Prognostic Value of Copy Number Alteration Burden in Early-Stage Breast Cancer and the Construction of an 11-Gene Copy Number Alteration Model
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics
2.2. Study Design
2.3. Genomic Information Acquisition
2.4. CNAB11 Modeling Method
2.5. Statistical Analysis
3. Results
3.1. Patient Characteristics
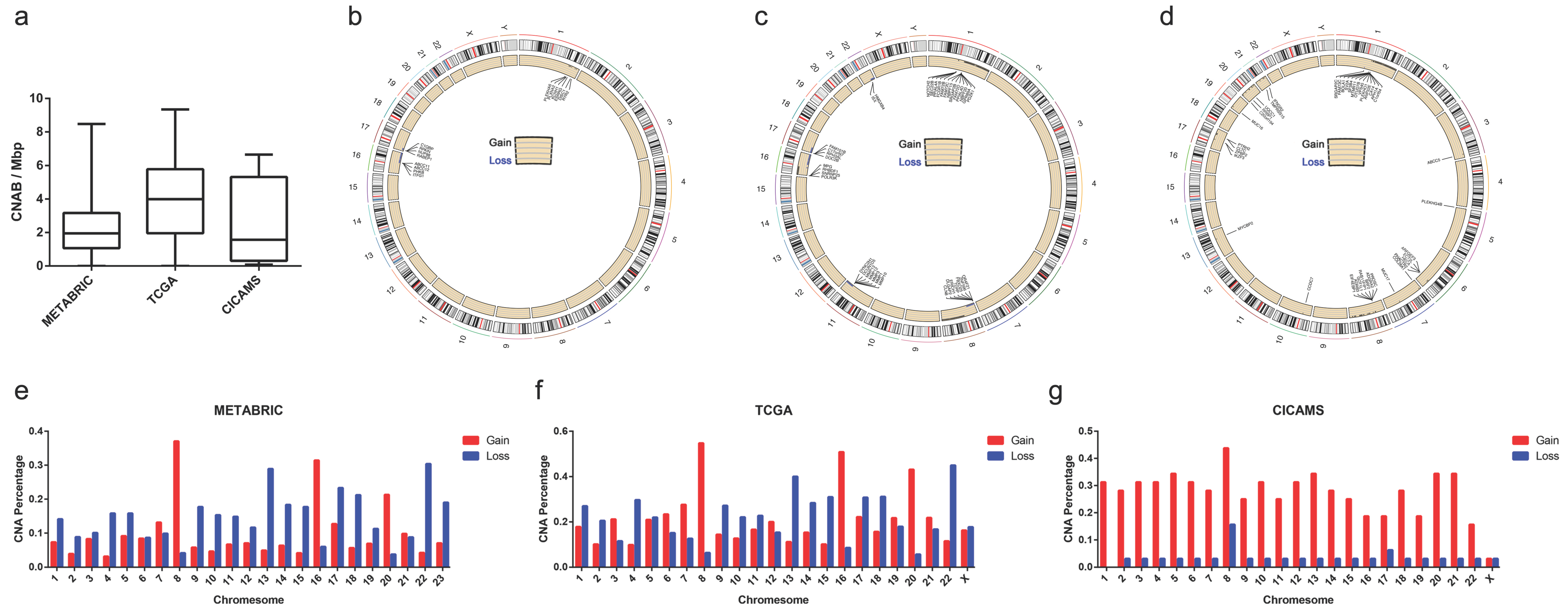
3.2. Differences in Clinical Characteristics between Different CNAB Subgroups
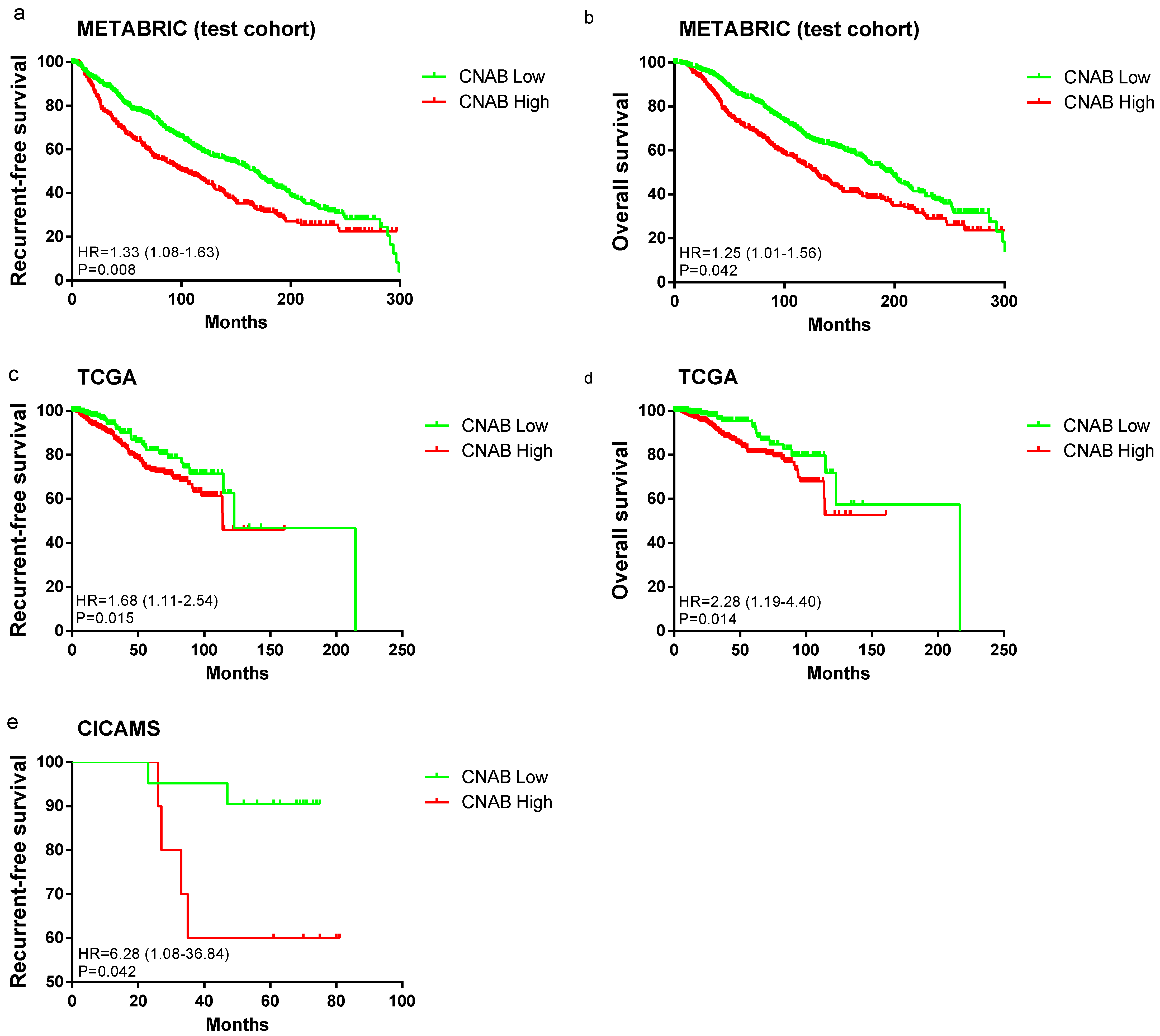
3.3. Survival Analysis between CNAB Groups
3.4. Combined Survival Analysis of TMB and CNAB
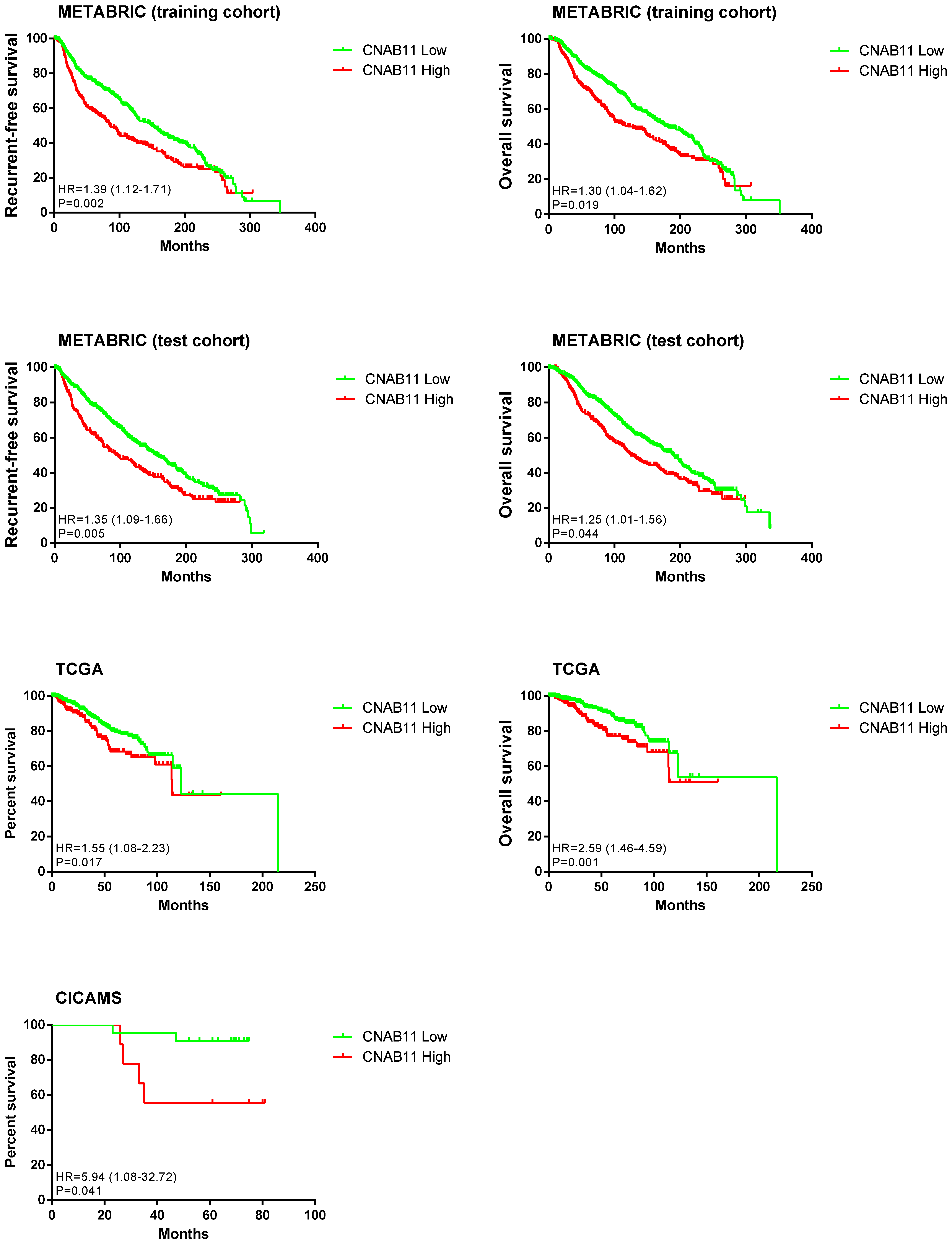
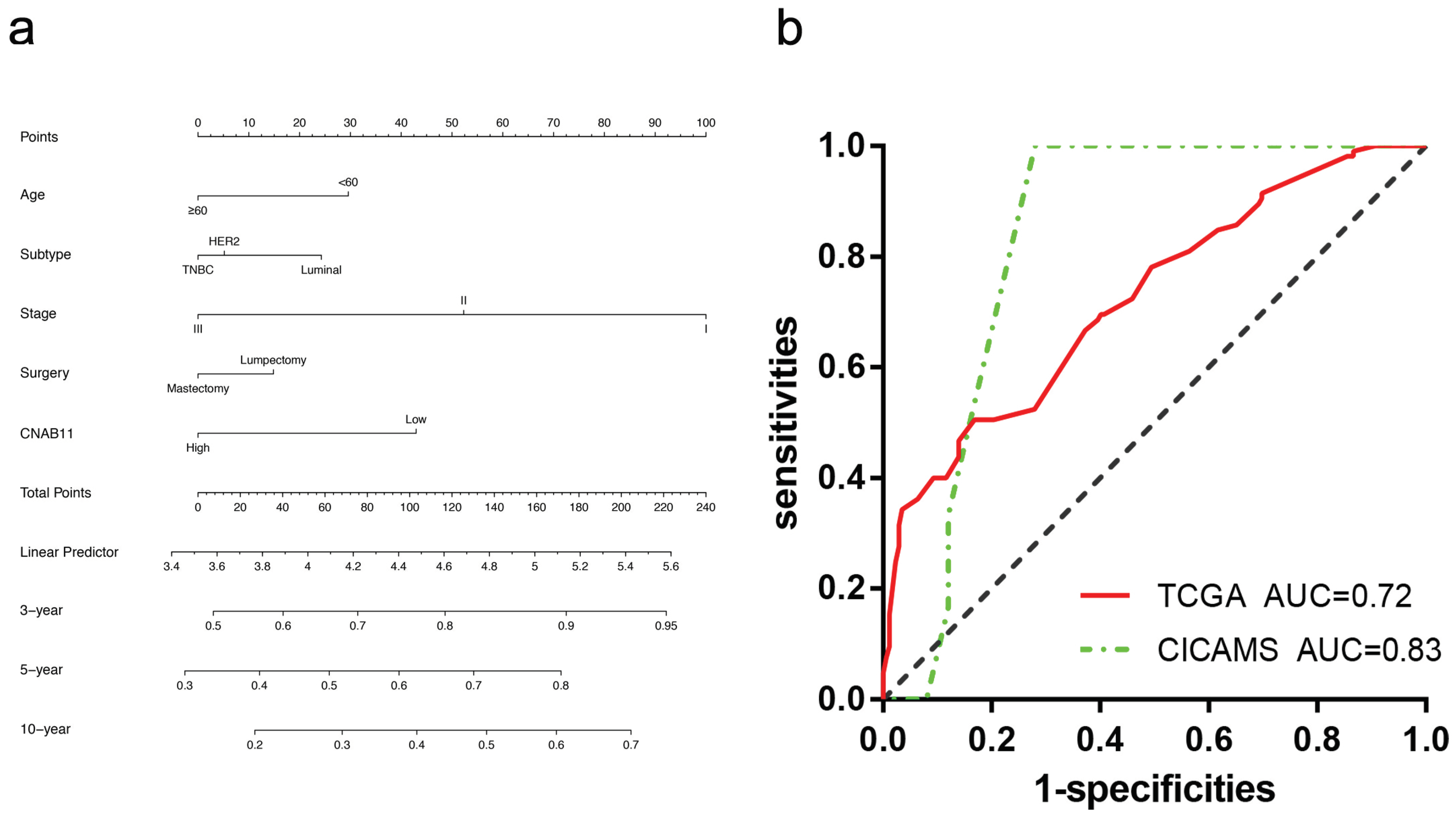
3.5. Construction and Validation of the CNAB11 Model
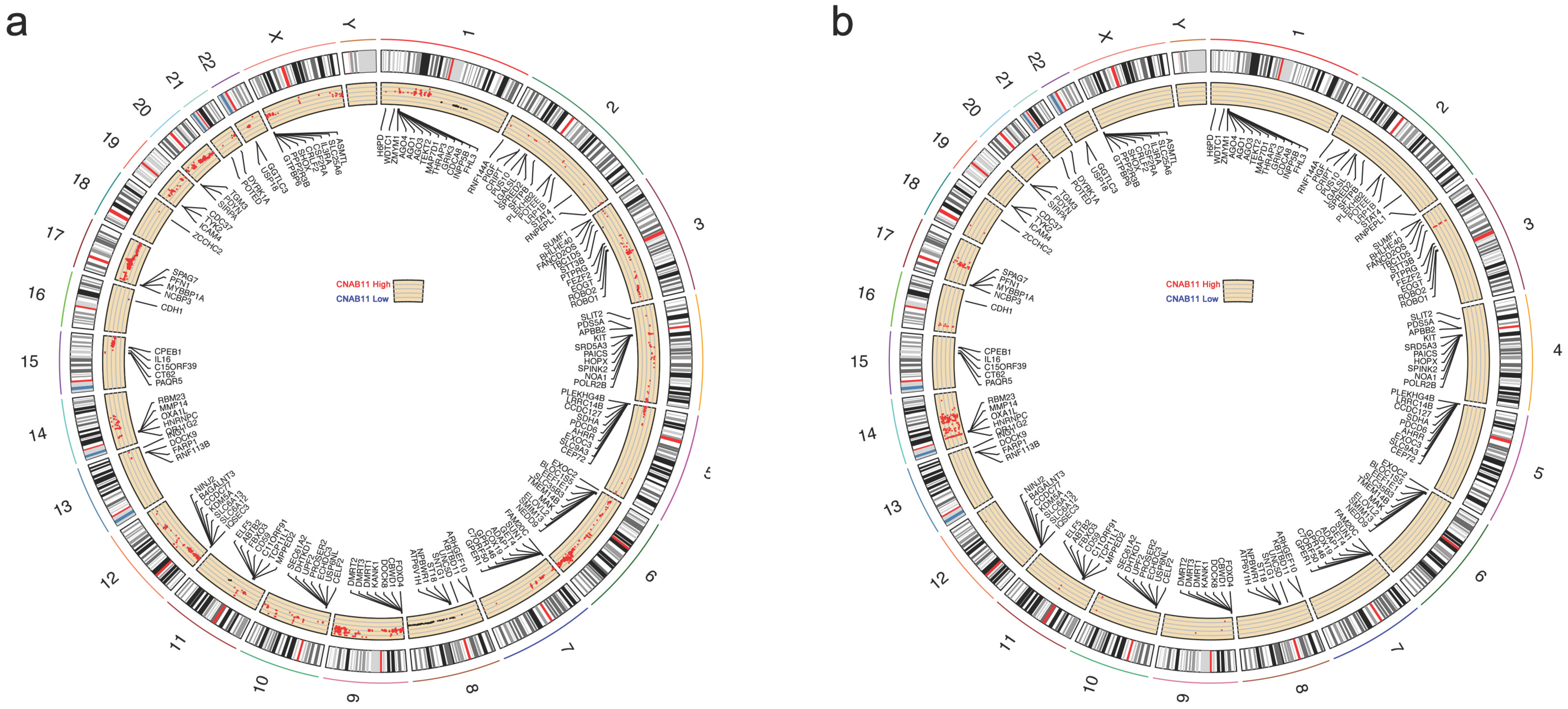
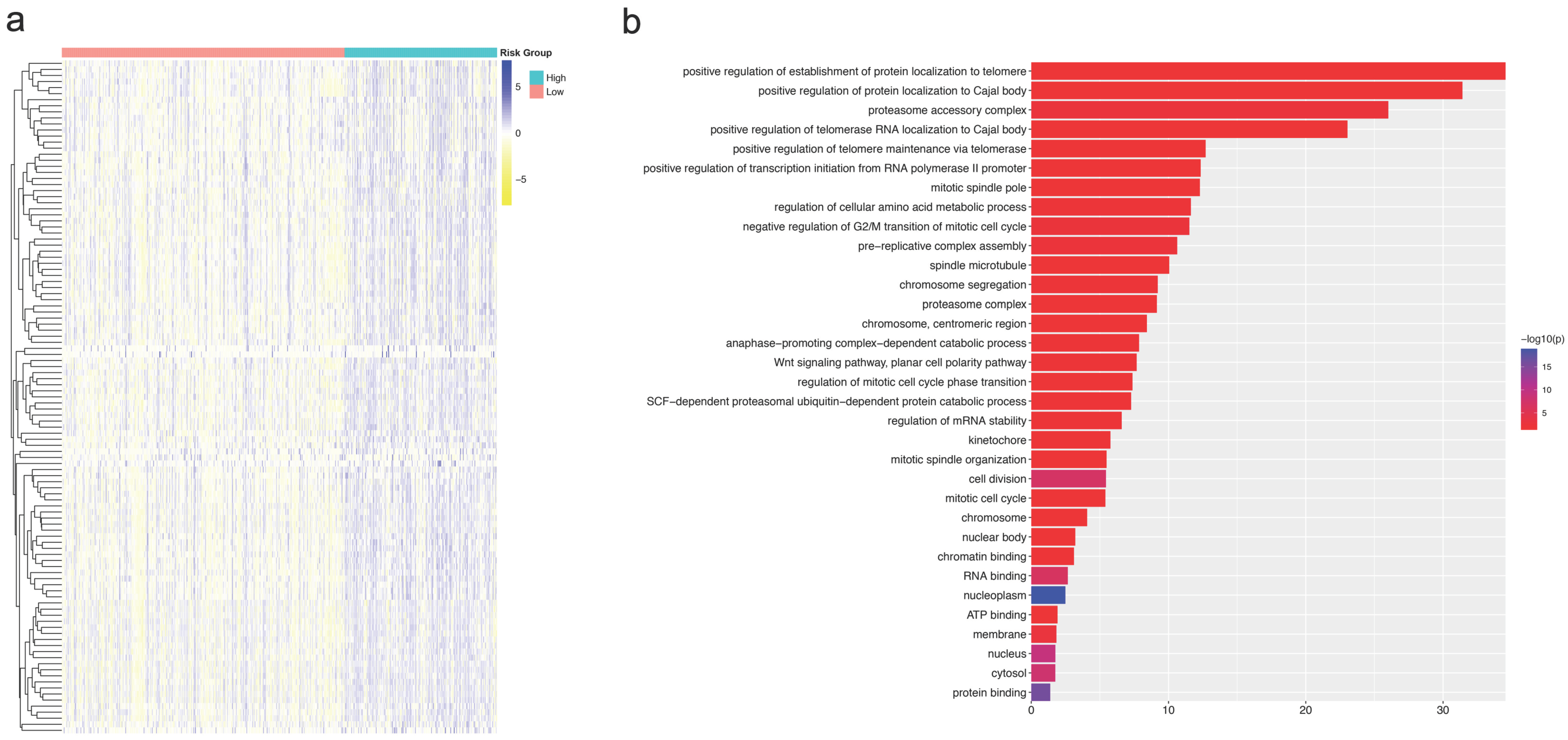
3.6. Differences in CNA and Expression Profiles of Different CNAB11 Groups
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, S.; Sun, K.; Zheng, R.; Zeng, H.; Wang, S.; Chen, R.; Wei, W.; He, J. Cancer incidence and mortality in China, 2015. J. Natl. Cancer Cent. 2021, 1, 2–11. [Google Scholar] [CrossRef]
- Zhang, L.; Feizi, N.; Chi, C.; Hu, P. Association Analysis of Somatic Copy Number Alteration Burden With Breast Cancer Survival. Front. Genet. 2018, 9, 421. [Google Scholar] [CrossRef] [PubMed]
- Glas, A.M.; Floore, A.; Delahaye, L.J.M.J.; Witteveen, A.T.; Pover, R.C.F.; Bakx, N.; Lahti-Domenici, J.S.T.; Bruinsma, T.J.; Warmoes, M.O.; Bernards, R.; et al. Converting a breast cancer microarray signature into a high-throughput diagnostic test. BMC Genomics. 2006, 7, 278. [Google Scholar] [CrossRef]
