Preservation of the Hypoxic Transcriptome in Glioblastoma Patient-Derived Cell Lines Maintained at Lowered Oxygen Tension
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Subjects
2.2. Materials
2.3. GBM Tissue Processing, GBM Cell Cultures Establishment and Maintenance
2.4. Cell Treatment and Viability Assay
2.5. Flow Cytometry
2.6. Nucleic Acids Isolation
2.7. RNA Sequencing and Analysis
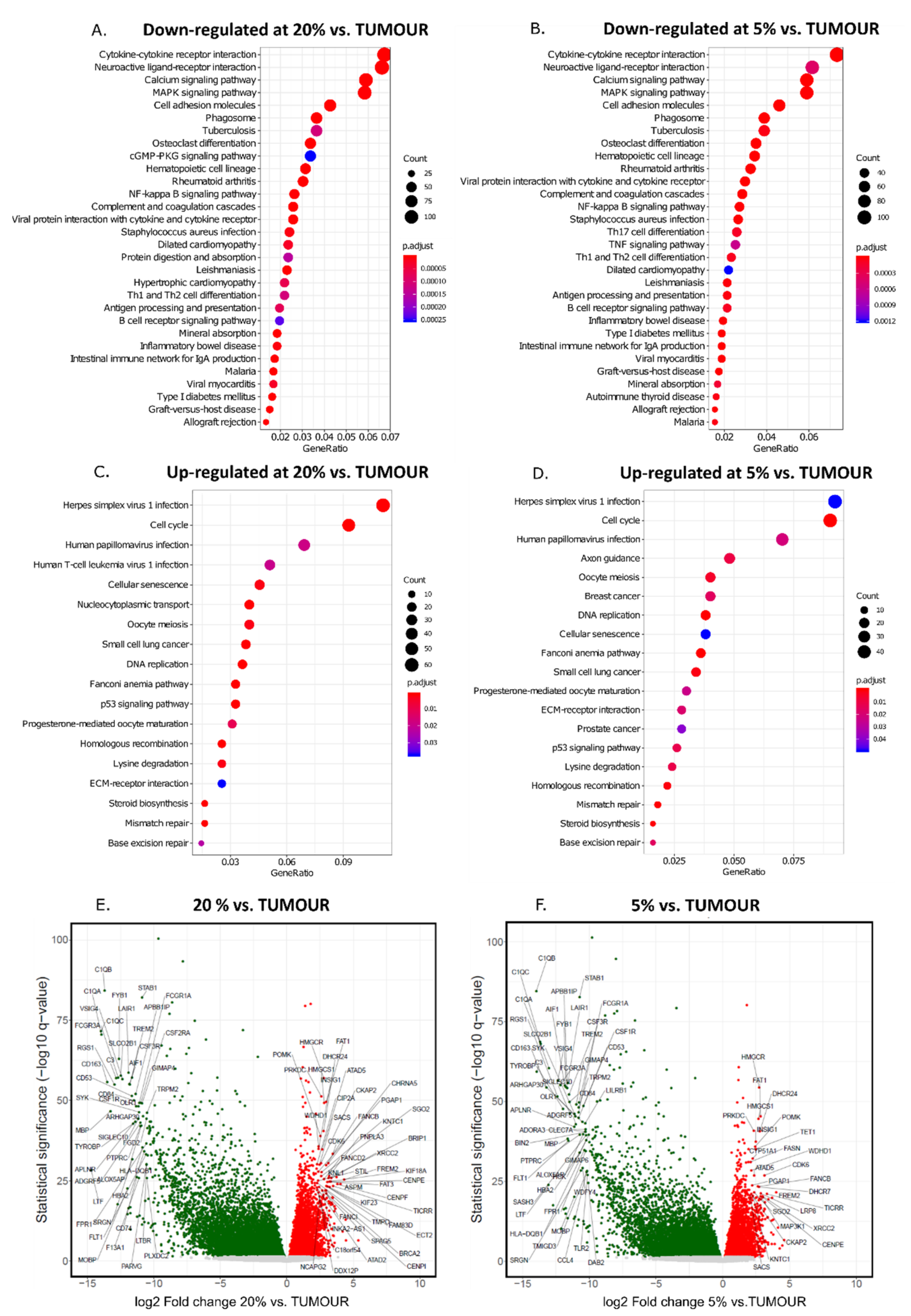
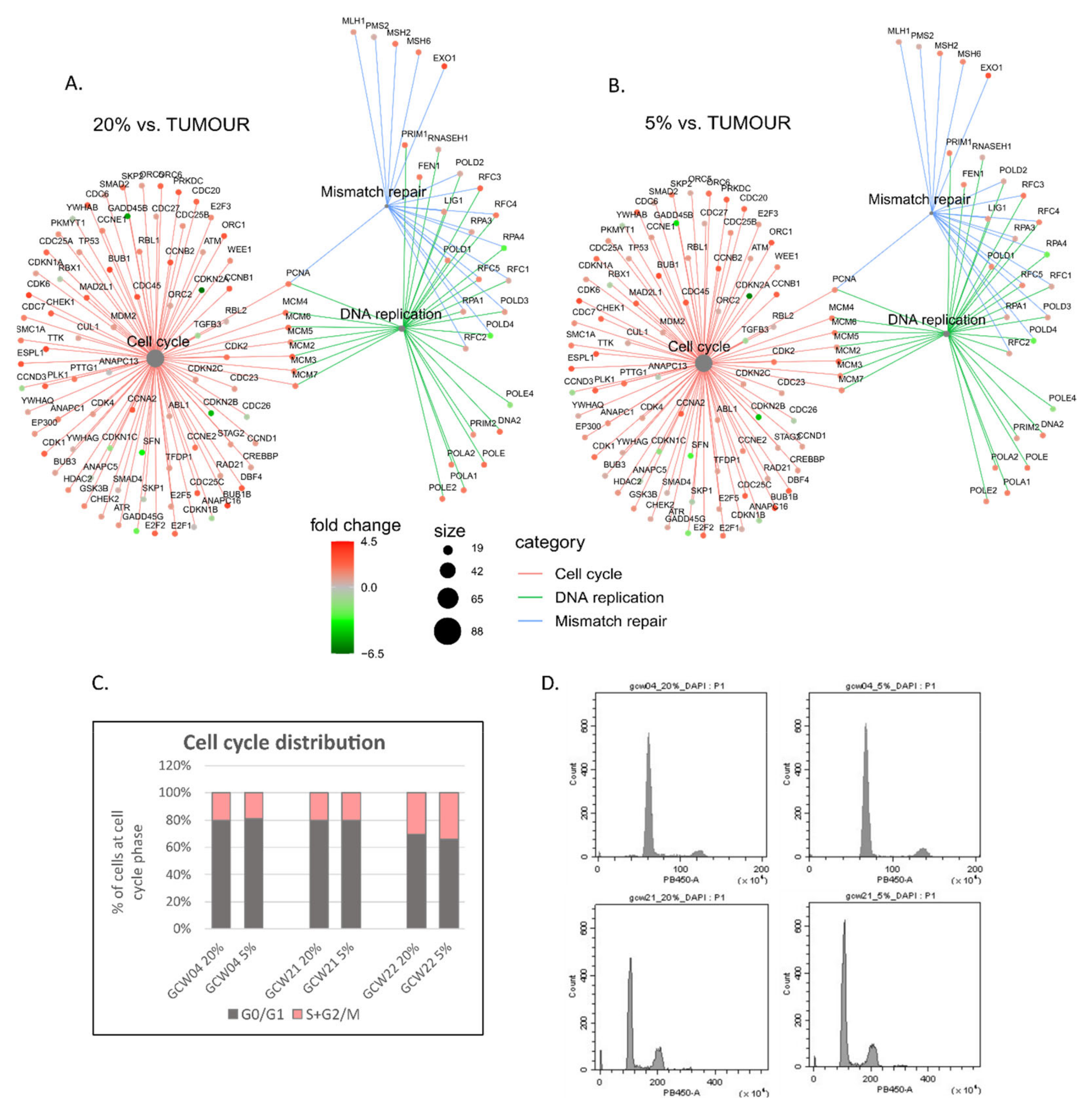
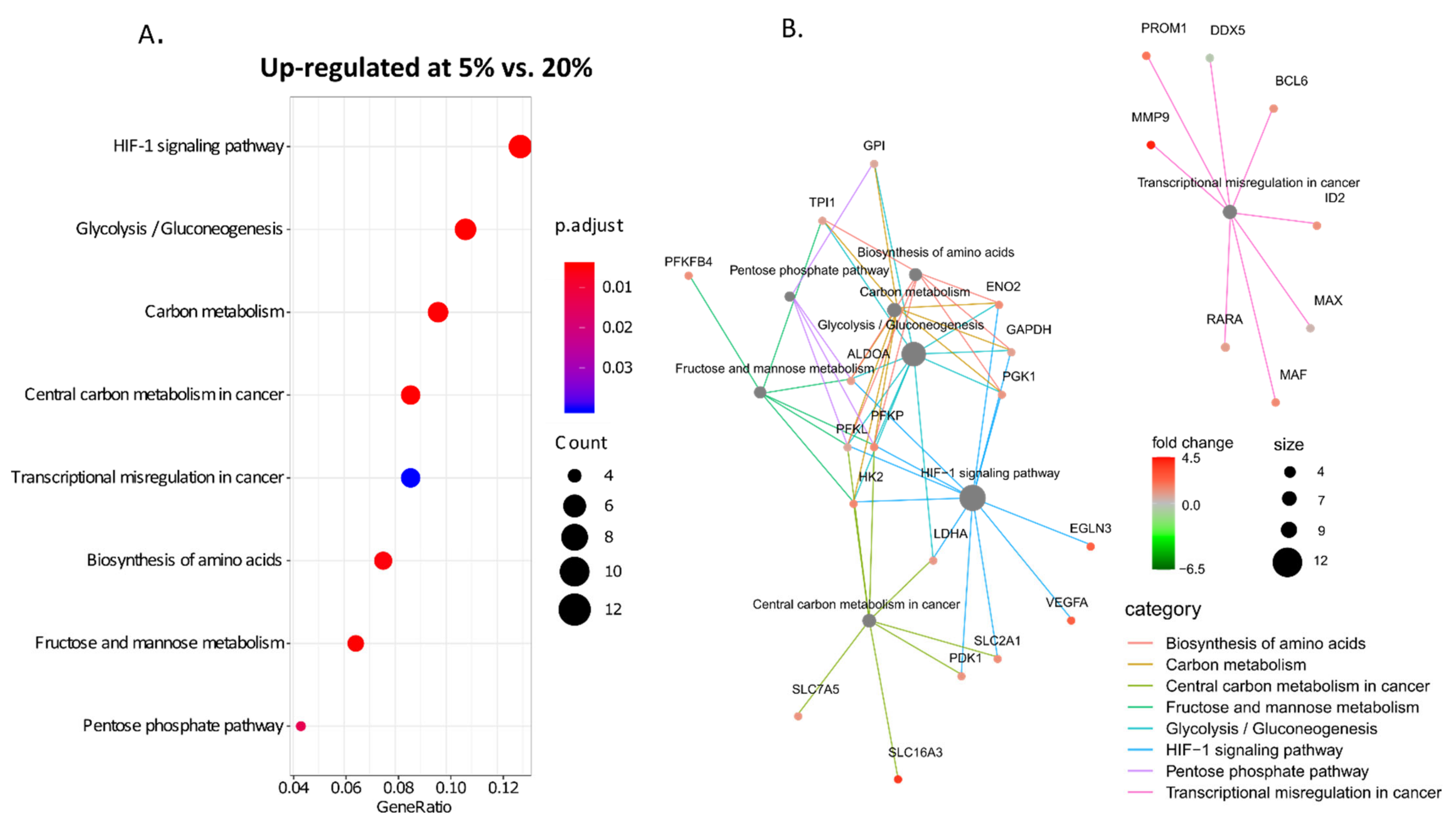
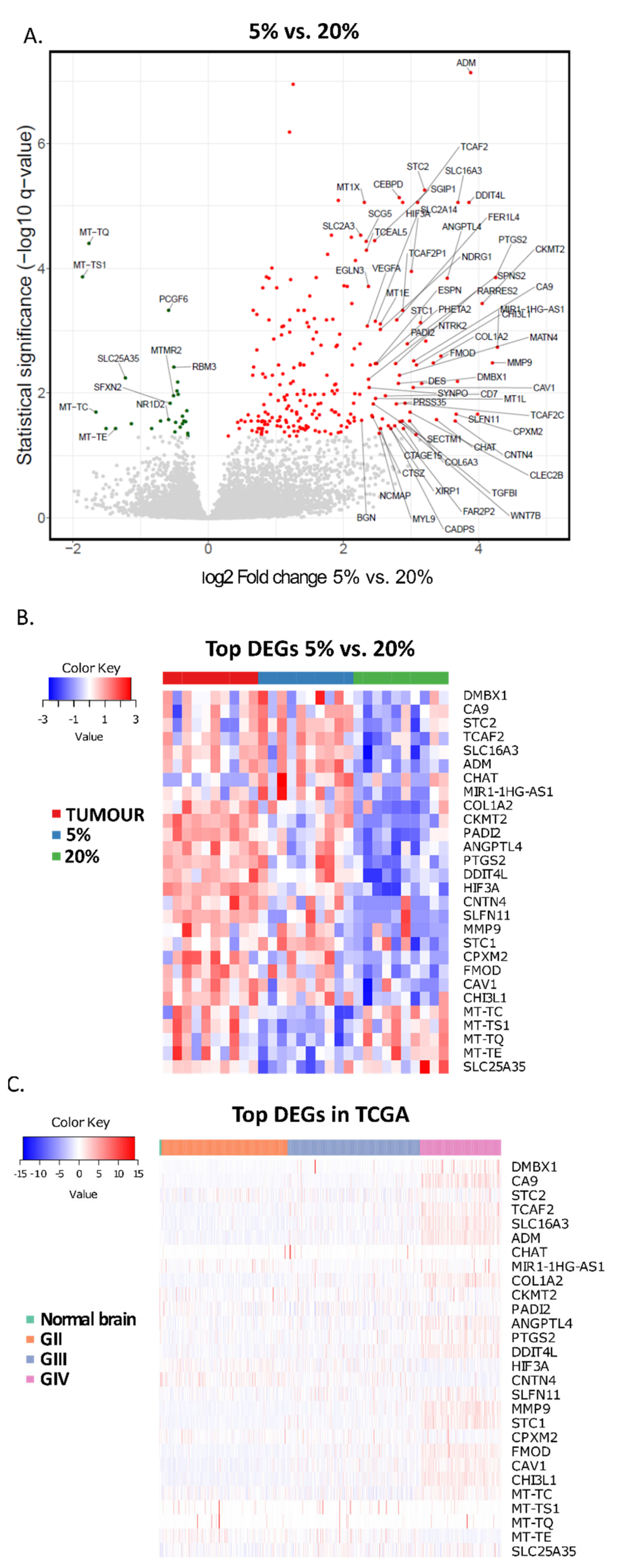
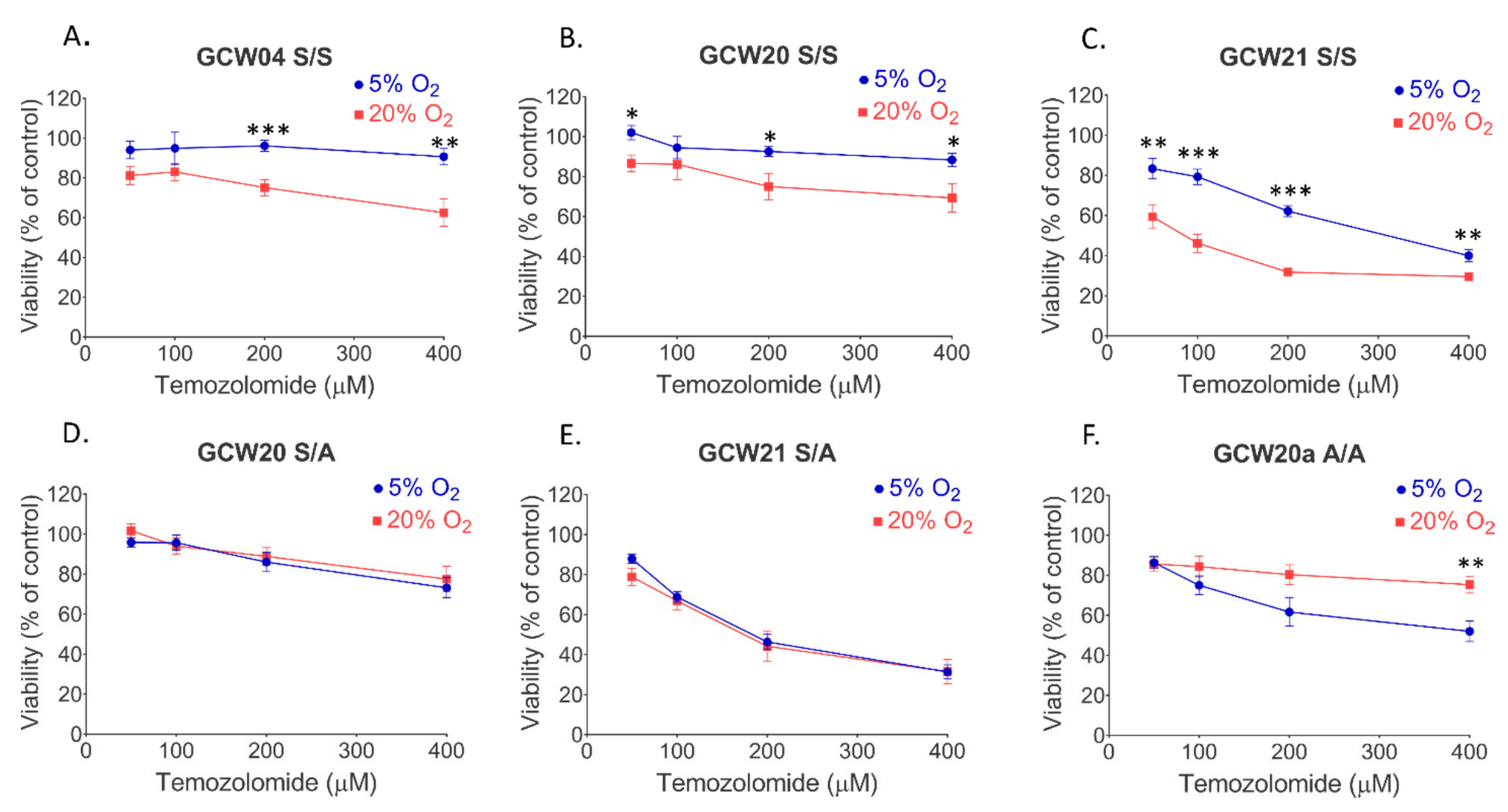
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wen, P.Y.; Weller, M.; Lee, E.Q.; Alexander, B.M.; Barnholtz-Sloan, J.S.; Barthel, F.P.; Batchelor, T.T.; Bindra, R.S.; Chang, S.M.; Chiocca, E.A.; et al. Glioblastoma in Adults: A Society for Neuro-Oncology (SNO) and European Society of Neuro-Oncology (EANO) Consensus Review on Current Management and Future Directions. Neuro-Oncol. 2020, 22, 1073–1113. [Google Scholar] [CrossRef]
- Di Nunno, V.; Franceschi, E.; Tosoni, A.; Gatto, L.; Lodi, R.; Bartolini, S.; Brandes, A.A. Glioblastoma: Emerging Treatments and Novel Trial Designs. Cancers 2021, 13, 3750. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-Cell RNA-Seq Highlights Intratumoral Heterogeneity in Primary Glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef] [PubMed]
- Sottoriva, A.; Spiteri, I.; Piccirillo, S.G.M.; Touloumis, A.; Collins, V.P.; Marioni, J.C.; Curtis, C.; Watts, C.; Tavare, S. Intratumor Heterogeneity in Human Glioblastoma Reflects Cancer Evolutionary Dynamics. Proc. Natl. Acad. Sci. USA 2013, 110, 4009–4014. [Google Scholar] [CrossRef]
- Dirkse, A.; Golebiewska, A.; Buder, T.; Nazarov, P.V.; Muller, A.; Poovathingal, S.; Brons, N.H.C.; Leite, S.; Sauvageot, N.; Sarkisjan, D.; et al. Stem Cell-Associated Heterogeneity in Glioblastoma Results from Intrinsic Tumor Plasticity Shaped by the Microenvironment. Nat. Commun. 2019, 10, 1787. [Google Scholar] [CrossRef]
- Becker, A.; Sells, B.; Haque, S.; Chakravarti, A. Tumor Heterogeneity in Glioblastomas: From Light Microscopy to Molecular Pathology. Cancers 2021, 13, 761. [Google Scholar] [CrossRef]
