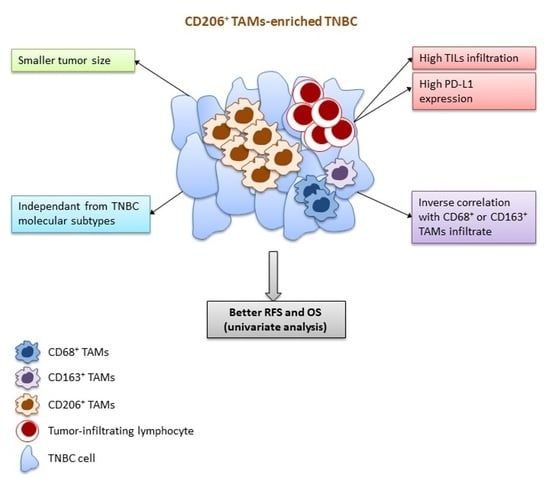
Association of CD206 Protein Expression with Immune Infiltration and Prognosis in Patients with Triple-Negative Breast Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
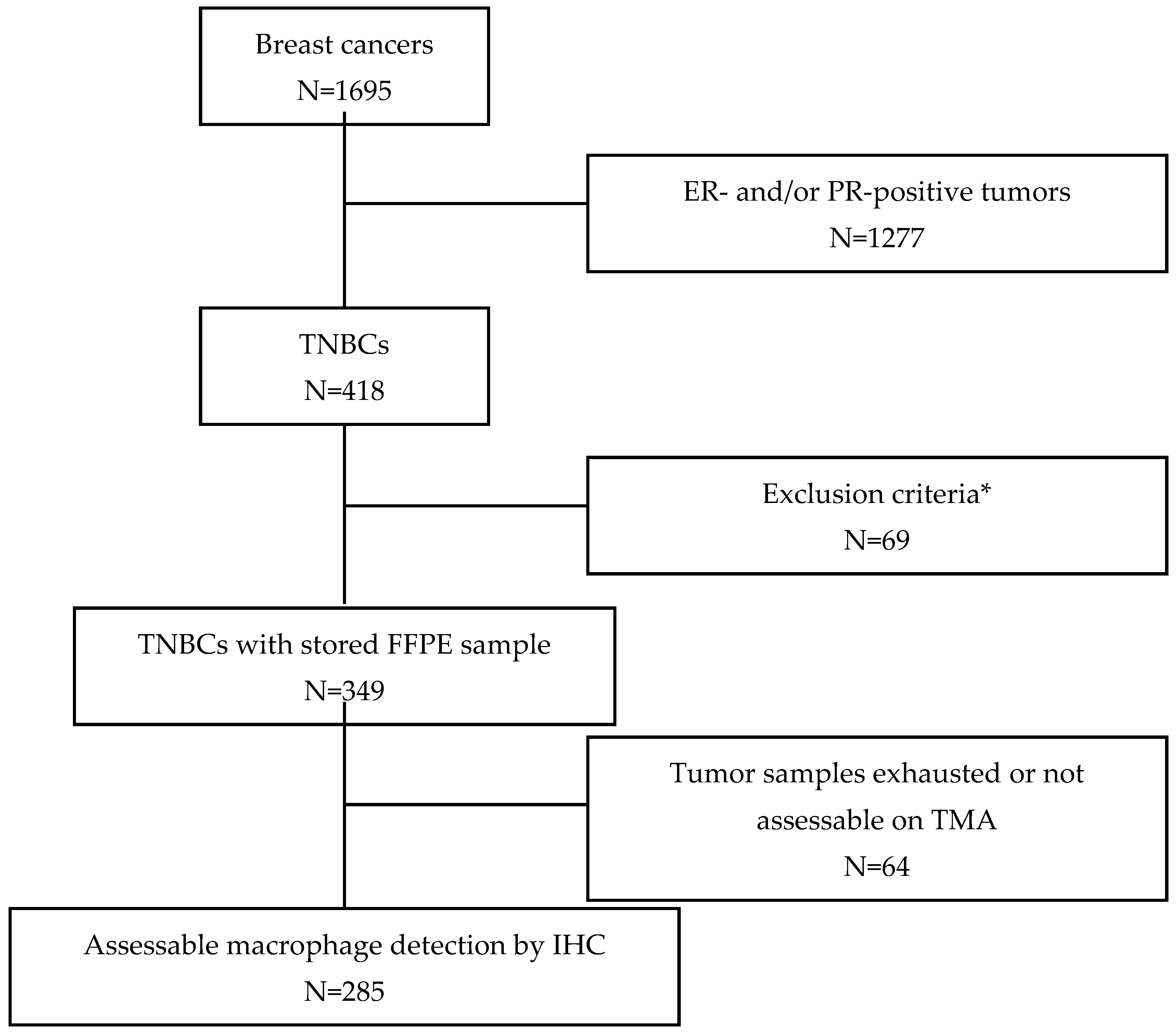
2.1. Patients and Tumor Samples
2.2. Tissue Microarrays (TMA) and Immunohistochemistry
2.3. Analysis of TAM Marker Expression
2.4. TIL Assessment
2.5. Statistical Analysis
3. Results
3.1. Patient and Tumor Characteristics
3.2. TAM Characterization and Quantification
3.3. Association of TAM Markers with TNBC Clinicopathological Features
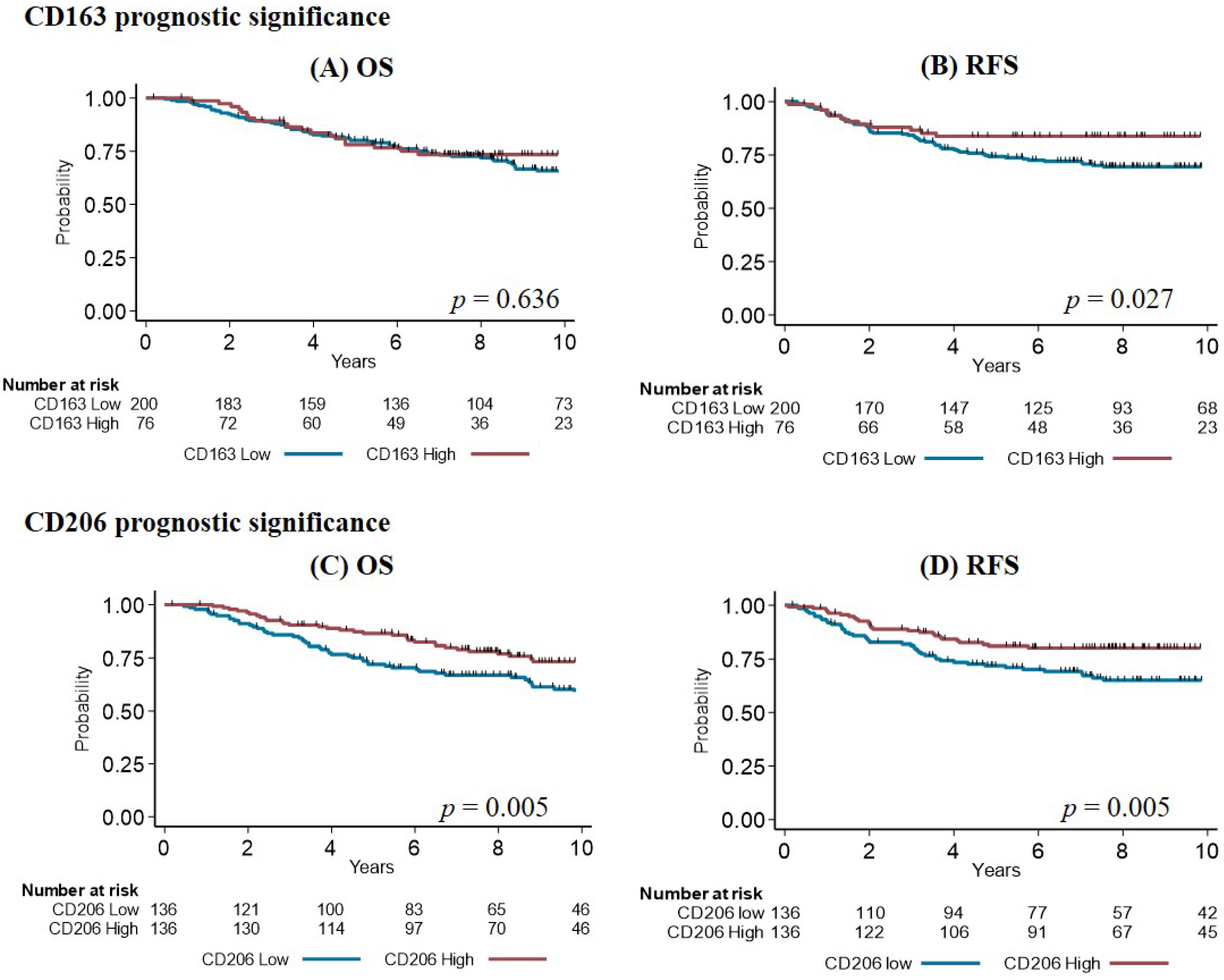
3.4. Survival Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AR | androgen receptor |
| CI | confidence interval |
| CK 5/6 | cytokeratin 5/6 |
| EGFR | epidermal growth factor receptor |
| ER | estrogen receptor |
| FoxA1 | forkhead box protein A1 |
| HES | hematoxylin-eosin-saffron |
| HER2 | human epidermal growth factor receptor 2 |
| HR | hazard ratio |
| IHC | immunohistochemistry |
| IRF8 | interferon regulatory factor 8 |
| LAR | luminal androgen receptor |
| MRC-1 | mannose receptor 1 |
| OS | overall survival |
| PD-1 | programmed cell death 1 |
| PD-L1 | programmed cell death ligand 1 |
| PR | progesterone receptor |
| RFS | relapse-free survival |
| TAM | tumor-associated macrophage |
| TIL | tumor-infiltrating lymphocyte |
| TMA | tissue microarray |
| TNBC | triple-negative breast cancer |
References
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Dent, R.; Trudeau, M.; Pritchard, K.I.; Hanna, W.M.; Kahn, H.K.; Sawka, C.A.; Lickley, L.A.; Rawlinson, E.; Sun, P.; Narod, S.A. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin. Cancer Res. 2007, 13, 4429–4434. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.; Wang, M.; Gralow, J.; Dickler, M.; Cobleigh, M.; Perez, E.A.; Shenkier, T.; Cella, D.; Davidson, N.E. Paclitaxel plus Bevacizumab versus Paclitaxel Alone for Metastatic Breast Cancer. N. Engl. J. Med. 2007, 357, 2666–2676. [Google Scholar] [CrossRef] [PubMed]
- Schmid, P.; Adams, S.; Rugo, H.S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Diéras, V.; Hegg, R.; Im, S.-A.; Shaw Wright, G.; et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N. Engl. J. Med. 2018, 379, 2108–2121. [Google Scholar] [CrossRef]
- Robson, M.; Im, S.-A.; Senkus, E.; Xu, B.; Domchek, S.M.; Masuda, N.; Delaloge, S.; Li, W.; Tung, N.; Armstrong, A.; et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N. Engl. J. Med. 2017, 377, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Litton, J.K.; Rugo, H.S.; Ettl, J.; Hurvitz, S.A.; Gonçalves, A.; Lee, K.-H.; Fehrenbacher, L.; Yerushalmi, R.; Mina, L.A.; Martin, M.; et al. Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. N. Engl. J. Med. 2018, 379, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Jovanović, B.; Chen, X.; Estrada, M.V.; Johnson, K.N.; Shyr, Y.; Moses, H.L.; Sanders, M.E.; Pietenpol, J.A. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS ONE 2016, 11, e0157368. [Google Scholar] [CrossRef]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef]