- Cobleigh, M.A.; Tabesh, B.; Bitterman, P.; Baker, J.; Cronin, M.; Liu, M.-L.; Borchik, R.; Mosquera, J.-M.; Walker, M.G.; Shak, S. Tumor gene expression and prognosis in breast cancer patients with 10 or more positive lymph nodes. Clin. Cancer Res. 2005, 11 Pt 1, 8623–8631. [Google Scholar] [CrossRef] [PubMed]
- Franch-Expósito, S.; Bassaganyas, L.; Vila-Casadesús, M.; Hernández-Illán, E.; Esteban-Fabró, R.; Díaz-Gay, M.; Lozano, J.J.; Castells, A.; Llovet, J.M.; Castellví-Bel, S.; et al. CNApp, a tool for the quantification of copy number alterations and integrative analysis revealing clinical implications. eLife 2020, 9, e50267. [Google Scholar] [CrossRef] [PubMed]
- Hieronymus, H.; Murali, R.; Tin, A.; Yadav, K.; Abida, W.; Moller, H.; Berney, D.; Scher, H.; Carver, B.; Scardino, P.; et al. Tumor copy number alteration burden is a pan-cancer prognostic factor associated with recurrence and death. eLife 2018, 7, e37294. [Google Scholar] [CrossRef] [PubMed]
- Sansregret, L.; Vanhaesebroeck, B.; Swanton, C. Determinants and clinical implications of chromosomal instability in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 139–150. [Google Scholar] [CrossRef]
- Smith, J.C.; Sheltzer, J.M. Systematic identification of mutations and copy number alterations associated with cancer patient prognosis. eLife 2018, 7, e39217. [Google Scholar] [CrossRef]
- Stopsack, K.H.; Whittaker, C.A.; Gerke, T.A.; Loda, M.; Kantoff, P.W.; Mucci, L.A.; Amon, A. Aneuploidy drives lethal progression in prostate cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 11390–11395. [Google Scholar] [CrossRef]