- Vartanian, A.; Singh, S.K.; Agnihotri, S.; Jalali, S.; Burrell, K.; Aldape, K.D.; Zadeh, G. GBM’s Multifaceted Landscape: Highlighting Regional and Microenvironmental Heterogeneity. Neuro-Oncol. 2014, 16, 1167–1175. [Google Scholar] [CrossRef]
- Boyd, N.H.; Tran, A.N.; Bernstock, J.D.; Etminan, T.; Jones, A.B.; Gillespie, G.Y.; Friedman, G.K.; Hjelmeland, A.B. Glioma Stem Cells and Their Roles within the Hypoxic Tumor Microenvironment. Theranostics 2021, 11, 665–683. [Google Scholar] [CrossRef]
- Gérard, M.; Corroyer-Dulmont, A.; Lesueur, P.; Collet, S.; Chérel, M.; Bourgeois, M.; Stefan, D.; Limkin, E.J.; Perrio, C.; Guillamo, J.-S.; et al. Hypoxia Imaging and Adaptive Radiotherapy: A State-of-the-Art Approach in the Management of Glioma. Front. Med. 2019, 6, 117. [Google Scholar] [CrossRef]
- Evans, S.M.; Judy, K.D.; Dunphy, I.; Jenkins, W.T.; Hwang, W.-T.; Nelson, P.T.; Lustig, R.A.; Jenkins, K.; Magarelli, D.P.; Hahn, S.M.; et al. Hypoxia Is Important in the Biology and Aggression of Human Glial Brain Tumors. Clin. Cancer Res. 2004, 10, 8177–8184. [Google Scholar] [CrossRef] [PubMed]
- Gerstner, E.R.; Zhang, Z.; Fink, J.R.; Muzi, M.; Hanna, L.; Greco, E.; Prah, M.; Schmainda, K.M.; Mintz, A.; Kostakoglu, L.; et al. ACRIN 6684: Assessment of Tumor Hypoxia in Newly Diagnosed Glioblastoma Using 18 F-FMISO PET and MRI. Clin. Cancer Res. 2016, 22, 5079–5086. [Google Scholar] [CrossRef]
- Ahir, B.K.; Engelhard, H.H.; Lakka, S.S. Tumor Development and Angiogenesis in Adult Brain Tumor: Glioblastoma. Mol. Neurobiol. 2020, 57, 2461–2478. [Google Scholar] [CrossRef] [PubMed]
- Graham, K.; Unger, E. Overcoming Tumor Hypoxia as a Barrier to Radiotherapy, Chemotherapy and Immunotherapy in Cancer Treatment. Int. J. Nanomed. 2018, 13, 6049–6058. [Google Scholar] [CrossRef]
- Bindra, R.S.; Chalmers, A.J.; Evans, S.; Dewhirst, M. GBM Radiosensitizers: Dead in the Water…or Just the Beginning? J. Neurooncol. 2017, 134, 513–521. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Domènech, M.; Hernández, A.; Plaja, A.; Martínez-Balibrea, E.; Balañà, C. Hypoxia: The Cornerstone of Glioblastoma. Int. J. Mol. Sci. 2021, 22, 12608. [Google Scholar] [CrossRef]
- Chédeville, A.L.; Madureira, P.A. The Role of Hypoxia in Glioblastoma Radiotherapy Resistance. Cancers 2021, 13, 542. [Google Scholar] [CrossRef]
- Mendichovszky, I.; Jackson, A. Imaging Hypoxia in Gliomas. Br. J. Radiol. 2011, 84, S145–S158. [Google Scholar] [CrossRef]
