Identification of Monobenzone as a Novel Potential Anti-Acute Myeloid Leukaemia Agent That Inhibits RNR and Suppresses Tumour Growth in Mouse Xenograft Model
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition and Gene Set Enrichment Analysis (GSEA)
2.2. Similarity Search and Molecular Docking
2.3. Compounds, Antibodies and shRNAs
2.4. Preparation of the Recombinant RNR Protein and Activity Assays
2.5. Microscale Thermophoresis (MST) Assays
2.6. Cell Lines and Drug Resistance Induction
2.7. MTT Cell Viability Assays and Analyses of the Effects of Drug Combinations
2.8. Flow Cytometry Measurements
2.9. Quantitative Real-Time PCR and Western Blotting
2.10. Tyrosinase Activity Assay
2.11. Mouse Tumour Xenograft Experiments
2.12. Statistical Analysis
3. Results
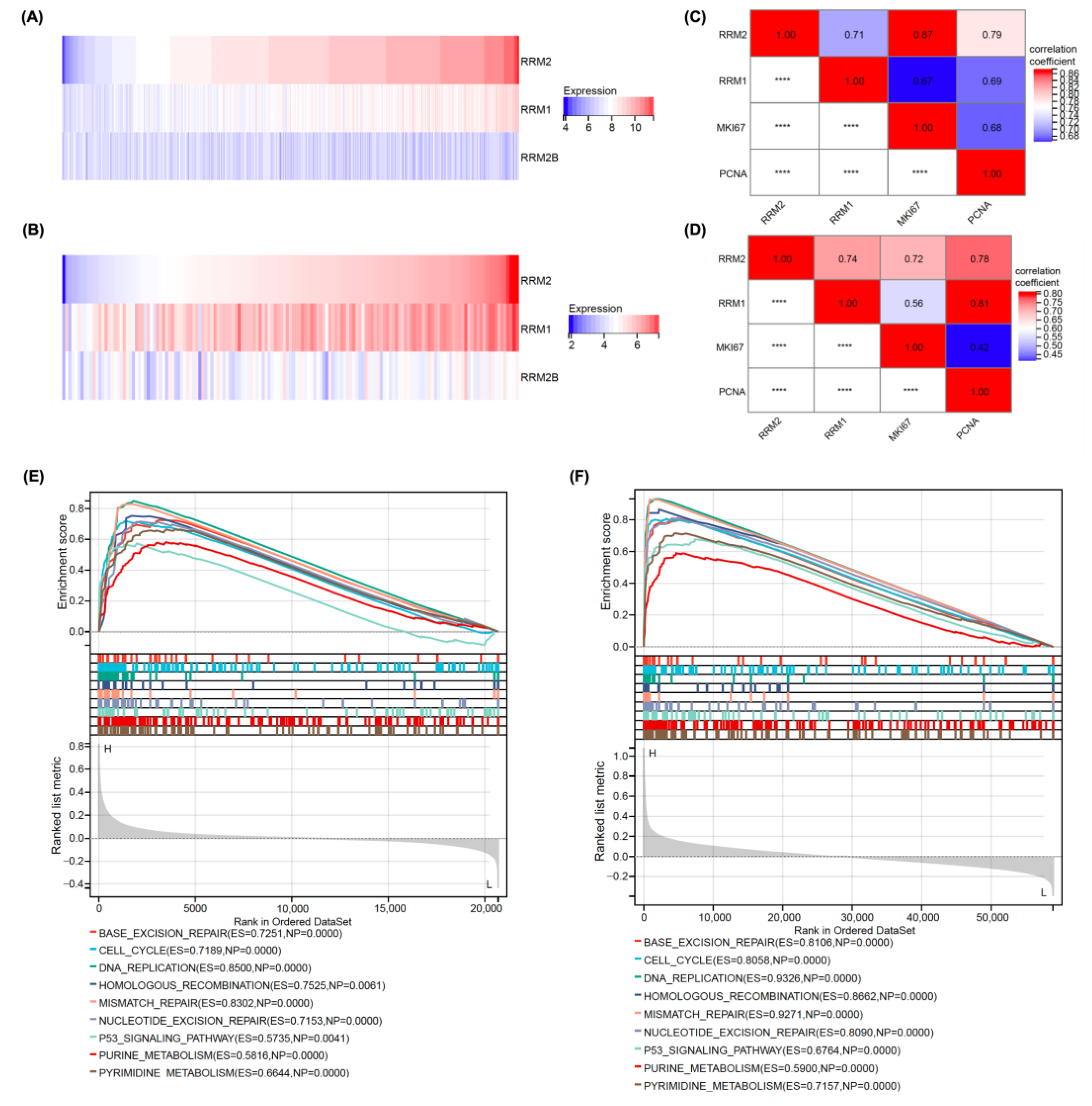
3.1. RRM2 Expression Was Positively Correlated with Malignant Proliferation in Patients with AML
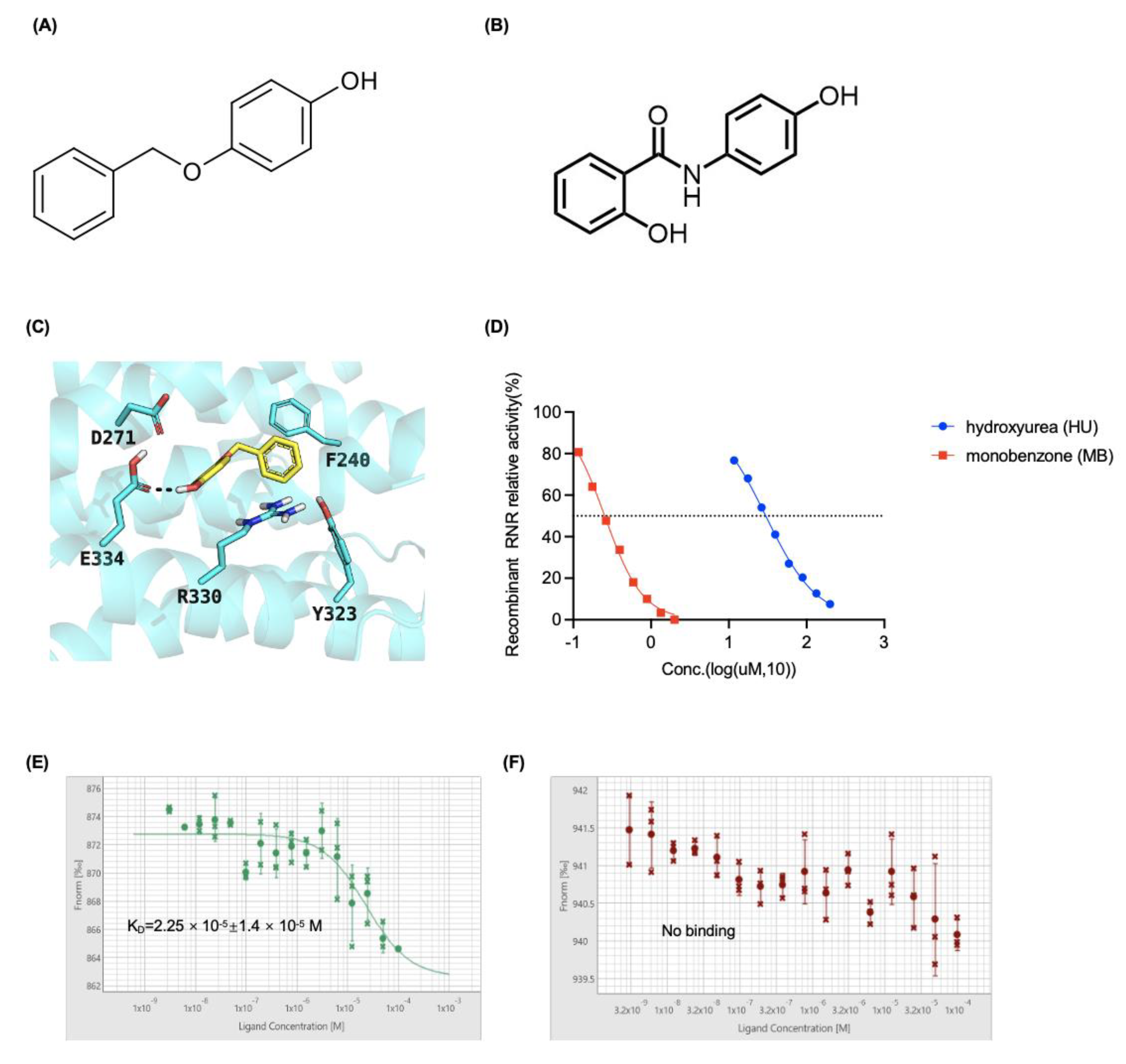
3.2. MB Potently Inhibited RNR Enzymatic Activity by Interacting with the RRM2 Protein
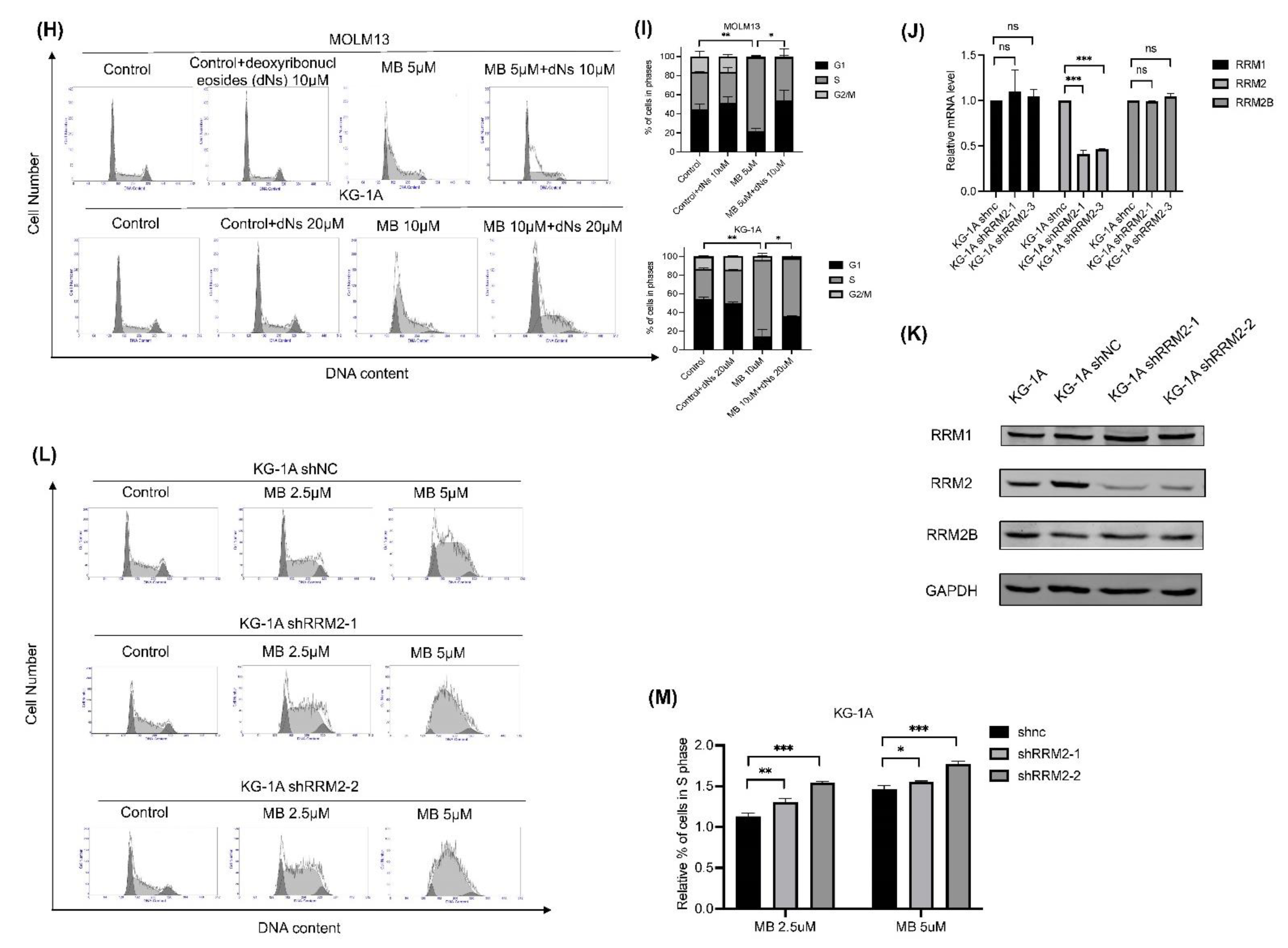
3.3. MB Effectively Inhibited Cell Growth and DNA Synthesis by Inhibiting RNR Enzymatic Activity in AML Cells
3.4. MB Overcame the Drug Resistance to Ara-C, DOX and HU in AML Cells
3.5. Combination of MB with the BCL-2 Inhibitor ABT-737 Resulted in Synergistic Inhibitory Effects on AML Cells
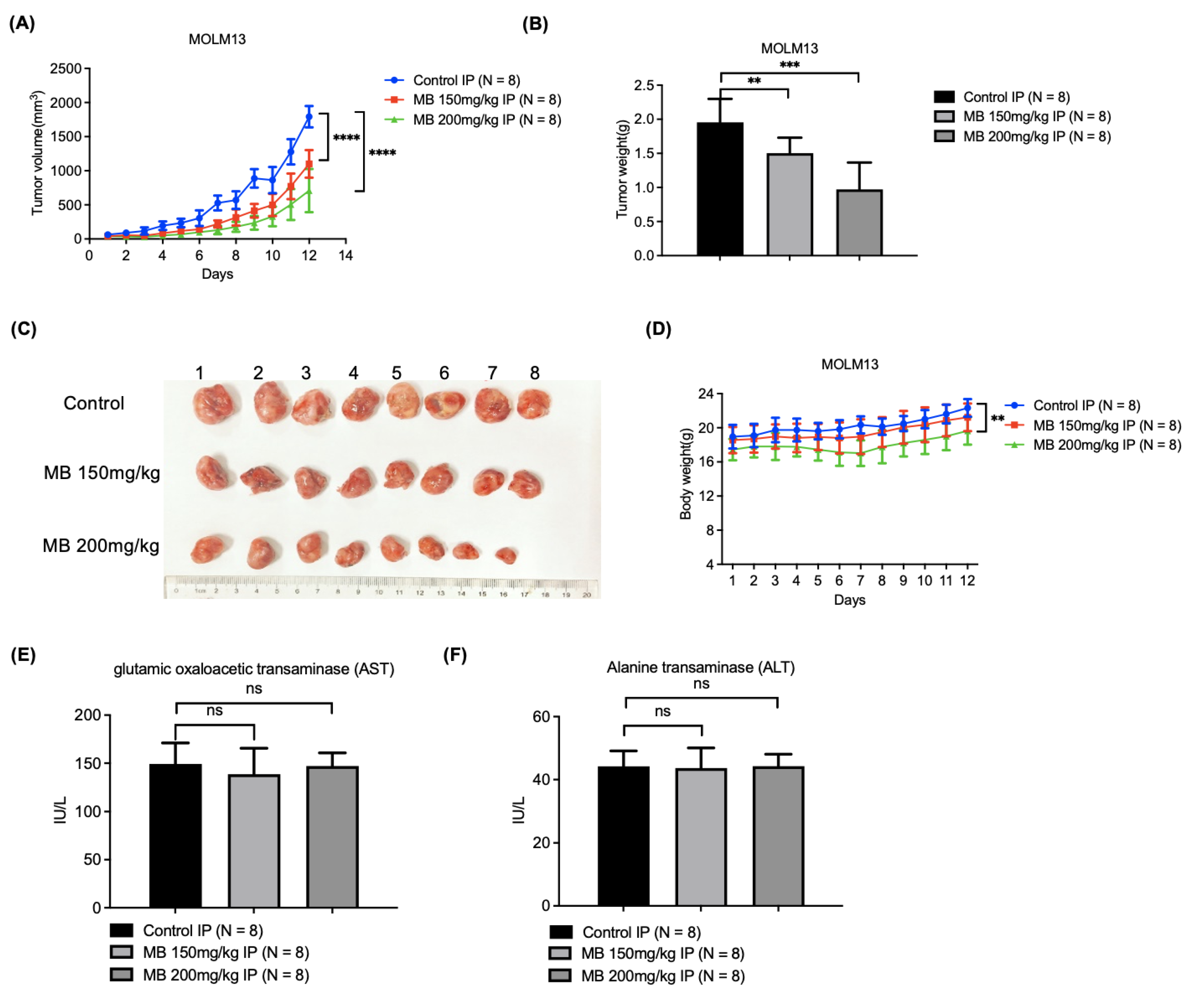
3.6. MB Effectively Inhibited AML Cell Xenograft Growth in Nude Mice with Relatively Low Toxicity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Short, N.J.; Rytting, M.E.; Cortes, J.E. Acute myeloid leukaemia. Lancet 2018, 392, 593–606. [Google Scholar] [CrossRef]
- Shallis, R.M.; Wang, R.; Davidoff, A.; Ma, X.; Zeidan, A.M. Epidemiology of acute myeloid leukemia: Recent progress and enduring challenges. Blood Rev. 2019, 36, 70–87. [Google Scholar] [CrossRef] [PubMed]