- Littman, D.R. Releasing the Brakes on Cancer Immunotherapy. Cell 2015, 162, 1186–1190. [Google Scholar] [CrossRef] [PubMed]
- Narang, P.; Chen, M.; Sharma, A.A.; Anderson, K.S.; Wilson, M.A. The neoepitope landscape of breast cancer: Implications for immunotherapy. BMC Cancer 2019, 19, 200. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; von Minckwitz, G.; Darb-Esfahani, S.; Lederer, B.; Heppner, B.I.; Weber, K.E.; Budczies, J.; Huober, J.; Klauschen, F.; Furlanetto, J.; et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: A pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018, 19, 40–50. [Google Scholar] [CrossRef]
- Filipovic, A.; Miller, G.; Bolen, J. Progress Toward Identifying Exact Proxies for Predicting Response to Immunotherapies. Front. Cell Dev. Biol. 2020, 8, 155. [Google Scholar] [CrossRef]
- Planes-Laine, G.; Rochigneux, P.; Bertucci, F.; Chrétien, A.-S.; Viens, P.; Sabatier, R.; Gonçalves, A. PD-1/PD-L1 Targeting in Breast Cancer: The First Clinical Evidences are Emerging—A Literature Review. Cancers 2019, 11, 1033. [Google Scholar] [CrossRef]
- Schmid, P.; Cortes, J.; Pusztai, L.; McArthur, H.; Kümmel, S.; Bergh, J.; Denkert, C.; Park, Y.H.; Hui, R.; Harbeck, N.; et al. Pembrolizumab for Early Triple-Negative Breast Cancer. N. Engl. J. Med. 2020, 382, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Cortes, J.; Cescon, D.W.; Rugo, H.S.; Nowecki, Z.; Im, S.-A.; Yusof, M.M.; Gallardo, C.; Lipatov, O.; Barrios, C.H.; Holgado, E.; et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): A randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet 2020, 396, 1817–1828. [Google Scholar] [CrossRef]
- Stovgaard, E.S.; Nielsen, D.; Hogdall, E.; Balslev, E. Triple negative breast cancer—prognostic role of immune-related factors: A systematic review. Acta Oncol. 2018, 57, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Yeong, J.; Thike, A.A.; Lim, J.C.T.; Lee, B.; Li, H.; Wong, S.-C.; Hue, S.S.S.; Tan, P.H.; Iqbal, J. Higher densities of Foxp3+ regulatory T cells are associated with better prognosis in triple-negative breast cancer. Breast Cancer Res. Treat. 2017, 163, 21–35. [Google Scholar] [CrossRef]
- Liu, S.; Foulkes, W.D.; Leung, S.; Gao, D.; Lau, S.; Kos, Z.; Nielsen, T.O. Prognostic significance of FOXP3+ tumor-infiltrating lymphocytes in breast cancer depends on estrogen receptor and human epidermal growth factor receptor-2 expression status and concurrent cytotoxic T-cell infiltration. Breast Cancer Res. 2014, 16, 432. [Google Scholar] [CrossRef]
- Gentles, A.J.; Newman, A.M.; Liu, C.L.; Bratman, S.V.; Feng, W.; Kim, D.; Nair, V.S.; Xu, Y.; Khuong, A.; Hoang, C.D.; et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015, 21, 938–945. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.K.; Mantovani, A. Macrophage plasticity and interaction with lymphocyte subsets: Cancer as a paradigm. Nat. Immunol. 2010, 11, 889–896. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Sozzani, S.; Locati, M.; Allavena, P.; Sica, A. Macrophage polarization: Tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002, 23, 549–555. [Google Scholar] [CrossRef]