- Carreau, A.; Hafny-Rahbi, B.E.; Matejuk, A.; Grillon, C.; Kieda, C. Why Is the Partial Oxygen Pressure of Human Tissues a Crucial Parameter? Small Molecules and Hypoxia. J. Cell. Mol. Med. 2011, 15, 1239–1253. [Google Scholar] [CrossRef]
- Jin, X.; Kim, L.J.Y.; Wu, Q.; Wallace, L.C.; Prager, B.C.; Sanvoranart, T.; Gimple, R.C.; Wang, X.; Mack, S.C.; Miller, T.E.; et al. Targeting Glioma Stem Cells through Combined BMI1 and EZH2 Inhibition. Nat. Med. 2017, 23, 1352–1361. [Google Scholar] [CrossRef]
- Bronisz, A.; Salińska, E.; Chiocca, E.A.; Godlewski, J. Hypoxic Roadmap of Glioblastoma—Learning about Directions and Distances in the Brain Tumor Environment. Cancers 2020, 12, 1213. [Google Scholar] [CrossRef]
- Bekaert, L.; Valable, S.; Lechapt-Zalcman, E.; Ponte, K.; Collet, S.; Constans, J.-M.; Levallet, G.; Bordji, K.; Petit, E.; Branger, P.; et al. [18F]-FMISO PET Study of Hypoxia in Gliomas before Surgery: Correlation with Molecular Markers of Hypoxia and Angiogenesis. Eur. J. Nucl. Med. Mol. Imaging 2017, 44, 1383–1392. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, D.C.J.; Smerdon, G.R.; Harries, L.W.; Dodd, N.J.F.; Murphy, M.P.; Curnow, A.; Winyard, P.G. Altered Cellular Redox Homeostasis and Redox Responses under Standard Oxygen Cell Culture Conditions versus Physioxia. Free Radic. Biol. Med. 2018, 126, 322–333. [Google Scholar] [CrossRef] [PubMed]
- Timpano, S.; Guild, B.D.; Specker, E.J.; Melanson, G.; Medeiros, P.J.; Sproul, S.L.J.; Uniacke, J. Physioxic Human Cell Culture Improves Viability, Metabolism, and Mitochondrial Morphology While Reducing DNA Damage. FASEB J. 2019, 33, 5716–5728. [Google Scholar] [CrossRef] [PubMed]
- McCord, A.M.; Jamal, M.; Shankavarum, U.T.; Lang, F.F.; Camphausen, K.; Tofilon, P.J. Physiologic Oxygen Concentration Enhances the Stem-Like Properties of CD133 + Human Glioblastoma Cells In Vitro. Mol. Cancer Res. 2009, 7, 489–497. [Google Scholar] [CrossRef]
- Platet, N.; Liu, S.Y.; Atifi, M.E.; Oliver, L.; Vallette, F.M.; Berger, F.; Wion, D. Influence of Oxygen Tension on CD133 Phenotype in Human Glioma Cell Cultures. Cancer Lett. 2007, 258, 286–290. [Google Scholar] [CrossRef]