- Jin, X.; Yan, J.; Chen, C.; Chen, Y.; Huang, W.-K. Integrated Analysis of Copy Number Variation, Microsatellite Instability, and Tumor Mutation Burden Identifies an 11-Gene Signature Predicting Survival in Breast Cancer. Front. Cell Dev. Biol. 2021, 9, 721505. [Google Scholar] [CrossRef]
- Ueno, T.; Emi, M.; Sato, H.; Ito, N.; Muta, M.; Kuroi, K.; Toi, M. Genome-wide copy number analysis in primary breast cancer. Expert Opin. Ther. Targets 2012, 16 (Suppl. 1), S31–S35. [Google Scholar] [CrossRef] [PubMed]
- Budczies, J.; Denkert, C.; Győrffy, B.; Schirmacher, P.; Stenzinger, A. Chromosome 9p copy number gains involving PD-L1 are associated with a specific proliferation and immune-modulating gene expression program active across major cancer types. BMC Med. Genomics 2017, 10, 74. [Google Scholar] [CrossRef] [PubMed]
- Ciriello, G.; Gatza, M.L.; Beck, A.H.; Wilkerson, M.D.; Rhie, S.K.; Pastore, A.; Zhang, H.; McLellan, M.; Yau, C.; Kandoth, C.; et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell. 2015, 163, 506–519. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Bai, X.; Wang, J.; Tang, X.-R.; Wu, D.-H.; Du, S.-S.; Du, X.-J.; Zhang, Y.-W.; Zhu, H.-B.; Fang, Y.; et al. Combination of TMB and CNA Stratifies Prognostic and Predictive Responses to Immunotherapy Across Metastatic Cancer. Clin. Cancer Res. 2019, 25, 7413–7423. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Fang, J.-Y.; Xu, J. Gastric cancer and gene copy number variation: Emerging cancer drivers for targeted therapy. Oncogene 2016, 35, 1475–1482. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liang, L.; Fang, J.-Y.; Xu, J. Somatic gene copy number alterations in colorectal cancer: New quest for cancer drivers and biomarkers. Oncogene 2016, 35, 2011–2019. [Google Scholar] [CrossRef] [PubMed]