- De Kouchkovsky, I.; Abdul-Hay, M. Acute myeloid leukemia: A comprehensive review and 2016 update. Blood Cancer J. 2016, 6, e441. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Weisdorf, D.J.; Bloomfield, C.D. Acute Myeloid Leukemia. N. Engl. J. Med. 2015, 373, 1136–1152. [Google Scholar] [CrossRef]
- Schimmer, A.D. Novel Mitochondrial Mechanisms of Cytarabine Resistance in Primary AML Cells. Cancer Discov. 2017, 7, 670–672. [Google Scholar] [CrossRef]
- Löwenberg, B. Sense and nonsense of high-dose cytarabine for acute myeloid leukemia. Blood 2013, 121, 26–28. [Google Scholar] [CrossRef]
- Löwenberg, B.; Pabst, T.; Vellenga, E.; van Putten, W.; Schouten, H.C.; Graux, C.; Ferrant, A.; Sonneveld, P.; Biemond, B.J.; Gratwohl, A.; et al. Cytarabine dose for acute myeloid leukemia. N. Engl. J. Med. 2011, 364, 1027–1036. [Google Scholar] [CrossRef]
- Estey, E.H. Acute myeloid leukemia: 2014 update on risk-stratification and management. Am. J. Hematol. 2014, 89, 1063–1081. [Google Scholar] [CrossRef]
- Ramos, N.R.; Mo, C.C.; Karp, J.E.; Hourigan, C.S. Current Approaches in the Treatment of Relapsed and Refractory Acute Myeloid Leukemia. J. Clin. Med. 2015, 4, 665–695. [Google Scholar] [CrossRef]
- Kornblau, S.M.; Estey, E.; Madden, T.; Tran, H.T.; Zhao, S.; Consoli, U.; Snell, V.; Sanchez-Williams, G.; Kantarjian, H.; Keating, M.; et al. Phase I study of mitoxantrone plus etoposide with multidrug blockade by SDZ PSC-833 in relapsed or refractory acute myelogenous leukemia. J. Clin. Oncol. 1997, 15, 1796–1802. [Google Scholar] [CrossRef]
- Arbelbide, J.; García Rivello, H.; Tacchi, M.C.; Fantl, D.; Cardenas, M.P.; Penchasky, D.; Viñuales, S.; Morandi, A.; Nucifora, E.M. Prognostic value of the expression of MDR-1 in acute myeloid leukemia. Medicina 2003, 63, 277–282. [Google Scholar]
- Hahn, C.N.; Venugopal, P.; Scott, H.S.; Hiwase, D.K. Splice factor mutations and alternative splicing as drivers of hematopoietic malignancy. Immunol. Rev. 2015, 263, 257–278. [Google Scholar] [CrossRef]