- Soeda, A.; Park, M.; Lee, D.; Mintz, A.; Androutsellis-Theotokis, A.; McKay, R.D.; Engh, J.; Iwama, T.; Kunisada, T.; Kassam, A.B.; et al. Hypoxia Promotes Expansion of the CD133-Positive Glioma Stem Cells through Activation of HIF-1α. Oncogene 2009, 28, 3949–3959. [Google Scholar] [CrossRef]
- Nishikawa, M.; Inoue, A.; Ohnishi, T.; Yano, H.; Ozaki, S.; Kanemura, Y.; Suehiro, S.; Ohtsuka, Y.; Kohno, S.; Ohue, S.; et al. Hypoxia-Induced Phenotypic Transition from Highly Invasive to Less Invasive Tumors in Glioma Stem-like Cells: Significance of CD44 and Osteopontin as Therapeutic Targets in Glioblastoma. Transl. Oncol. 2021, 14, 101137. [Google Scholar] [CrossRef]
- Bar, E.E.; Lin, A.; Mahairaki, V.; Matsui, W.; Eberhart, C.G. Hypoxia Increases the Expression of Stem-Cell Markers and Promotes Clonogenicity in Glioblastoma Neurospheres. Am. J. Pathol. 2010, 177, 1491–1502. [Google Scholar] [CrossRef]
- Leelatian, N.; Doxie, D.B.; Greenplate, A.R.; Sinnaeve, J.; Ihrie, R.A.; Irish, J.M. Preparing Viable Single Cells from Human Tissue and Tumors for Cytomic Analysis. Curr. Protoc. Mol. Biol. 2017, 118, 25C.1.1–25C.1.23. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. ClusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, M.; Barthel, F.P.; Malta, T.M.; Sabedot, T.S.; Salama, S.R.; Murray, B.A.; Morozova, O.; Newton, Y.; Radenbaugh, A.; Pagnotta, S.M.; et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell 2016, 164, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Verhaak, R.G.W.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated Genomic Analysis Identifies Clinically Relevant Subtypes of Glioblastoma Characterized by Abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef]
- Galli, R.; Binda, E.; Orfanelli, U.; Cipelletti, B.; Gritti, A.; De Vitis, S.; Fiocco, R.; Foroni, C.; Dimeco, F.; Vescovi, A. Isolation and Characterization of Tumorigenic, Stem-like Neural Precursors from Human Glioblastoma. Cancer Res. 2004, 64, 7011–7021. [Google Scholar] [CrossRef]
- Yuan, X.; Curtin, J.; Xiong, Y.; Liu, G.; Waschsmann-Hogiu, S.; Farkas, D.L.; Black, K.L.; Yu, J.S. Isolation of Cancer Stem Cells from Adult Glioblastoma Multiforme. Oncogene 2004, 23, 9392–9400. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kotliarova, S.; Kotliarov, Y.; Li, A.; Su, Q.; Donin, N.M.; Pastorino, S.; Purow, B.W.; Christopher, N.; Zhang, W.; et al. Tumor Stem Cells Derived from Glioblastomas Cultured in BFGF and EGF More Closely Mirror the Phenotype and Genotype of Primary Tumors than Do Serum-Cultured Cell Lines. Cancer Cell 2006, 9, 391–403. [Google Scholar] [CrossRef]