- Rőszer, T. Understanding the Mysterious M2 Macrophage through Activation Markers and Effector Mechanisms. Mediat. Inflamm. 2015, 2015, 816460. [Google Scholar] [CrossRef] [PubMed]
- Strack, E.; Rolfe, P.A.; Fink, A.F.; Bankov, K.; Schmid, T.; Solbach, C.; Savai, R.; Sha, W.; Pradel, L.; Hartmann, S.; et al. Identification of tumor-associated macrophage subsets that are associated with breast cancer prognosis. Clin. Transl. Med. 2020, 10, e239. [Google Scholar] [CrossRef]
- Huang, Y.-K.; Wang, M.; Sun, Y.; Di Costanzo, N.; Mitchell, C.; Achuthan, A.; Hamilton, J.A.; Busuttil, R.A.; Boussioutas, A. Macrophage spatial heterogeneity in gastric cancer defined by multiplex immunohistochemistry. Nat. Commun. 2019, 10, 3928. [Google Scholar] [CrossRef]
- Dong, P.; Ma, L.; Liu, L.; Zhao, G.; Zhang, S.; Dong, L.; Xue, R.; Chen, S. CD86+/CD206+, Diametrically Polarized Tumor-Associated Macrophages, Predict Hepatocellular Carcinoma Patient Prognosis. Int. J. Mol. Sci. 2016, 17, 320. [Google Scholar] [CrossRef]
- Masumi, A.; Tamaoki, S.; Wang, I.M.; Ozato, K.; Komuro, K. IRF-8/ICSBP and IRF-1 cooperatively stimulate mouse IL-12 promoter activity in macrophages. FEBS Lett. 2002, 531, 348–353. [Google Scholar] [CrossRef]
- Günthner, R.; Anders, H.-J. Interferon-regulatory factors determine macrophage phenotype polarization. Mediat. Inflamm. 2013, 2013, 731023. [Google Scholar] [CrossRef]
- Gupta, M.; Shin, D.-M.; Ramakrishna, L.; Goussetis, D.J.; Platanias, L.C.; Xiong, H.; Morse Iii, H.C.; Ozato, K. IRF8 directs stress-induced autophagy in macrophages and promotes clearance of Listeria monocytogenes. Nat. Commun. 2015, 6, 6379. [Google Scholar] [CrossRef]
- O’Meara, T.; Marczyk, M.; Blenman, K.; Yaghoobi, V.; Pelenkanou, V.; Rimm, D.; Pusztai, L. Immunological differences between immune-rich estrogen receptor-positive and -negative breast cancers. Ann. Oncol. 2019, 30, v60–v61. [Google Scholar] [CrossRef]
- Mahmoud, S.M.A.; Lee, A.H.S.; Paish, E.C.; Macmillan, R.D.; Ellis, I.O.; Green, A.R. Tumour-infiltrating macrophages and clinical outcome in breast cancer. J. Clin. Pathol. 2012, 65, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Medrek, C.; Pontén, F.; Jirström, K.; Leandersson, K. The presence of tumor associated macrophages in tumor stroma as a prognostic marker for breast cancer patients. BMC Cancer 2012, 12, 306. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.-Y.; Luo, R.-Z.; Peng, R.-J.; Wang, S.-S.; Xue, C. High infiltration of tumor-associated macrophages in triple-negative breast cancer is associated with a higher risk of distant metastasis. Onco Targets Ther. 2014, 7, 1475–1480. [Google Scholar] [CrossRef]
- Jamiyan, T.; Kuroda, H.; Yamaguchi, R.; Abe, A.; Hayashi, M. CD68- and CD163-positive tumor-associated macrophages in triple negative cancer of the breast. Virchows Arch. 2020, 477, 767–775. [Google Scholar] [CrossRef]
- Pelekanou, V.; Villarroel-Espindola, F.; Schalper, K.A.; Pusztai, L.; Rimm, D.L. CD68, CD163, and matrix metalloproteinase 9 (MMP-9) co-localization in breast tumor microenvironment predicts survival differently in ER-positive and -negative cancers. Breast Cancer Res. 2018, 20, 154. [Google Scholar] [CrossRef]
