Expression of MUC16/CA125 Is Associated with Impaired Survival in Patients with Surgically Resected Cholangiocarcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Database and Study Population
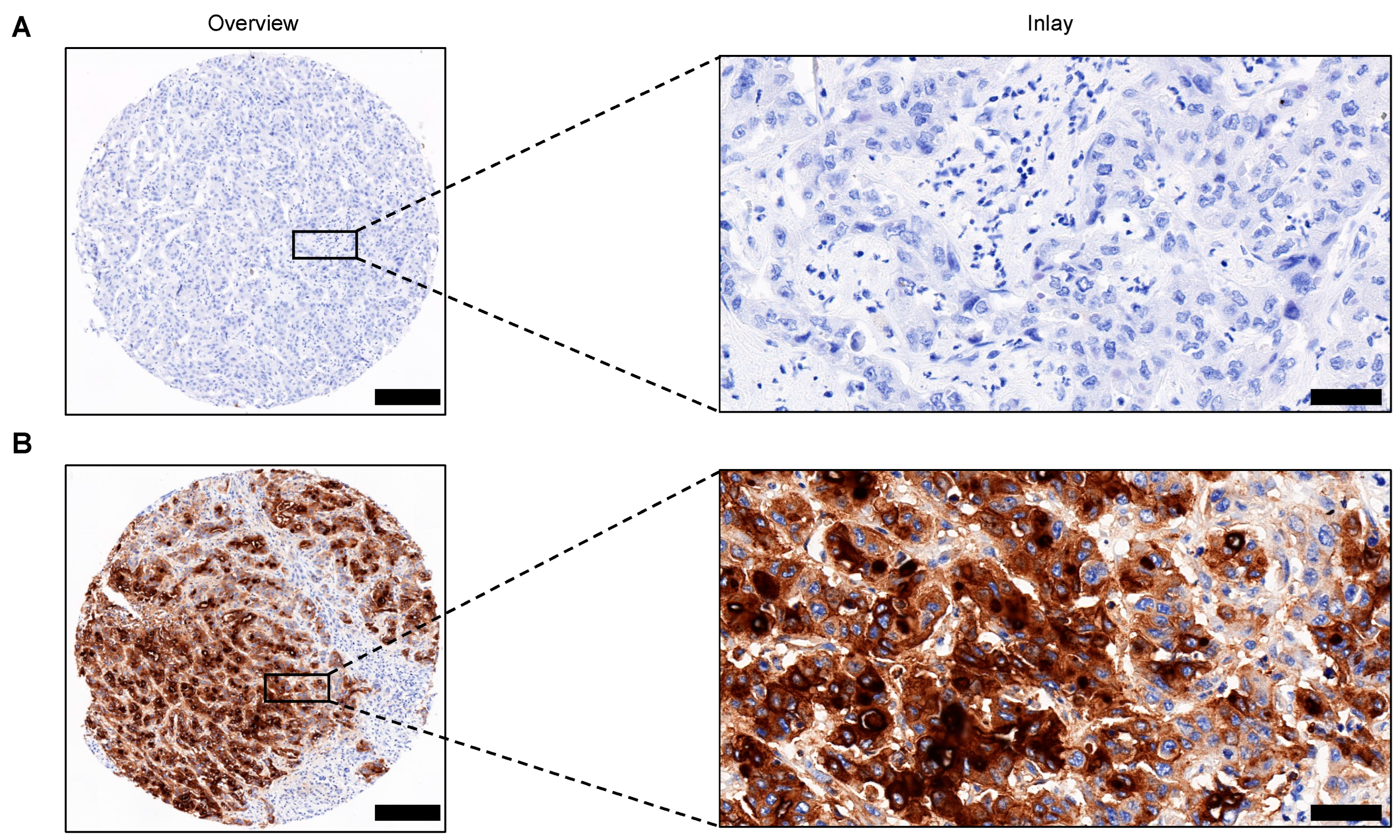
2.2. Tissue Microarray (TMA) Construction and Immunohistochemistry (IHC) Analysis
2.3. Ribonucleic Acid (RNA) Isolation and Immune Exhaustion Expression Analysis
2.4. Statistical Analysis
3. Results
3.1. Patients and Clinical Characteristics
3.2. Impact of MUC16 Expression on Overall Survival
3.3. Risk Factors Correlating with OS in CCA Patients
3.4. MUC16 Expressing iCCA Display a Differential Expression Profile
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bertuccio, P.; Bosetti, C.; Levi, F.; Decarli, A.; Negri, E.; la Vecchia, C. A comparison of trends in mortality from primary liver cancer and intrahepatic cholangiocarcinoma in Europe. Ann. Oncol. 2013, 24, 1667–1674. [Google Scholar] [CrossRef] [PubMed]
- Shaib, Y.H.; Davila, J.A.; McGlynn, K.; El-Serag, H.B. Rising incidence of intrahepatic cholangiocarcinoma in the United States: A true increase? J. Hepatol. 2004, 40, 472–477. [Google Scholar] [CrossRef]
- Koerkamp, B.G.; Fong, Y. Outcomes in biliary malignancy. J. Surg. Oncol. 2014, 110, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Dhanisha, S.S.; Guruvayoorappan, C.; Drishya, S.; Abeesh, P. Mucins: Structural diversity, biosynthesis, its role in pathogenesis and as possible therapeutic targets. Crit. Rev. Oncol. 2018, 122, 98–122. [Google Scholar] [CrossRef]
- Kufe, D.W. Mucins in cancer: Function, prognosis and therapy. Nat. Cancer 2009, 9, 874–885. [Google Scholar] [CrossRef]
- Lakshmanan, I.; Ponnusamy, M.P.; Das, S.; Chakraborty, S.; Haridas, D.; Mukhopadhyay, P.; Lele, S.M.; Batra, S.K. MUC16 induced rapid G2/M transition via interactions with JAK2 for increased proliferation and anti-apoptosis in breast cancer cells. Oncogene 2012, 31, 805–817. [Google Scholar] [CrossRef]
- Okudaira, K.; Kakar, S.; Cun, L.; Choi, E.; Decamillis, R.W.; Miura, S.; Sleisenger, M.H.; Kim, Y.S.; Deng, G. MUC2 gene promoter methylation in mucinous and non-mucinous colorectal cancer tissues. Int. J. Oncol. 2010, 36, 765–775. [Google Scholar] [PubMed]