- Chen, J.; Li, Y.; Yu, T.-S.; McKay, R.M.; Burns, D.K.; Kernie, S.G.; Parada, L.F. A Restricted Cell Population Propagates Glioblastoma Growth after Chemotherapy. Nature 2012, 488, 522–526. [Google Scholar] [CrossRef]
- Bao, S.; Wu, Q.; McLendon, R.E.; Hao, Y.; Shi, Q.; Hjelmeland, A.B.; Dewhirst, M.W.; Bigner, D.D.; Rich, J.N. Glioma Stem Cells Promote Radioresistance by Preferential Activation of the DNA Damage Response. Nature 2006, 444, 756–760. [Google Scholar] [CrossRef]
- Liu, G.; Yuan, X.; Zeng, Z.; Tunici, P.; Ng, H.; Abdulkadir, I.R.; Lu, L.; Irvin, D.; Black, K.L.; Yu, J.S. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol. Cancer 2006, 5, 67. [Google Scholar] [CrossRef]
- Garcion, E. In Vitro Expansion of Human Glioblastoma Cells at Non-Physiological Oxygen Tension Irreversibly Alters Subsequent in Vivo Aggressiveness and AC133 Expression. Int. J. Oncol. 2011, 40, 1220–1229. [Google Scholar] [CrossRef] [PubMed]
- Iwadate, Y. Epithelial-Mesenchymal Transition in Glioblastoma Progression. Oncol. Lett. 2016, 11, 1615–1620. [Google Scholar] [CrossRef] [PubMed]
- Joseph, J.V.; Conroy, S.; Pavlov, K.; Sontakke, P.; Tomar, T.; Eggens-Meijer, E.; Balasubramaniyan, V.; Wagemakers, M.; den Dunnen, W.F.A.; Kruyt, F.A.E. Hypoxia Enhances Migration and Invasion in Glioblastoma by Promoting a Mesenchymal Shift Mediated by the HIF1α–ZEB1 Axis. Cancer Lett. 2015, 359, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Depner, C.; zum Buttel, H.; Böğürcü, N.; Cuesta, A.M.; Aburto, M.R.; Seidel, S.; Finkelmeier, F.; Foss, F.; Hofmann, J.; Kaulich, K.; et al. EphrinB2 Repression through ZEB2 Mediates Tumour Invasion and Anti-Angiogenic Resistance. Nat. Commun. 2016, 7, 12329. [Google Scholar] [CrossRef]
- Hambardzumyan, D.; Gutmann, D.H.; Kettenmann, H. The Role of Microglia and Macrophages in Glioma Maintenance and Progression. Nat. Neurosci. 2016, 19, 20–27. [Google Scholar] [CrossRef]
- Gieryng, A.; Pszczolkowska, D.; Walentynowicz, K.A.; Rajan, W.D.; Kaminska, B. Immune Microenvironment of Gliomas. Lab. Investig. 2017, 97, 498–518. [Google Scholar] [CrossRef]