- Nibourel, O.; Guihard, S.; Roumier, C.; Pottier, N.; Terre, C.; Paquet, A.; Peyrouze, P.; Geffroy, S.; Quentin, S.; Alberdi, A.; et al. Copy-number analysis identified new prognostic marker in acute myeloid leukemia. Leukemia 2017, 31, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Pladsen, A.V.; Nilsen, G.; Rueda, O.M.; Aure, M.R.; Borgan, Ø.; Liestøl, K.; Vitelli, V.; Frigessi, A.; Langerød, A.; Mathelier, A.; et al. DNA copy number motifs are strong and independent predictors of survival in breast cancer. Commun. Biol. 2020, 3, 153. [Google Scholar] [CrossRef]
- The American Joint Committee on Cancer. AJCC Cancer Staging Manual, 7th ed.; Springer: New York, NY, USA, 2010. [Google Scholar]
- WHO. World Organization of Classification of Tumours: Pathology and Genetics of Tumors of the Breast and Female Genital Organs, 3rd ed.; IARC Press: Lyon, France, 2003. [Google Scholar]
- Beroukhim, R.; Getz, G.; Nghiemphu, L.; Barretina, J.; Hsueh, T.; Linhart, D.; Vivanco, I.; Lee, J.C.; Huang, J.H.; Alexander, S.; et al. Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma. Proc. Natl. Acad. Sci. USA 2007, 104, 20007–20012. [Google Scholar] [CrossRef]
- Tolaney, S.M.; Garrett-Mayer, E.; White, J.; Blinder, V.S.; Foster, J.C.; Amiri-Kordestani, L.; Hwang, E.S.; Bliss, J.M.; Rakovitch, E.; Perlmutter, J.; et al. Updated Standardized Definitions for Efficacy End Points (STEEP) in Adjuvant Breast Cancer Clinical Trials: STEEP Version 2.0. J. Clin. Oncol. 2021, 39, 2720–2731. [Google Scholar] [CrossRef]