- Spencer, D.H.; Russler-Germain, D.A.; Ketkar, S.; Helton, N.M.; Lamprecht, T.L.; Fulton, R.S.; Fronick, C.C.; O’Laughlin, M.; Heath, S.E.; Shinawi, M.; et al. CpG Island Hypermethylation Mediated by DNMT3A Is a Consequence of AML Progression. Cell 2017, 168, 801–816.e813. [Google Scholar] [CrossRef]
- Tanaka, H.; Arakawa, H.; Yamaguchi, T.; Shiraishi, K.; Fukuda, S.; Matsui, K.; Takei, Y.; Nakamura, Y. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature 2000, 404, 42–49. [Google Scholar] [CrossRef]
- Shao, J.; Zhou, B.; Zhu, L.; Qiu, W.; Yuan, Y.C.; Xi, B.; Yen, Y. In vitro characterization of enzymatic properties and inhibition of the p53R2 subunit of human ribonucleotide reductase. Cancer Res. 2004, 64, 1–6. [Google Scholar] [CrossRef]
- Kang, G.; Taguchi, A.T.; Stubbe, J.; Drennan, C.L. Structure of a trapped radical transfer pathway within a ribonucleotide reductase holocomplex. Science 2020, 368, 424–427. [Google Scholar] [CrossRef]
- Aye, Y.; Li, M.; Long, M.J.; Weiss, R.S. Ribonucleotide reductase and cancer: Biological mechanisms and targeted therapies. Oncogene 2015, 34, 2011–2021. [Google Scholar] [CrossRef]
- Lou, M.; Liu, Q.; Ren, G.; Zeng, J.; Xiang, X.; Ding, Y.; Lin, Q.; Zhong, T.; Liu, X.; Zhu, L.; et al. Physical interaction between human ribonucleotide reductase large subunit and thioredoxin increases colorectal cancer malignancy. J. Biol. Chem. 2017, 292, 9136–9149. [Google Scholar] [CrossRef]
- Liu, Q.; Guo, L.; Qi, H.; Lou, M.; Wang, R.; Hai, B.; Xu, K.; Zhu, L.; Ding, Y.; Li, C.; et al. A MYBL2 complex for RRM2 transactivation and the synthetic effect of MYBL2 knockdown with WEE1 inhibition against colorectal cancer. Cell Death Dis. 2021, 12, 683. [Google Scholar] [CrossRef]
- Nocentini, G. Ribonucleotide reductase inhibitors: New strategies for cancer chemotherapy. Crit. Rev. Oncol. Hematol. 1996, 22, 89–126. [Google Scholar] [CrossRef]