- Musah-Eroje, A.; Watson, S. Adaptive Changes of Glioblastoma Cells Following Exposure to Hypoxic (1% Oxygen) Tumour Microenvironment. Int. J. Mol. Sci. 2019, 20, 2091. [Google Scholar] [CrossRef]
- Li, Z.; Bao, S.; Wu, Q.; Wang, H.; Eyler, C.; Sathornsumetee, S.; Shi, Q.; Cao, Y.; Lathia, J.; McLendon, R.E.; et al. Hypoxia-Inducible Factors Regulate Tumorigenic Capacity of Glioma Stem Cells. Cancer Cell 2009, 15, 501–513. [Google Scholar] [CrossRef]
- Heddleston, J.M.; Li, Z.; McLendon, R.E.; Hjelmeland, A.B.; Rich, J.N. The Hypoxic Microenvironment Maintains Glioblastoma Stem Cells and Promotes Reprogramming towards a Cancer Stem Cell Phenotype. Cell Cycle 2009, 8, 3274–3284. [Google Scholar] [CrossRef]
- Richards, R.; Jenkinson, M.D.; Haylock, B.J.; See, V. Cell Cycle Progression in Glioblastoma Cells Is Unaffected by Pathophysiological Levels of Hypoxia. PeerJ 2016, 4, e1755. [Google Scholar] [CrossRef]
- Phillips, H.S.; Kharbanda, S.; Chen, R.; Forrest, W.F.; Soriano, R.H.; Wu, T.D.; Misra, A.; Nigro, J.M.; Colman, H.; Soroceanu, L.; et al. Molecular Subclasses of High-Grade Glioma Predict Prognosis, Delineate a Pattern of Disease Progression, and Resemble Stages in Neurogenesis. Cancer Cell 2006, 9, 157–173. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Bergström, T.; Jiang, Y.; Johansson, P.; Marinescu, V.D.; Lindberg, N.; Segerman, A.; Wicher, G.; Niklasson, M.; Baskaran, S.; et al. The Human Glioblastoma Cell Culture Resource: Validated Cell Models Representing All Molecular Subtypes. EBioMedicine 2015, 2, 1351–1363. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.; Shen, Y.; Poon, C.C.; Luchman, H.A.; Stechishin, O.D.; Pontifex, C.S.; Wu, W.; Kelly, J.J.; Blough, M.D. Comparative Genomic and Genetic Analysis of Glioblastoma-Derived Brain Tumor-Initiating Cells and Their Parent Tumors. Neuro-Oncol. 2016, 18, 350–360. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, Q.; Hu, B.; Hu, X.; Kim, H.; Squatrito, M.; Scarpace, L.; deCarvalho, A.C.; Lyu, S.; Li, P.; Li, Y.; et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 2017, 32, 42–56. [Google Scholar] [CrossRef]