- Jacot, W.; Mollevi, C.; Fina, F.; Lopez-Crapez, E.; Martin, P.-M.; Colombo, P.-E.; Bibeau, F.; Romieu, G.; Lamy, P.-J. High EGFR protein expression and exon 9 PIK3CA mutations are independent prognostic factors in triple negative breast cancers. BMC Cancer 2015, 15, 986. [Google Scholar] [CrossRef]
- Guiu, S.; Mollevi, C.; Charon-Barra, C.; Boissière, F.; Crapez, E.; Chartron, E.; Lamy, P.-J.; Gutowski, M.; Bourgier, C.; Romieu, G.; et al. Prognostic value of androgen receptor and FOXA1 co-expression in non-metastatic triple negative breast cancer and correlation with other biomarkers. Br. J. Cancer 2018, 119, 76–79. [Google Scholar] [CrossRef]
- Jacot, W.; Lopez-Crapez, E.; Mollevi, C.; Boissière-Michot, F.; Simony-Lafontaine, J.; Ho-Pun-Cheung, A.; Chartron, E.; Theillet, C.; Lemoine, A.; Saffroy, R.; et al. BRCA1 Promoter Hypermethylation is Associated with Good Prognosis and Chemosensitivity in Triple-Negative Breast Cancer. Cancers 2020, 12, 828. [Google Scholar] [CrossRef]
- Jacot, W.; Lopez-Crapez, E.; Thezenas, S.; Senal, R.; Fina, F.; Bibeau, F.; Romieu, G.; Lamy, P.-J. Lack of EGFR-activating mutations in European patients with triple-negative breast cancer could emphasise geographic and ethnic variations in breast cancer mutation profiles. Breast Cancer Res. 2011, 13, R133. [Google Scholar] [CrossRef]
- Jacot, W.; Gutowski, M.; Azria, D.; Romieu, G. Adjuvant early breast cancer systemic therapies according to daily used technologies. Crit. Rev. Oncol. Hematol. 2012, 82, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Boissière-Michot, F.; Chabab, G.; Mollevi, C.; Guiu, S.; Lopez-Crapez, E.; Ramos, J.; Bonnefoy, N.; Lafont, V.; Jacot, W. Clinicopathological Correlates of γδ T Cell Infiltration in Triple-Negative Breast Cancer. Cancers 2021, 13, 765. [Google Scholar] [CrossRef] [PubMed]
- Boissière-Michot, F.; Jacot, W.; Massol, O.; Mollevi, C.; Lazennec, G. CXCR2 Levels Correlate with Immune Infiltration and a Better Prognosis of Triple-Negative Breast Cancers. Cancers 2021, 13, 2328. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, H.; Alcaraz, L.B.; Mollevi, C.; Mallavialle, A.; Jacot, W.; Boissière-Michot, F.; Simony-Lafontaine, J.; Laurent-Matha, V.; Roger, P.; Liaudet-Coopman, E.; et al. Co-Expression of Androgen Receptor and Cathepsin D Defines a Triple-Negative Breast Cancer Subgroup with Poorer Overall Survival. Cancers 2020, 12, 1244. [Google Scholar] [CrossRef]
- Salgado, R.; Denkert, C.; Demaria, S.; Sirtaine, N.; Klauschen, F.; Pruneri, G.; Wienert, S.; Van den Eynden, G.; Baehner, F.L.; Penault-Llorca, F.; et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: Recommendations by an International TILs Working Group 2014. Ann. Oncol. 2015, 26, 259–271. [Google Scholar] [CrossRef]
- Vikas, P.; Borcherding, N.; Zhang, W. The Clinical Promise of Immunotherapy in Triple-Negative Breast Cancer. Available online: https://www.dovepress.com/the-clinical-promise-of-immunotherapy-in-triple-negative-breast-cancer-peer-reviewed-article-CMAR (accessed on 23 September 2019).