- Smith, B.D.; Karp, J.E. Ribonucleotide reductase: An old target with new potential. Leuk Res. 2003, 27, 1075–1076. [Google Scholar] [CrossRef]
- Shao, J.; Liu, X.; Zhu, L.; Yen, Y. Targeting ribonucleotide reductase for cancer therapy. Expert Opin. Ther. Targets 2013, 17, 1423–1437. [Google Scholar] [CrossRef] [PubMed]
- Sterkers, Y.; Preudhomme, C.; Laï, J.L.; Demory, J.L.; Caulier, M.T.; Wattel, E.; Bordessoule, D.; Bauters, F.; Fenaux, P. Acute myeloid leukemia and myelodysplastic syndromes following essential thrombocythemia treated with hydroxyurea: High proportion of cases with 17p deletion. Blood 1998, 91, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Graser-Loescher, G.; Schoenhuber, A.; Ciglenec, C.; Eberl, S.; Krupitza, G.; Mader, R.M.; Jadav, S.S.; Jayaprakash, V.; Fritzer-Szekeres, M.; Szekeres, T.; et al. Thiosemicarbazone derivatives, thiazolyl hydrazones, effectively inhibit leukemic tumor cell growth: Down-regulation of ribonucleotide reductase activity and synergism with arabinofuranosylcytosine. Food Chem. Toxicol. 2017, 108 Pt A, 53–62. [Google Scholar] [CrossRef]
- Ghanem, H.; Kantarjian, H.; Ohanian, M.; Jabbour, E. The role of clofarabine in acute myeloid leukemia. Leuk Lymphoma 2013, 54, 688–698. [Google Scholar] [CrossRef]
- Kunos, C.A.; Chu, E.; Beumer, J.H.; Sznol, M.; Ivy, S.P. Phase I trial of daily triapine in combination with cisplatin chemotherapy for advanced-stage malignancies. Cancer Chemother. Pharm. 2017, 79, 201–207. [Google Scholar] [CrossRef]
- Fang, X.; Yin, H.; Zhang, H.; Wu, F.; Liu, Y.; Fu, Y.; Yu, D.; Zong, L. p53 mediates hydroxyurea resistance in aneuploid cells of colon cancer. Exp. Cell Res. 2019, 376, 39–48. [Google Scholar] [CrossRef]
- Oliver, E.A.; Schwartz, L.; Warren, L.H. Occupational Leukoderma: Preliminary Report. J. Am. Med. Assoc. 1939, 113, 927–928. [Google Scholar] [CrossRef]