- Chauhan, S.C.; Vannatta, K.; Ebeling, M.C.; Vinayek, N.; Watanabe, A.; Pandey, K.K.; Bell, M.C.; Koch, M.D.; Aburatani, H.; Lio, Y.; et al. Expression and functions of transmembrane mucin MUC13 in ovarian cancer. Cancer Res. 2009, 69, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Danese, E.; Ruzzenente, A.; Montagnana, M.; Lievens, P.M. Current and future roles of mucins in cholangiocarcinoma-recent evidences for a possible interplay with bile acids. Ann. Transl. Med. 2018, 6, 333. [Google Scholar] [CrossRef]
- Bast, R.C., Jr.; Spriggs, D.R. More than a biomarker: CA125 may contribute to ovarian cancer pathogenesis. Gynecol. Oncol. 2011, 121, 429–430. [Google Scholar] [CrossRef]
- Wu, Y.M.; Nowack, D.D.; Omenn, G.S.; Haab, B.B. Mucin glycosylation is altered by pro-inflammatory signaling in pancreatic-cancer cells. J. Proteome Res. 2009, 8, 1876–1886. [Google Scholar] [CrossRef]
- Chen, Y.; Huang, Y.; Kanwal, M.; Li, G.; Yang, J.; Niu, H.; Li, Z.; Ding, X. MUC16 in non-small cell lung cancer patients affected by familial lung cancer and indoor air pollution: Clinical characteristics and cell behaviors. Transl. Lung Cancer Res. 2019, 8, 476–488. [Google Scholar] [CrossRef]
- Liu, Z.; Gu, Y.; Li, X.; Zhou, L.; Cheng, X.; Jiang, H.; Huang, Y.; Zhang, Y.; Xu, T.; Yang, W.; et al. Mucin 16 Promotes Colorectal Cancer Development and Progression Through Activation of Janus Kinase 2. Am. J. Dig. Dis. 2022, 67, 2195–2208. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.L.; Ou, Y.J.; Dai, H.S.; Wan, K.; Bie, P.; Chen, Z.Y.; Zhang, L.D.; Zhang, C.C. Elevated preoperative CA125 levels predicts poor prognosis of hilar cholangiocarcinoma receiving radical surgery. Clin. Res. Hepatol. Gastroenterol. 2021, 45, 101695. [Google Scholar] [CrossRef] [PubMed]
- Fang, T.; Wang, H.; Wang, Y.; Lin, X.; Cui, Y.; Wang, Z. Clinical Significance of Preoperative Serum CEA, CA125, and CA19-9 Levels in Predicting the Resectability of Cholangiocarcinoma. Dis. Markers 2019, 2019, 6016931. [Google Scholar] [CrossRef] [PubMed]