- Pariyar, M.; Johns, A.; Thorne, R.F.; Scott, R.J.; Avery-Kiejda, K.A. Copy number variation in triple negative breast cancer samples associated with lymph node metastasis. Neoplasia 2021, 23, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Schrank, T.P.; Lenze, N.; Landess, L.P.; Hoyle, A.; Parker, J.; Lal, A.; Sheth, S.; Chera, B.S.; Patel, S.N.; Hackman, T.G.; et al. Genomic heterogeneity and copy number variant burden are associated with poor recurrence-free survival and 11q loss in human papillomavirus-positive squamous cell carcinoma of the oropharynx. Cancer 2021, 127, 2788–2800. [Google Scholar] [CrossRef] [PubMed]
- Bassaganyas, L.; Pinyol, R.; Esteban-Fabró, R.; Torrens, L.; Torrecilla, S.; Willoughby, C.E.; Franch-Expósito, S.; Vila-Casadesús, M.; Salaverria, I.; Montal, R.; et al. Copy-Number Alteration Burden Differentially Impacts Immune Profiles and Molecular Features of Hepatocellular Carcinoma. Clin. Cancer Res. 2020, 26, 6350–6361. [Google Scholar] [CrossRef] [PubMed]
- Sparano, J.A.; Gray, R.J.; Makower, D.F.; Pritchard, K.I.; Albain, K.S.; Hayes, D.F.; Geyer, C.E.; Dees, E.C.; Perez, E.A.; Olson, J.A.; et al. Prospective Validation of a 21-Gene Expression Assay in Breast Cancer. New Engl. J. Med. 2015, 373, 2005–2014. [Google Scholar] [CrossRef]
- Pessoa, L.S.; Heringer, M.; Ferrer, V.P. ctDNA as a cancer biomarker: A broad overview. Crit. Rev. Oncol. /Hematol. 2020, 155, 103109. [Google Scholar] [CrossRef]
- Clatot, F. Review ctDNA and Breast Cancer. Recent Results Cancer Res. 2020, 215, 231–252. [Google Scholar] [CrossRef]
- Wang, Z.; Duan, J.; Cai, S.; Han, M.; Dong, H.; Zhao, J.; Zhu, B.; Wang, S.; Zhuo, M.; Sun, J.; et al. Assessment of Blood Tumor Mutational Burden as a Potential Biomarker for Immunotherapy in Patients With Non-Small Cell Lung Cancer With Use of a Next-Generation Sequencing Cancer Gene Panel. JAMA Oncol. 2019, 5, 696–702. [Google Scholar] [CrossRef]