- Harris, J.E. Chemical-Induced Vitiligo. Dermatol. Clin. 2017, 35, 151–161. [Google Scholar] [CrossRef]
- Boissy, R.E.; Manga, P. On the etiology of contact/occupational vitiligo. Pigment Cell Res. 2004, 17, 208–214. [Google Scholar] [CrossRef]
- McGuire, J.; Hendee, J. Biochemical basis for depigmentation of skin by phenolic germicides. J. Investig. Dermatol. 1971, 57, 256–261. [Google Scholar] [CrossRef][Green Version]
- Cooksey, C.J.; Jimbow, K.; Land, E.J.; Riley, P.A. Reactivity of orthoquinones involved in tyrosinase-dependent cytotoxicity: Differences between alkylthio- and alkoxy-substituents. Melanoma Res. 1992, 2, 283–293. [Google Scholar] [CrossRef]
- Nazih, A.; Benezra, C.; Lepoittevin, J.P. Bihaptens with 5- and 6-methyl-substituted alkylcatechols and methylene lactone functional groups: Tools for hapten (allergen or tolerogen)-protein interaction studies. Chem. Res. Toxicol. 1993, 6, 215–222. [Google Scholar] [CrossRef]
- Naish, S.; Riley, P.A. Studies on the kinetics of oxidation of 4-hydroxyanisole by tyrosinase. Biochem. Pharmacol. 1989, 38, 1103–1107. [Google Scholar] [CrossRef]
- van den Boorn, J.G.; Picavet, D.I.; van Swieten, P.F.; van Veen, H.A.; Konijnenberg, D.; van Veelen, P.A.; van Capel, T.; Jong, E.C.; Reits, E.A.; Drijfhout, J.W.; et al. Skin-depigmenting agent monobenzone induces potent T-cell autoimmunity toward pigmented cells by tyrosinase haptenation and melanosome autophagy. J. Investig. Dermatol. 2011, 131, 1240–1251. [Google Scholar] [CrossRef]
- Teulings, H.E.; Tjin, E.P.M.; Willemsen, K.J.; van der Kleij, S.; Ter Meulen, S.; Kemp, E.H.; Krebbers, G.; van Noesel, C.J.M.; Franken, C.; Drijfhout, J.W.; et al. Anti-Melanoma immunity and local regression of cutaneous metastases in melanoma patients treated with monobenzone and imiquimod; a phase 2 a trial. Oncoimmunology 2018, 7, e1419113. [Google Scholar] [CrossRef]