- Gangoso, E.; Southgate, B.; Bradley, L.; Rus, S.; Galvez-Cancino, F.; McGivern, N.; Güç, E.; Kapourani, C.-A.; Byron, A.; Ferguson, K.M.; et al. Glioblastomas Acquire Myeloid-Affiliated Transcriptional Programs via Epigenetic Immunoediting to Elicit Immune Evasion. Cell 2021, 184, 2454–2470. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, Z.; Li, J.; Huang, T.; Wang, Y.; Chang, L.; Zheng, W.; Ma, Y.; Chen, F.; Gong, X.; et al. Genomic Analysis of Primary and Recurrent Gliomas Reveals Clinical Outcome Related Molecular Features. Sci. Rep. 2019, 9, 16058. [Google Scholar] [CrossRef]
- Ohnishi, K.; Tani, T.; Bando, S.-I.; Kubota, N.; Fujii, Y.; Hatano, O.; Harada, H. Plastic Induction of CD133AC133-Positive Cells in the Microenvironment of Glioblastoma Spheroids. Int. J. Oncol. 2014, 45, 581–586. [Google Scholar] [CrossRef][Green Version]
- Hsieh, C.H.; Lee, C.H.; Liang, J.A.; Yu, C.Y.; Shyu, W.C. Cycling Hypoxia Increases U87 Glioma Cell Radioresistance via ROS Induced Higher and Long-Term HIF-1 Signal Transduction Activity. Oncol. Rep. 2010, 24, 1629–1636. [Google Scholar] [CrossRef]
- Fedele, M.; Cerchia, L.; Pegoraro, S.; Sgarra, R.; Manfioletti, G. Proneural-Mesenchymal Transition: Phenotypic Plasticity to Acquire Multitherapy Resistance in Glioblastoma. Int. J. Mol. Sci. 2019, 20, 2746. [Google Scholar] [CrossRef]
- Schmitt, M.J.; Company, C.; Dramaretska, Y.; Barozzi, I.; Göhrig, A.; Kertalli, S.; Großmann, M.; Naumann, H.; Sanchez-Bailon, M.P.; Hulsman, D.; et al. Phenotypic Mapping of Pathologic Cross-Talk between Glioblastoma and Innate Immune Cells by Synthetic Genetic Tracing. Cancer Discov. 2021, 11, 754–777. [Google Scholar] [CrossRef]
- Pistollato, F.; Abbadi, S.; Rampazzo, E.; Persano, L.; Puppa, A.D.; Frasson, C.; Sarto, E.; Scienza, R.; D’Avella, D.; Basso, G. Intratumoral Hypoxic Gradient Drives Stem Cells Distribution and MGMT Expression in Glioblastoma. Stem Cells 2010, 28, 851–862. [Google Scholar] [CrossRef] [PubMed]
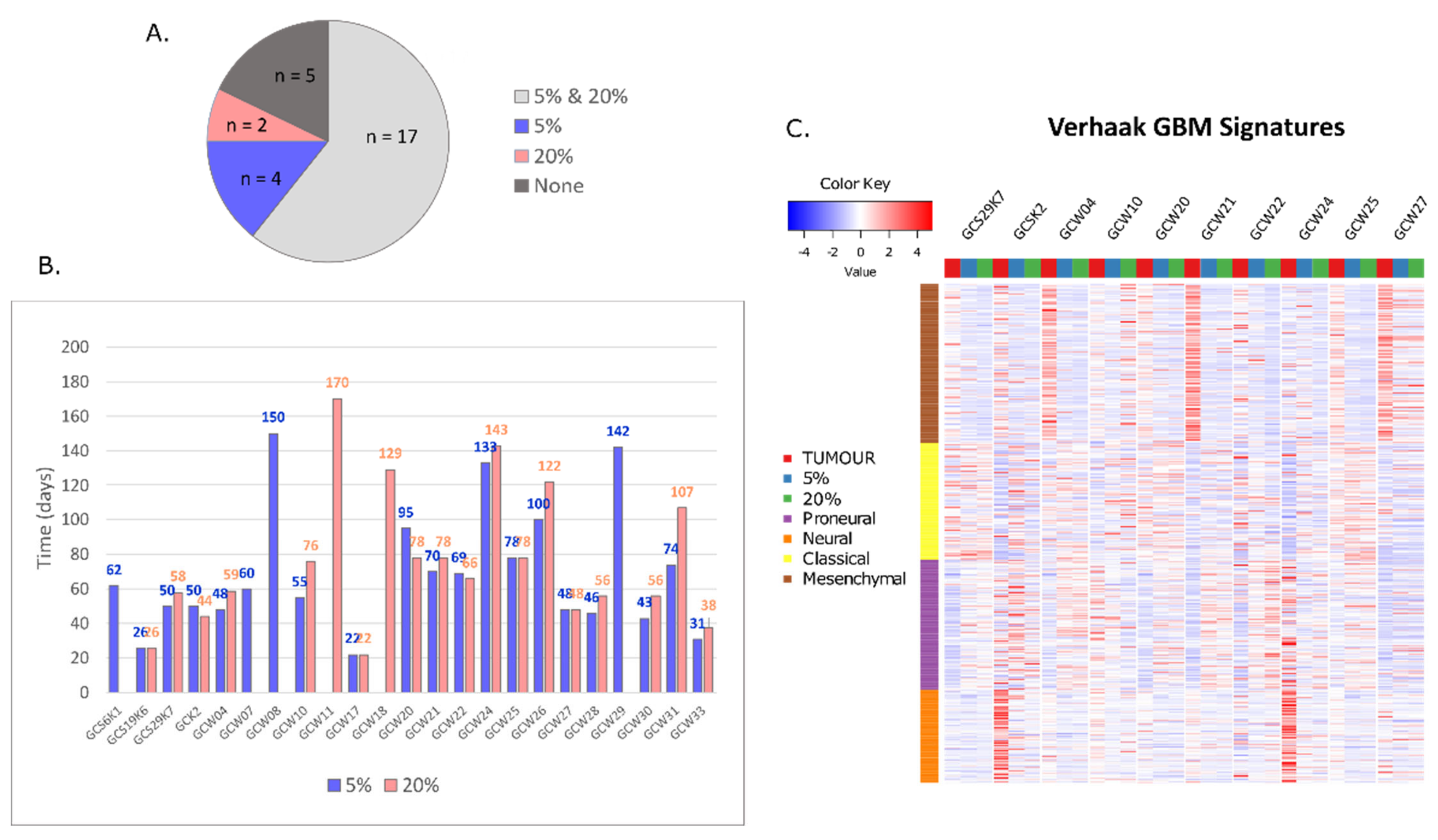

| # | Sample ID | Patient Sex and Age | Cell Line | Culture Type |
|---|---|---|---|---|
| 1 | GCW01 | M 62 | None | n.a. |
| 2 | GCS6K1 | F 66 | 5% | spheroid |
| 3 | GCSK2 | F 45 | 5% and 20% | spheroid |
| 4 | GCS7K3 | F 55 | None | n.a. |
| 5 | GCW04 | M 41 | 5% and 20% | spheroid |
| 6 | GCW07 | M 62 | 5% | spheroid |
| 7 | GCW08 | M 61 | 5% | spheroid |
| 8 | GCS19K6 | F 51 | 5% and 20% | adherent |
| 9 | GCW10 | M 48 | 5% and 20% | spheroid |