- Kong, X.; Duan, Y.; Sang, Y.; Li, Y.; Zhang, H.; Liang, Y.; Liu, Y.; Zhang, N.; Yang, Q. LncRNA-CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. J. Cell. Physiol. 2019, 234, 9105–9117. [Google Scholar] [CrossRef]
- Mahadevappa, R.; Neves, H.; Yuen, S.M.; Bai, Y.; McCrudden, C.M.; Yuen, H.F.; Wen, Q.; Zhang, S.-D.; Kwok, H.F. The prognostic significance of Cdc6 and Cdt1 in breast cancer. Sci. Rep. 2017, 7, 985. [Google Scholar] [CrossRef]
- Nakano, T.; Shimizu, K.; Kawashima, O.; Kamiyoshihara, M.; Kakegawa, S.; Sugano, M.; Ibe, T.; Nagashima, T.; Kaira, K.; Sunaga, N.; et al. Establishment of a human lung cancer cell line with high metastatic potential to multiple organs: Gene expression associated with metastatic potential in human lung cancer. Oncol. Rep. 2012, 28, 1727–1735. [Google Scholar] [CrossRef] [Green Version]
- Mao, X.; Gauche, C.; Coughtrie, M.; Bui, C.; Gulberti, S.; Merhi-Soussi, F.; Ramalanjaona, N.; Bertin-Jung, I.; Diot, A.; Dumas, D.; et al. The heparan sulfate sulfotransferase 3-OST3A (HS3ST3A) is a novel tumor regulator and a prognostic marker in breast cancer. Oncogene 2016, 35, 5043–5055. [Google Scholar] [CrossRef] [PubMed]
- Garcia, J.; Lizcano, F. KDM4C Activity Modulates Cell Proliferation and Chromosome Segregation in Triple-Negative Breast Cancer. Breast Cancer Basic Clin. Res. 2016, 10, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Li, R.; Song, Q.; Zhang, C.; Jia, R.; Han, Z.; Zhou, L.; Sui, H.; Liu, X.; Zhu, H.; et al. JMJD2C promotes colorectal cancer metastasis via regulating histone methylation of MALAT1 promoter and enhancing β-catenin signaling pathway. J. Exp. Clin. Cancer Res. 2019, 38, 435. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Fang, R.; Yue, C.; Chang, G.; Li, P.; Guo, Q.; Wang, J.; Zhou, A.; Zhang, S.; Fuller, G.N.; et al. Wnt-Induced Stabilization of KDM4C Is Required for Wnt/β-Catenin Target Gene Expression and Glioblastoma Tumorigenesis. Cancer Res. 2020, 80, 1049–1063. [Google Scholar] [CrossRef]
- Sreevalsan, S.; Döring, M.; Paszkowski-Rogacz, M.; Brux, M.; Blanck, C.; Meyer, M.; Momburg, F.; Buchholz, F.; Theis, M. MLLT6 maintains PD-L1 expression and mediates tumor immune resistance. EMBO Rep. 2020, 21, e50155. [Google Scholar] [CrossRef]
- Marsh, T.; Debnath, J. Autophagy suppresses breast cancer metastasis by degrading NBR1. Autophagy 2020, 16, 1164–1165. [Google Scholar] [CrossRef]
| METABRIC | TCGA | CICAMS | ||||
|---|---|---|---|---|---|---|
| Cases | % | Cases | % | Cases | % | |
| Overall | 1427 | 837 | 31 | |||
| Age | ||||||
| <60 | 647 | 45.34% | 445 | 53.17% | 22 | 70.97% |
| ≥60 | 780 | 54.66% | 392 | 46.83% | 9 | 29.03% |
| Subtype | ||||||
| HoR † +HER2 ‡ − | 1025 | 71.83% | 527 | 62.96% | 15 | 48.39% |
| HER2+ | 177 | 12.4% | 174 | 20.79% | 14 | 45.16% |
| TNBC § | 225 | 15.77% | 136 | 16.25% | 2 | 6.45% |
| Grade | ||||||
| I | 116 | 8.13% | 0 | 0% | ||
| II | 549 | 38.47% | 17 | 54.84% | ||
| III | 715 | 50.11% | 14 | 45.16% | ||
| Stage | ||||||
| I | 496 | 34.76% | 147 | 17.56% | 0 | 0% |
| II | 816 | 57.18% | 494 | 59.02% | 16 | 51.61% |
| III | 115 | 8.06% | 196 | 23.42% | 15 | 48.39% |
| Radiotherapy | ||||||
| Yes | 942 | 66.01% | 439 | 52.45% | 13 | 41.94% |