- Nehme, A.; Dakik, H.; Picou, F.; Cheok, M.; Preudhomme, C.; Dombret, H.; Lambert, J.; Gyan, E.; Pigneux, A.; Récher, C.; et al. Horizontal meta-analysis identifies common deregulated genes across AML subgroups providing a robust prognostic signature. Blood Adv. 2020, 4, 5322–5335. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Chen, X.; Xu, Z.; Zhang, L.; Liu, H.; Liu, X.; Lou, M.; Zhu, L.; Huang, B.; Yang, C.G.; Zhu, W.; et al. The conserved Lys-95 charged residue cluster is critical for the homodimerization and enzyme activity of human ribonucleotide reductase small subunit M2. J. Biol. Chem. 2014, 289, 909–920. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xu, Z.; Hou, C.; Wang, M.; Chen, X.; Lin, Q.; Song, R.; Lou, M.; Zhu, L.; Qiu, Y.; et al. Inhibition of hepatitis B virus replication by targeting ribonucleotide reductase M2 protein. Biochem. Pharmacol. 2016, 103, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Kim, S.Y.; Chung, J.H.; Kim, K.H.; Eun, H.C.; Park, K.C. Delayed ERK activation by ceramide reduces melanin synthesis in human melanocytes. Cell Signal. 2002, 14, 779–785. [Google Scholar] [CrossRef]
- Ma, P.; Jia, G.; Song, Z. Monobenzone, a Novel and Potent KDM1A Inhibitor, Suppresses Migration of Gastric Cancer Cells. Front. Pharmacol. 2021, 12, 640949. [Google Scholar] [CrossRef]
- Ding, Y.; Zhong, T.; Wang, M.; Xiang, X.; Ren, G.; Jia, Z.; Lin, Q.; Liu, Q.; Dong, J.; Li, L.; et al. Integrative Analysis Reveals Across-Cancer Expression Patterns and Clinical Relevance of Ribonucleotide Reductase in Human Cancers. Front. Oncol. 2019, 9, 956. [Google Scholar] [CrossRef]
- Grant, S. Ara-C: Cellular and molecular pharmacology. Adv. Cancer Res. 1998, 72, 197–233. [Google Scholar]
- Zhang, Y.; Li, H.; Zhang, C.; An, X.; Liu, L.; Stubbe, J.; Huang, M. Conserved electron donor complex Dre2-Tah18 is required for ribonucleotide reductase metallocofactor assembly and DNA synthesis. Proc. Natl. Acad. Sci. USA 2014, 111, E1695–E1704. [Google Scholar] [CrossRef]