- Cassetta, L.; Pollard, J.W. Targeting macrophages: Therapeutic approaches in cancer. Nat. Rev. Drug Discov. 2018, 17, 887–904. [Google Scholar] [CrossRef]
- Yang, M.; Li, Z.; Ren, M.; Li, S.; Zhang, L.; Zhang, X.; Liu, F. Stromal Infiltration of Tumor-Associated Macrophages Conferring Poor Prognosis of Patients with Basal-Like Breast Carcinoma. J. Cancer 2018, 9, 2308–2316. [Google Scholar] [CrossRef]
- Mohammed, Z.M.; Going, J.J.; Edwards, J.; Elsberger, B.; Doughty, J.C.; McMillan, D.C. The relationship between components of tumour inflammatory cell infiltrate and clinicopathological factors and survival in patients with primary operable invasive ductal breast cancer. Br. J. Cancer 2012, 107, 864–873. [Google Scholar] [CrossRef]
- Barros, M.H.M.; Hauck, F.; Dreyer, J.H.; Kempkes, B.; Niedobitek, G. Macrophage polarisation: An immunohistochemical approach for identifying M1 and M2 macrophages. PLoS ONE 2013, 8, e80908. [Google Scholar] [CrossRef]
- Bowman, R.L.; Klemm, F.; Akkari, L.; Pyonteck, S.M.; Sevenich, L.; Quail, D.F.; Dhara, S.; Simpson, K.; Gardner, E.E.; Iacobuzio-Donahue, C.A.; et al. Macrophage Ontogeny Underlies Differences in Tumor-Specific Education in Brain Malignancies. Cell Rep. 2016, 17, 2445–2459. [Google Scholar] [CrossRef]
- Zhu, Y.; Herndon, J.M.; Sojka, D.K.; Kim, K.-W.; Knolhoff, B.L.; Zuo, C.; Cullinan, D.R.; Luo, J.; Bearden, A.R.; Lavine, K.J.; et al. Tissue-Resident Macrophages in Pancreatic Ductal Adenocarcinoma Originate from Embryonic Hematopoiesis and Promote Tumor Progression. Immunity 2017, 47, 323–338.e6. [Google Scholar] [CrossRef] [PubMed]
- Ni, C.; Yang, L.; Xu, Q.; Yuan, H.; Wang, W.; Xia, W.; Gong, D.; Zhang, W.; Yu, K. CD68- and CD163-positive tumor infiltrating macrophages in non-metastatic breast cancer: A retrospective study and meta-analysis. J. Cancer 2019, 10, 4463–4472. [Google Scholar] [CrossRef]
- Gwak, J.M.; Jang, M.H.; Kim, D.I.; Seo, A.N.; Park, S.Y. Prognostic Value of Tumor-Associated Macrophages According to Histologic Locations and Hormone Receptor Status in Breast Cancer. PLoS ONE 2015, 10, e0125728. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Qu, J.; Sun, Y.; Wang, J.; Liu, X.; Wang, F. Prognostic significance of tumor-associated macrophages in breast cancer: A meta-analysis of the literature. Oncotarget 2017, 8, 30576–30586. [Google Scholar] [CrossRef] [PubMed]
- Nixon, B.G.; Kuo, F.; Liu, M.; Capistrano, K.; Do, M.; Franklin, R.A.; Wu, X.; Kansler, E.R.; Srivastava, R.M.; Purohit, T.A.; et al. IRF8 Governs Tumor-Associated Macrophage Control of T Cell Exhaustion. bioRxiv 2020. [Google Scholar] [CrossRef]
- Muhitch, J.B.; Hoffend, N.C.; Azabdaftari, G.; Miller, A.; Bshara, W.; Morrison, C.D.; Schwaab, T.; Abrams, S.I. Tumor-associated macrophage expression of interferon regulatory Factor-8 (IRF8) is a predictor of progression and patient survival in renal cell carcinoma. J. Immunother. Cancer 2019, 7, 155. [Google Scholar] [CrossRef] [PubMed]
- Chevrier, S.; Levine, J.H.; Zanotelli, V.R.T.; Silina, K.; Schulz, D.; Bacac, M.; Ries, C.H.; Ailles, L.; Jewett, M.A.S.; Moch, H.; et al. An Immune Atlas of Clear Cell Renal Cell Carcinoma. Cell 2017, 169, 736–749.e18. [Google Scholar] [CrossRef]
- Biswas, S.K.; Allavena, P.; Mantovani, A. Tumor-associated macrophages: Functional diversity, clinical significance, and open questions. Semin. Immunopathol. 2013, 35, 585–600. [Google Scholar] [CrossRef]
- Yang, M.; McKay, D.; Pollard, J.W.; Lewis, C.E. Diverse Functions of Macrophages in Different Tumor Microenvironments. Cancer Res. 2018, 78, 5492–5503. [Google Scholar] [CrossRef]
- Larionova, I.; Tuguzbaeva, G.; Ponomaryova, A.; Stakheyeva, M.; Cherdyntseva, N.; Pavlov, V.; Choinzonov, E.; Kzhyshkowska, J. Tumor-Associated Macrophages in Human Breast, Colorectal, Lung, Ovarian and Prostate Cancers. Front. Oncol. 2020, 10, 566511. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, L.; Gong, C.; Shi, H.; Zeng, Y.; Wang, X.; Zhao, Y.; Wei, Y. Prognostic significance of tumor-associated macrophages in solid tumor: A meta-analysis of the literature. PLoS ONE 2012, 7, e50946. [Google Scholar] [CrossRef] [PubMed]