| 10 | GCW11 | M 70 | 20% | spheroid |
| 11 | GCW12 | F 74 | None | n.a. |
| 12 | GCW14 | F 57 | None | n.a. |
| 13 | GCW16 | F48 | None | n.a. |
| 14 | GCW17 | M 68 | 5% and 20% | adherent |
| 15 | GCW18 | F 49 | 20% | adherent |
| 16 | GCW20 | M 61 | 5% and 20% | spheroid |
| 17 | GCW21 | M 68 | 5% and 20% | spheroid |
| 18 | GCW22 | F 59 | 5% and 20% | spheroid |
| 19 | GCS29K7 | F 55 | 5% and 20% | spheroid |
| 20 | GCW24 | M 71 | 5% and 20% | spheroid |
| 21 | GCW25 | M 57 | 5% and 20% | spheroid |
| 22 | GCW26 | M 61 | 5% and 20% | spheroid |
| 23 | GCW27 | F 66 | 5% and 20% | spheroid |
| 24 | GCW28 | M 70 | 5% and 20% | spheroid |
| 26 | GCW29 | F 63 | 5% | spheroid |
| 27 | GCW30 | M 60 | 5% and 20% | spheroid |
| 28 | GCW31 | M 63 | 5% and 20% | spheroid |
| 29 | GCW33 | M 34 | 5% and 20% | adherent |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gozdz, A.; Wojtaś, B.; Szpak, P.; Szadkowska, P.; Czernicki, T.; Marchel, A.; Wójtowicz, K.; Kaspera, W.; Ladzinski, P.; Szopa, W.; et al. Preservation of the Hypoxic Transcriptome in Glioblastoma Patient-Derived Cell Lines Maintained at Lowered Oxygen Tension. Cancers 2022, 14, 4852. https://doi.org/10.3390/cancers14194852
Gozdz A, Wojtaś B, Szpak P, Szadkowska P, Czernicki T, Marchel A, Wójtowicz K, Kaspera W, Ladzinski P, Szopa W, et al. Preservation of the Hypoxic Transcriptome in Glioblastoma Patient-Derived Cell Lines Maintained at Lowered Oxygen Tension. Cancers. 2022; 14(19):4852. https://doi.org/10.3390/cancers14194852
Chicago/Turabian StyleGozdz, Agata, Bartosz Wojtaś, Patrycja Szpak, Paulina Szadkowska, Tomasz Czernicki, Andrzej Marchel, Katarzyna Wójtowicz, Wojciech Kaspera, Piotr Ladzinski, Wojciech Szopa, and et al. 2022. "Preservation of the Hypoxic Transcriptome in Glioblastoma Patient-Derived Cell Lines Maintained at Lowered Oxygen Tension" Cancers 14, no. 19: 4852. https://doi.org/10.3390/cancers14194852
APA StyleGozdz, A., Wojtaś, B., Szpak, P., Szadkowska, P., Czernicki, T., Marchel, A., Wójtowicz, K., Kaspera, W., Ladzinski, P., Szopa, W., Niedbala, M., Nawrocki, S., Kaminska, B., & Kalaszczynska, I. (2022). Preservation of the Hypoxic Transcriptome in Glioblastoma Patient-Derived Cell Lines Maintained at Lowered Oxygen Tension. Cancers, 14(19), 4852. https://doi.org/10.3390/cancers14194852