- Lassmann, G.; Thelander, L.; Gräslund, A. EPR stopped-flow studies of the reaction of the tyrosyl radical of protein R2 from ribonucleotide reductase with hydroxyurea. Biochem. Biophys. Res. Commun. 1992, 188, 879–887. [Google Scholar] [CrossRef]
- Swarts, J.C.; Aquino, M.A.; Han, J.Y.; Lam, K.Y.; Sykes, A.G. Kinetic studies on the reduction of the tyrosyl radical of the R2 subunit of E. coli ribonucleotide reductase. Biochim. Biophys. Acta 1995, 1247, 215–224. [Google Scholar] [CrossRef]
- Karlsson, M.; Sahlin, M.; Sjöberg, B.M. Escherichia coli ribonucleotide reductase. Radical susceptibility to hydroxyurea is dependent on the regulatory state of the enzyme. J. Biol. Chem. 1992, 267, 12622–12626. [Google Scholar] [CrossRef]
- Jiang, W.; Xie, J.; Varano, P.T.; Krebs, C.; Bollinger, J.M., Jr. Two distinct mechanisms of inactivation of the class Ic ribonucleotide reductase from Chlamydia trachomatis by hydroxyurea: Implications for the protein gating of intersubunit electron transfer. Biochemistry 2010, 49, 5340–5349. [Google Scholar] [CrossRef]
- Konopleva, M.; Contractor, R.; Tsao, T.; Samudio, I.; Ruvolo, P.P.; Kitada, S.; Deng, X.; Zhai, D.; Shi, Y.X.; Sneed, T.; et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell 2006, 10, 375–388. [Google Scholar] [CrossRef]
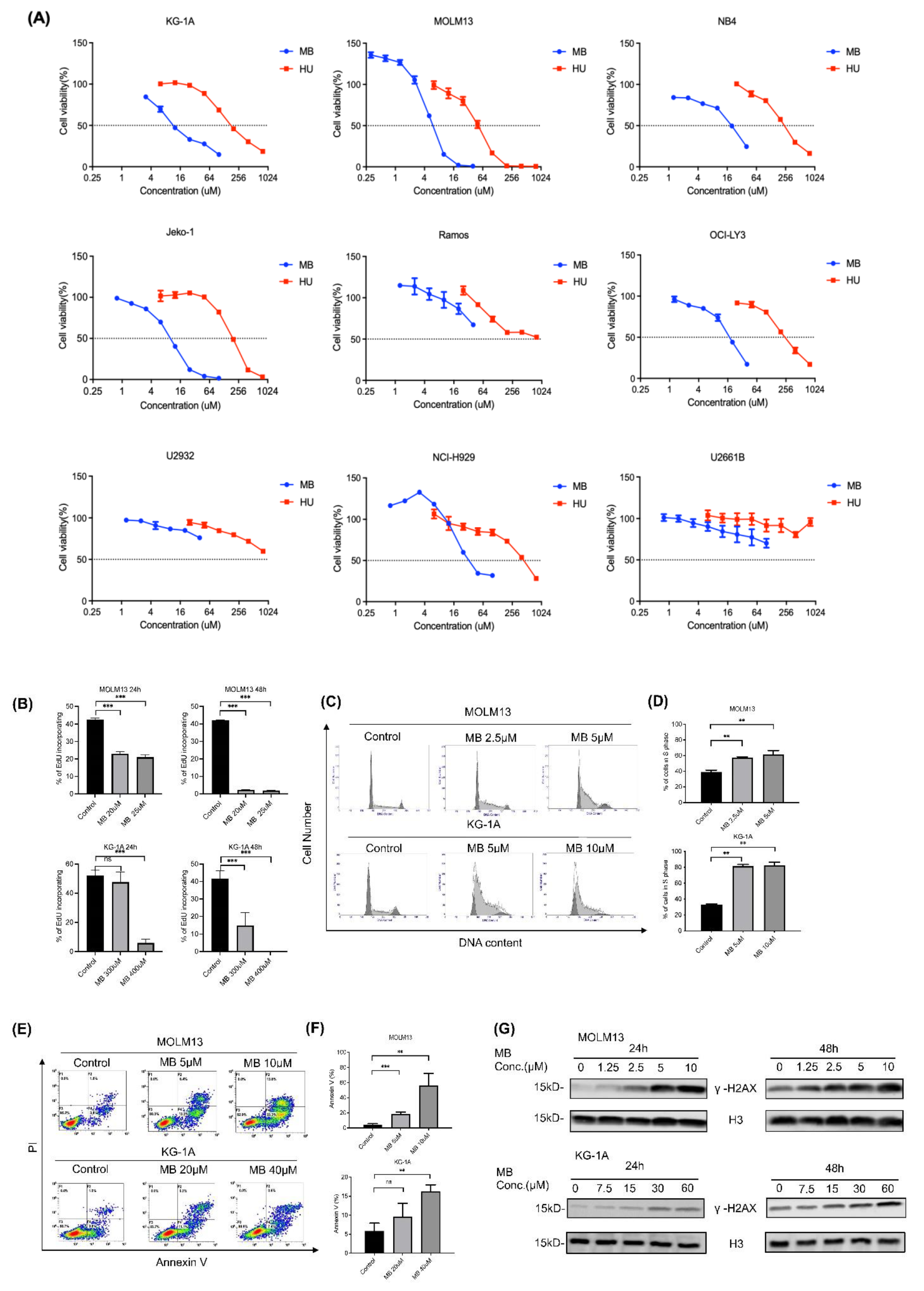


| IC50 (μM) | MB | HU |
|---|---|---|
| KG-1A | 13.9 ± 1.64 | 201.05 ± 16.75 |
| MOLM13 | 6.07 ± 1.55 | 48.85 ± 3.38 |
| NB4 | 18.04 ± 3.36 | 242.6 ± 14.2 |
| Jeko-1 | 9.6 ± 0.44 | 192.85 ± 11.45 |
| Ramos | 62.95 ± 29.02 | 715.65 ± 327.35 |
| U2932 | 302.3 ± 156 | 1517 ± 309 |
| OCI-LY3 | 16.96 ± 1.57 | 249.85 ± 16.55 |
| NCI-H929 | 42.86 ± 14.68 | 422.85 ± 67.95 |
| U2661B | >100.00 | >800.00 |
| IC50 (μM) | Parental | Resistant | Resistant Ratio |
|---|---|---|---|
| Ara-C | 0.32 ± 0.03 | 33.04 ± 14.36 | 103.25 |
| DOX | 0.12 ± 0.02 | 1.53 ± 0.1 | 12.64 |