- Koru-Sengul, T.; Santander, A.M.; Miao, F.; Sanchez, L.G.; Jorda, M.; Glück, S.; Ince, T.A.; Nadji, M.; Chen, Z.; Penichet, M.L.; et al. Breast cancers from black women exhibit higher numbers of immunosuppressive macrophages with proliferative activity and of crown-like structures associated with lower survival compared to non-black Latinas and Caucasians. Breast Cancer Res. Treat. 2016, 158, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Gruosso, T.; Gigoux, M.; Manem, V.S.K.; Bertos, N.; Zuo, D.; Perlitch, I.; Saleh, S.M.I.; Zhao, H.; Souleimanova, M.; Johnson, R.M.; et al. Spatially distinct tumor immune microenvironments stratify triple-negative breast cancers. J. Clin Investig. 2019, 129, 1785–1800. [Google Scholar] [CrossRef]
- Kennedy, B.C.; Showers, C.R.; Anderson, D.E.; Anderson, L.; Canoll, P.; Bruce, J.N.; Anderson, R.C.E. Tumor-associated macrophages in glioma: Friend or foe? J. Oncol. 2013, 2013, 486912. [Google Scholar] [CrossRef]
- Kuang, D.-M.; Zhao, Q.; Peng, C.; Xu, J.; Zhang, J.-P.; Wu, C.; Zheng, L. Activated monocytes in peritumoral stroma of hepatocellular carcinoma foster immune privilege and disease progression through PD-L1. J. Exp. Med. 2009, 206, 1327–1337. [Google Scholar] [CrossRef]
- Liu, C.-Q.; Xu, J.; Zhou, Z.-G.; Jin, L.-L.; Yu, X.-J.; Xiao, G.; Lin, J.; Zhuang, S.-M.; Zhang, Y.-J.; Zheng, L. Expression patterns of programmed death ligand 1 correlate with different microenvironments and patient prognosis in hepatocellular carcinoma. Br. J. Cancer 2018, 119, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Pollari, M.; Brück, O.; Pellinen, T.; Vähämurto, P.; Karjalainen-Lindsberg, M.-L.; Mannisto, S.; Kallioniemi, O.; Kellokumpu-Lehtinen, P.-L.; Mustjoki, S.; Leivonen, S.-K.; et al. PD-L1+ tumor-associated macrophages and PD-1+ tumor-infiltrating lymphocytes predict survival in primary testicular lymphoma. Haematologica 2018, 103, 1908–1914. [Google Scholar] [CrossRef] [PubMed]
- Sepesi, B.; Cuentas, E.P.; Canales, J.R.; Behrens, C.; Correa, A.M.; Vaporciyan, A.; Weissferdt, A.; Kalhor, N.; Moran, C.; Swisher, S.; et al. Programmed Death Cell Ligand 1 (PD-L1) Is Associated With Survival in Stage I Non–Small Cell Lung Cancer. Semin. Thorac. Cardiovasc. Surg. 2017, 29, 408–415. [Google Scholar] [CrossRef]
- Bouchard, A.; Sikner, H.; Baverel, V.; Garnier, A.-R.; Monterrat, M.; Moreau, M.; Limagne, E.; Garrido, C.; Kohli, E.; Collin, B.; et al. The GRP94 Inhibitor PU-WS13 Decreases M2-like Macrophages in Murine TNBC Tumors: A Pharmaco-Imaging Study with 99mTc-Tilmanocept SPECT. Cells 2021, 10, 3393. [Google Scholar] [CrossRef]
- Tsujikawa, T.; Kumar, S.; Borkar, R.N.; Azimi, V.; Thibault, G.; Chang, Y.H.; Balter, A.; Kawashima, R.; Choe, G.; Sauer, D.; et al. Quantitative Multiplex Immunohistochemistry Reveals Myeloid-Inflamed Tumor-Immune Complexity Associated with Poor Prognosis. Cell Rep. 2017, 19, 203–217. [Google Scholar] [CrossRef]
- Jackson, H.W.; Fischer, J.R.; Zanotelli, V.R.T.; Ali, H.R.; Mechera, R.; Soysal, S.D.; Moch, H.; Muenst, S.; Varga, Z.; Weber, W.P.; et al. The single-cell pathology landscape of breast cancer. Nature 2020, 578, 615–620. [Google Scholar] [CrossRef] [PubMed]

| Variable | Number of Patients (n = 285) | % |
|---|---|---|
| Age (years), median [min–max] | 57.76 | [28.54–89.10] |
| <55 | 126 | 44.21 |
| ≥55 | 159 | 55.79 |
| Tumor size | ||
| T1 | 127 | 44.56 |
| T2 | 140 | 49.12 |
| T3/T4 | 18 | 6.32 |
| Nodal status | ||
| N− | 183 | 64.21 |
| N+ | 102 | 35.79 |
| Histological grade | 3 missing values | |
| 1–2 | 65 | 23.05 |
| 3 | 217 | 76.95 |
| Histology | 3 missing values | |
| Ductal | 236 | 83.69 |
| Lobular | 15 | 5.32 |
| Other | 31 | 10.99 |