| No | 485 | 33.99% | 398 | 47.55% | 18 | 58.06% |
| Drug therapy | ||||||
| Yes | 1034 | 72.46% | 65 | 7.77% | 27 | 87.1% |
| No | 393 | 27.54% | 772 | 92.23% | 4 | 12.9% |
| Chemotherapy | ||||||
| Yes | 308 | 21.58% | 24 | 77.42% | ||
| No | 1119 | 78.42% | 7 | 22.58% | ||
| Hormone Therapy | ||||||
| Yes | 875 | 61.32% | 14 | 45.16% | ||
| No | 552 | 38.68% | 17 | 54.84% | ||
| Surgery | ||||||
| Mastectomy | 818 | 57.32% | 382 | 45.64% | 31 | 100% |
| Lumpectomy | 609 | 42.68% | 194 | 23.18% | 0 | 0% |
| Unknown | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Gao, S.; Qian, H.; Yuan, P.; Zhang, B. Prognostic Value of Copy Number Alteration Burden in Early-Stage Breast Cancer and the Construction of an 11-Gene Copy Number Alteration Model. Cancers 2022, 14, 4145. https://doi.org/10.3390/cancers14174145
Wang D, Gao S, Qian H, Yuan P, Zhang B. Prognostic Value of Copy Number Alteration Burden in Early-Stage Breast Cancer and the Construction of an 11-Gene Copy Number Alteration Model. Cancers. 2022; 14(17):4145. https://doi.org/10.3390/cancers14174145
Chicago/Turabian StyleWang, Dingyuan, Songlin Gao, Haili Qian, Peng Yuan, and Bailin Zhang. 2022. "Prognostic Value of Copy Number Alteration Burden in Early-Stage Breast Cancer and the Construction of an 11-Gene Copy Number Alteration Model" Cancers 14, no. 17: 4145. https://doi.org/10.3390/cancers14174145
APA StyleWang, D., Gao, S., Qian, H., Yuan, P., & Zhang, B. (2022). Prognostic Value of Copy Number Alteration Burden in Early-Stage Breast Cancer and the Construction of an 11-Gene Copy Number Alteration Model. Cancers, 14(17), 4145. https://doi.org/10.3390/cancers14174145