| HU | 189.3 ± 34.3 | 1498 ± 179 | 7.90 |
| IC50 (μM) | MB | Resistant Ratio |
|---|---|---|
| KG-1A | 13.9 ± 1.64 | 1 |
| KG-1A-ARAC | 16.88 ± 3.08 | 1.20 |
| KG-1A-DOX | 24.25 ± 3.82 | 1.74 |
| KG-1A-HU | 37.04 ± 4.38 | 2.68 |
| Compound | IC50 of Cell Viability Inhibition(μM) | ||
|---|---|---|---|
| MOLM13 | KG-1A | ||
| Single | MB | 5.40 ± 0.22 | 13.66 ± 1.71 |
| ABT-737 | 1.41 ± 0.17 | 9.37 ± 0.97 | |
| Combination | MB | 2.52 | 9.69 |
| ABT-737 | 0.13 | 2.13 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, J.; Zhong, T.; Xu, Z.; Chen, H.; Wang, X.; Yang, L.; Lou, Z.; Xu, Y.; Hou, T.; Xu, R.; et al. Identification of Monobenzone as a Novel Potential Anti-Acute Myeloid Leukaemia Agent That Inhibits RNR and Suppresses Tumour Growth in Mouse Xenograft Model. Cancers 2022, 14, 4710. https://doi.org/10.3390/cancers14194710
Dong J, Zhong T, Xu Z, Chen H, Wang X, Yang L, Lou Z, Xu Y, Hou T, Xu R, et al. Identification of Monobenzone as a Novel Potential Anti-Acute Myeloid Leukaemia Agent That Inhibits RNR and Suppresses Tumour Growth in Mouse Xenograft Model. Cancers. 2022; 14(19):4710. https://doi.org/10.3390/cancers14194710
Chicago/Turabian StyleDong, Jingwen, Tingting Zhong, Zhijian Xu, Haiyi Chen, Xianjun Wang, Lili Yang, Zhiyuan Lou, Yuanling Xu, Tingjun Hou, Rongzhen Xu, and et al. 2022. "Identification of Monobenzone as a Novel Potential Anti-Acute Myeloid Leukaemia Agent That Inhibits RNR and Suppresses Tumour Growth in Mouse Xenograft Model" Cancers 14, no. 19: 4710. https://doi.org/10.3390/cancers14194710
APA StyleDong, J., Zhong, T., Xu, Z., Chen, H., Wang, X., Yang, L., Lou, Z., Xu, Y., Hou, T., Xu, R., Zhu, W., & Shao, J. (2022). Identification of Monobenzone as a Novel Potential Anti-Acute Myeloid Leukaemia Agent That Inhibits RNR and Suppresses Tumour Growth in Mouse Xenograft Model. Cancers, 14(19), 4710. https://doi.org/10.3390/cancers14194710