| Adjuvant chemotherapy | 1 missing values | |
| No | 71 | 25.00 |
| Yes | 213 | 75.00 |
| Basal-like phenotype | 2 missing values | |
| No (≤10%) | 103 | 36.40 |
| Yes | 180 | 63.60 |
| Molecular apocrine phenotype | 15 missing values | |
| No (<1%) | 156 | 57.78 |
| Yes (≥1%) | 114 | 42.22 |
| TILs | 5 missing values | |
| ≤5% | 174 | 62.14 |
| >5% | 106 | 37.86 |
| PD-L1+ tumor cells | 22 missing values | |
| <1% | 118 | 44.87 |
| ≥1% | 145 | 55.13 |
| PD-L1+ stromal cells | 25 missing values | |
| 0 | 45 | 17.31 |
| [0–10] | 86 | 33.07 |
| [10–50] | 71 | 27.31 |
| ≥50 | 58 | 22.31 |
| CD68 | IRF8 | CD163 | CD206 | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | High | p-Value | Low | High | p-Value | Low | High | p-Value | Low | High | p-Value | |||||||||
| N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | |||||
| Age (years) | ||||||||||||||||||||
| <55 | 70 | 40.46 | 52 | 55.32 | 0.020 | 47 | 38.84 | 78 | 50.0 | 0.064 | 84 | 42.0 | 40 | 52.63 | 0.113 | 54 | 39.71 | 65 | 47.79 | 0.179 |
| ≥55 | 103 | 59.54 | 42 | 44.68 | 74 | 61.16 | 78 | 50.0 | 116 | 58.0 | 36 | 47.37 | 82 | 60.29 | 71 | 52.21 | ||||
| Tumor size | ||||||||||||||||||||
| T1 | 77 | 44.51 | 44 | 46.81 | 0.364 | 62 | 44.60 | 61 | 44.20 | 0.716 | 89 | 44.50 | 36 | 47.37 | 0.695 | 45 | 33.09 | 79 | 58.09 | <0.001 |
| T2 | 83 | 47.98 | 47 | 50.00 | 68 | 48.92 | 71 | 51.45 | 97 | 48.50 | 37 | 48.68 | 77 | 56.62 | 53 | 38.97 | ||||
| T3/T4 | 13 | 7.51 | 3 | 3.19 | 9 | 6.47 | 6 | 4.35 | 14 | 7.00 | 3 | 3.95 | 14 | 10.29 | 4 | 2.94 | ||||
| Nodal status | ||||||||||||||||||||
| N− | 107 | 61.85 | 63 | 67.02 | 0.401 | 85 | 61.15 | 94 | 68.12 | 0.225 | 125 | 62.50 | 52 | 68.42 | 0.360 | 82 | 60.29 | 93 | 68.38 | 0.164 |
| N+ | 66 | 38.15 | 31 | 32.98 | 54 | 38.85 | 44 | 31.88 | 75 | 37.50 | 24 | 31.58 | 54 | 39.71 | 43 | 31.62 | ||||
| Histological grade | ||||||||||||||||||||
| 1–2 | 49 | 28.49 | 13 | 13.98 | 0.008 | 43 | 31.39 | 20 | 14.60 | 0.001 | 58 | 29.29 | 5 | 6.67 | <0.001 | 30 | 22.56 | 33 | 24.26 | 0.741 |
| 3 | 123 | 71.51 | 80 | 86.02 | 94 | 68.61 | 117 | 85.40 | 140 | 70.71 | 70 | 93.33 | 103 | 77.44 | 103 | 75.74 | ||||
| Basal-like | ||||||||||||||||||||
| No (≤10%) | 66 | 38.37 | 30 | 32.26 | 0.323 | 52 | 37.68 | 47 | 34.31 | 0.560 | 84 | 42.21 | 15 | 20.00 | 0.001 | 44 | 32.59 | 52 | 38.52 | 0.309 |
| Yes | 106 | 61.63 | 63 | 67.74 | 86 | 62.32 | 90 | 65.69 | 115 | 57.79 | 60 | 80.00 | 91 | 67.41 | 83 | 61.48 | ||||
| Molecular apocrine | ||||||||||||||||||||
| No (<1%) | 81 | 50.31 | 65 | 71.43 | <0.001 | 71 | 53.79 | 80 | 60.61 | 0.263 | 97 | 51.60 | 57 | 77.03 | <0.001 | 76 | 58.46 | 76 | 58.91 | 0.941 |
| Yes (≥1%) | 80 | 49.69 | 26 | 28.57 | 61 | 46.21 | 52 | 39.39 | 91 | 48.40 | 17 | 22.97 | 54 | 41.54 | 53 | 41.09 | ||||
| TILs | ||||||||||||||||||||
| ≤5% | 132 | 77.19 | 29 | 31.87 | <0.001 | 110 | 79.71 | 58 | 42.96 | <0.001 | 154 | 77.39 | 15 | 20.83 | <0.001 | 99 | 74.44 | 66 | 49.25 | <0.001 |
| >5% | 39 | 22.81 | 62 | 68.13 | 28 | 20.29 | 77 | 57.04 | 45 | 22.61 | 57 | 79.17 | 34 | 25.56 | 68 | 50.75 | ||||
| PD-L1 tumor cells | ||||||||||||||||||||
| <1% | 88 | 56.77 | 22 | 23.91 | <0.001 | 76 | 59.84 | 39 | 30.00 | <0.001 | 98 | 54.14 | 18 | 24.32 | <0.001 | 68 | 54.40 | 42 | 33.07 | 0.001 |
| ≥1% | 67 | 43.23 | 70 | 76.09 | 51 | 40.16 | 91 | 70.00 | 83 | 45.86 | 56 | 75.68 | 57 | 45.60 | 85 | 66.93 | ||||
| PD-L1 stromal cells | ||||||||||||||||||||
| 0 | 33 | 21.43 | 9 | 10.00 | <0.001 | 26 | 20.47 | 18 | 14.06 | <0.001 | 37 | 20.55 | 8 | 11.11 | 0.001 | 27 | 21.95 | 15 | 11.90 | 0.002 |
| [0–10] | 58 | 37.66 | 23 | 25.56 | 60 | 47.25 | 25 | 19.53 | 70 | 38.89 | 14 | 19.45 | 47 | 38.21 | 34 | 26.99 | ||||
| [10–50] | 38 | 24.68 | 27 | 30.00 | 27 | 21.26 | 41 | 32.03 | 41 | 22.78 | 26 | 36.11 | 32 | 26.02 | 38 | 30.16 | ||||
| ≥50 | 25 | 16.23 | 31 | 34.44 | 14 | 11.02 | 44 | 34.38 | 32 | 17.78 | 24 | 33.33 | 17 | 13.82 | 39 | 30.95 | ||||
