proliferation, Metastasis and Tumor-Infiltrating Immune Cells. Front. Oncol. 2022, 12, 809589. [Google Scholar] [CrossRef] [PubMed]
- Sartorelli, V.; Lauberth, S.M. Enhancer RNAs are an important regulatory layer of the epigenome. Nat. Struct. Mol. Biol. 2020, 27, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zeng, P.; Li, Y.H.; Zhang, Z.; Cui, Q. SRAMP: Prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016, 44, e91. [Google Scholar] [CrossRef]
- Carlisle, J.W.; Steuer, C.E.; Owonikoko, T.K.; Saba, N.F. An update on the immune landscape in lung and head and neck cancers. CA Cancer J. Clin. 2020, 70, 505–517. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Z. The history and advances in cancer immunotherapy: Understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell. Mol. Immunol. 2020, 17, 807–821. [Google Scholar] [CrossRef] [PubMed]
- Da, C.M.; Gong, C.Y.; Nan, W.; Zhou, K.S.; Wu, Z.L.; Zhang, H.H. The role of long non-coding RNA MIAT in cancers. Biomed. Pharmacother. 2020, 129, 110359. [Google Scholar] [CrossRef] [PubMed]
- Ghasemian, M.; Rajabibazl, M.; Sahebi, U.; Sadeghi, S.; Maleki, R.; Hashemnia, V.; Mirfakhraie, R. Long non-coding RNA MIR4435-2HG: A key molecule in progression of cancer and non-cancerous disorders. Cancer Cell Int. 2022, 22, 215. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chen, X.; Qiao, T. Long noncoding RNA MIR44352HG promotes the progression of head and neck squamous cell carcinoma by regulating the miR3835p/RBM3 axis. Oncol. Rep. 2021, 45, 99. [Google Scholar] [CrossRef]
- Zhang, Z.; Lee, J.H.; Ruan, H.; Ye, Y.; Krakowiak, J.; Hu, Q.; Xiang, Y.; Gong, J.; Zhou, B.; Wang, L.; et al. Transcriptional landscape and clinical utility of enhancer RNAs for eRNA-targeted therapy in cancer. Nat. Commun. 2019, 10, 4562. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liang, H. A High-Resolution Map of Human Enhancer RNA Loci Characterizes Super-enhancer Activities in Cancer. Cancer Cell 2020, 38, 701–715.e705. [Google Scholar] [CrossRef] [PubMed]
- Helmink, B.A.; Reddy, S.M.; Gao, J.; Zhang, S.; Basar, R.; Thakur, R.; Yizhak, K.; Sade-Feldman, M.; Blando, J.; Han, G.; et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 2020, 577, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Borst, J.; Ahrends, T.; Bąbała, N.; Melief, C.J.M.; Kastenmüller, W. CD4+ T cell help in cancer immunology and immunotherapy. Nat. Rev. Immunol. 2019, 18, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Philip, M.A.-O.; Schietinger, A.A.-O. CD8+ T cell differentiation and dysfunction in cancer. Nat. Rev. Immunol. 2022, 22, 209–223. [Google Scholar] [CrossRef]
- Kraehenbuehl, L.A.-O.; Weng, C.A.-O.; Eghbali, S.A.-O.; Wolchok, J.A.-O.; Merghoub, T.A.-O. Enhancing immunotherapy in cancer by targeting emerging immunomodulatory pathways. Nat. Rev. Clin. Oncol. 2021, 19, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Bedard, P.L.; Hyman, D.M.; Davids, M.S.; Siu, L.L. Small molecules, big impact: 20 years of targeted therapy in oncology. Lancet 2020, 395, 1011–1088. [Google Scholar] [CrossRef]
- Fasano, M.; Corte, C.M.D.; Liello, R.D.; Viscardi, G.; Sparano, F.; Iacovino, M.L.; Paragliola, F.; Piccolo, A.; Napolitano, S.; Martini, G.; et al. Immunotherapy for head and neck cancer: Present and future. Crit. Rev. Oncol. Hematol. 2022, 174, 103679. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.; Bonner, J.A.; Bredel, M. EGFR Mutations in Head and Neck Squamous Cell Carcinoma. Int. J. Mol. Sci. 2022, 23, 3818. [Google Scholar] [CrossRef] [PubMed]


| Id | Coefficient | HR | HR.95 Low | HR.95 High | p-Value |
|---|---|---|---|---|---|
| MIR4435-2HG | 0.354 | 1.425 | 1.015 | 2.002 | 0.041 |
| LINC00271 | −0.395 | 0.674 | 0.491 | 0.924 | 0.014 |
| COPDA1 | −0.113 | 0.893 | 0.792 | 1.006 | 0.063 |
| AC005562.1 | 0.391 | 1.478 | 1.118 | 1.955 | 0.006 |
| MIAT | −0.131 | 0.878 | 0.765 | 1.007 | 0.062 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, H.; Liang, J.; Jiang, Y.; Tan, R.; Hou, C.; Hou, J. Integrative Analysis of N6-Methyladenosine-Related Enhancer RNAs Identifies Distinct Prognosis and Tumor Immune Micro-Environment Patterns in Head and Neck Squamous Cell Carcinoma. Cancers 2022, 14, 4657. https://doi.org/10.3390/cancers14194657
Cai H, Liang J, Jiang Y, Tan R, Hou C, Hou J. Integrative Analysis of N6-Methyladenosine-Related Enhancer RNAs Identifies Distinct Prognosis and Tumor Immune Micro-Environment Patterns in Head and Neck Squamous Cell Carcinoma. Cancers. 2022; 14(19):4657. https://doi.org/10.3390/cancers14194657
Chicago/Turabian StyleCai, Hongshi, Jianfeng Liang, Yaoqi Jiang, Rukeng Tan, Chen Hou, and Jinsong Hou. 2022. "Integrative Analysis of N6-Methyladenosine-Related Enhancer RNAs Identifies Distinct Prognosis and Tumor Immune Micro-Environment Patterns in Head and Neck Squamous Cell Carcinoma" Cancers 14, no. 19: 4657. https://doi.org/10.3390/cancers14194657
APA StyleCai, H., Liang, J., Jiang, Y., Tan, R., Hou, C., & Hou, J. (2022). Integrative Analysis of N6-Methyladenosine-Related Enhancer RNAs Identifies Distinct Prognosis and Tumor Immune Micro-Environment Patterns in Head and Neck Squamous Cell Carcinoma. Cancers, 14(19), 4657. https://doi.org/10.3390/cancers14194657