| Variables | OS | RFS | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p-Value | HR | 95% CI | p-Value | |
| Age (years) | <0.001 | 0.067 | ||||
| <55 | 1 | 1 | ||||
| ≥55 | 2.10 | 1.33–3.31 | 1.55 | 0.96–2.51 | ||
| Tumor size | <0.001 | <0.001 | ||||
| T1 | 1 | 1 | ||||
| T2/T3/T4 | 2.78 | 1.71–4.50 | 2.44 | 1.46–4.09 | ||
| Nodal status | <0.001 | <0.001 | ||||
| N− | 1 | 1 | ||||
| N+ | 2.45 | 1.61–3.72 | 4.61 | 2.82–7.51 | ||
| Histological grade | 0.472 | 0.904 | ||||
| 1–2 | 1 | 1 | ||||
| 3 | 0.84 | 0.52–1.34 | 1.03 | 0.60–1.78 | ||
| Histology | 0.032 | 0.600 | ||||
| Ductal | 1 | 1 | ||||
| Other | 0.50 | 0.25–1.00 | 0.84 | 0.44–1.61 | ||
| Adjuvant chemotherapy | <0.001 | 0.002 | ||||
| No | 1 | 1 | ||||
| Yes | 0.34 | 0.22–0.51 | 0.46 | 0.29–0.73 | ||
| Basal-like phenotype | 0.697 | 0.550 | ||||
| No (≤10%) | 1 | 1 | ||||
| Yes | 1.09 | 0.70–1.69 | 0.87 | 0.54–1.39 | ||
| Molecular apocrine | ||||||
| No (<1%) | 1 | 0.041 | 1 | 0.032 | ||
| Yes (≥1%) | 1.56 | 1.02–2.39 | 1.67 | 1.04–2.66 | ||
| TILs | ||||||
| ≤5% | 1 | 0.005 | 1 | 0.001 | ||
| >5% | 0.51 | 0.32–0.83 | 0.42 | 0.24–0.74 | ||
| PD-L1 tumor cells | 0.090 | 0.055 | ||||
| <1% | 1 | 1 | ||||
| ≥1% | 0.69 | 0.45–1.06 | 0.63 | 0.39–1.01 | ||
| PD-L1 stromal cells | ||||||
| 0 | 1 | 1 | ||||
| [0–10] | 1.42 | 0.75–2.69 | 0.191 | 1.33 | 0.68–2.61 | 0.069 |
| [10–50] | 0.85 | 0.42–1.74 | 0.56 | 0.25–1.26 | ||
| ≥50 | 0.82 | 0.38–1.74 | 0.81 | 0.37–1.79 | ||
| CD68 | ||||||
| Low | 1 | 0.852 | 1 | 0.299 | ||
| High | 0.96 | 0.61–1.50 | 0.77 | 0.46–1.28 | ||
| IRF8 | ||||||
| Low | 1 | 0.495 | 1 | 0.456 | ||
| High | 0.86 | 0.56–1.32 | 0.84 | 0.52–1.34 | ||
| CD163 | ||||||
| Low | 1 | 1 | ||||
| High | 0.89 | 0.54–1.46 | 0.636 | 0.52 | 0.28–0.97 | 0.027 |
| CD206 | ||||||
| Low | 1 | 1 | ||||
| High | 0.54 | 0.35–0.83 | 0.005 | 0.51 | 0.31–0.82 | 0.005 |
| Variables | OS | RFS | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p-Value | HR | 95% CI | p-Value | |
| Tumor size | 0.004 | |||||
| T1 | 1 | |||||
| T2/T3/T4 | 2.03 | 1.23–3.34 | ||||
| Nodal status | <0.001 | <0.001 | ||||
| N− | 1 | 1 | ||||
| N+ | 2.57 | 1.65–4.00 | 4.87 | 2.91–8.12 | ||
| Adjuvant chemotherapy | <0.001 | 0.004 | ||||
| No | 1 | 1 | ||||
| Yes | 0.34 | 0.22–0.53 | 0.48 | 0.29–0.80 | ||
| Histology | 0.002 | |||||
| Ductal | 1 | |||||
| Other | 0.37 | 0.18–0.76 | ||||
| TILs | 0.030 | |||||
| ≤5% | 1 | 0.028 | 1 | |||
| >5% | 0.59 | 0.36–0.96 | 0.45 | 0.22–0.93 | ||
| CD206 | 0.073 | |||||
| Low | 1 | |||||
| High | 0.63 | 0.33–1.04 | ||||
| CD163 | ||||||
| Low | 1 | 0.872 | ||||
| High | 1.07 | 0.49–2.34 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bobrie, A.; Massol, O.; Ramos, J.; Mollevi, C.; Lopez-Crapez, E.; Bonnefoy, N.; Boissière-Michot, F.; Jacot, W. Association of CD206 Protein Expression with Immune Infiltration and Prognosis in Patients with Triple-Negative Breast Cancer. Cancers 2022, 14, 4829. https://doi.org/10.3390/cancers14194829
Bobrie A, Massol O, Ramos J, Mollevi C, Lopez-Crapez E, Bonnefoy N, Boissière-Michot F, Jacot W. Association of CD206 Protein Expression with Immune Infiltration and Prognosis in Patients with Triple-Negative Breast Cancer. Cancers. 2022; 14(19):4829. https://doi.org/10.3390/cancers14194829
Chicago/Turabian StyleBobrie, Angélique, Océane Massol, Jeanne Ramos, Caroline Mollevi, Evelyne Lopez-Crapez, Nathalie Bonnefoy, Florence Boissière-Michot, and William Jacot. 2022. "Association of CD206 Protein Expression with Immune Infiltration and Prognosis in Patients with Triple-Negative Breast Cancer" Cancers 14, no. 19: 4829. https://doi.org/10.3390/cancers14194829
APA StyleBobrie, A., Massol, O., Ramos, J., Mollevi, C., Lopez-Crapez, E., Bonnefoy, N., Boissière-Michot, F., & Jacot, W. (2022). Association of CD206 Protein Expression with Immune Infiltration and Prognosis in Patients with Triple-Negative Breast Cancer. Cancers, 14(19), 4829. https://doi.org/10.3390/cancers14194829