- Higashi, M.; Yamada, N.; Yokoyama, S.; Kitamoto, S.; Tabata, K.; Koriyama, C.; Batra, S.K.; Yonezawa, S. Pathobiological implications of MUC16/CA125 expression in intrahepatic cholangiocarcinoma-mass forming type. Pathobiology 2012, 79, 101–106. [Google Scholar] [CrossRef]
- Takihata, Y.; Einama, T.; Kobayashi, K.; Suzuki, T.; Yonamine, N.; Fujinuma, I.; Tsunenari, T.; Yamagishi, Y.; Iwasaki, T.; Miyata, Y.; et al. Different role of MSLN and CA125 co-expression as a prognostic predictor between perihilar and distal bile duct carcinoma. Oncol. Lett. 2021, 21, 414. [Google Scholar] [CrossRef]
- Pietragalla, A.; Duranti, S.; Daniele, G.; Nero, C.; Ciccarone, F.; Lorusso, D.; Fagotti, A.; Scambia, G. Oregovomab: An investigational agent for the treatment of advanced ovarian cancer. Expert Opin. Investig. Drugs 2021, 30, 103–110. [Google Scholar] [CrossRef]
- Lin, C.; Verma, V.; Lazenby, A.; Ly, Q.P.; Berim, L.D.; Schwarz, J.K.; Madiyalakan, M.; Nicodemus, C.F.; Hollingsworth, M.A.; Meza, J.L.; et al. Phase I/II Trial of Neoadjuvant Oregovomab-based Chemoimmunotherapy Followed by Stereotactic Body Radiotherapy and Nelfinavir For Locally Advanced Pancreatic Adenocarcinoma. Am. J. Clin. Oncol. 2019, 42, 755–760. [Google Scholar] [CrossRef]
- Lee, D.H.; Choi, S.; Park, Y.; Jin, H.S. Mucin1 and Mucin16: Therapeutic Targets for Cancer Therapy. Pharmaceuticals 2021, 14, 1053. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Galon, J.; Mlecnik, B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics 2013, 29, 661–663. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Roh, S.J.; Kim, Y.N.; Kim, S.Z.; Park, H.S.; Jang, K.Y.; Chung, M.J.; Kang, M.J.; Lee, D.G.; Moon, W.S. Expression of MUC1, MUC2, MUC5AC and MUC6 in cholangiocarcinoma: Prognostic impact. Oncol. Rep. 2009, 22, 649–657. [Google Scholar]
- Smyth, E.C.; Fitzgerald, R.C. MUC16 Mutations and Prognosis in Gastric Cancer: A Little Goes a Long Way. JAMA Oncol. 2018, 4, 1698–1699. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Qin, Y.; Zhang, B.; Ji, S.; Shi, S.; Xu, W.; Liu, J.; Xiang, J.; Liang, D.; Hu, Q.; et al. Oncogenic KRAS Targets MUC16/CA125 in Pancreatic Ductal Adenocarcinoma. Mol. Cancer Res. 2017, 15, 201–212. [Google Scholar] [CrossRef]
- Matull, W.R.; Andreola, F.; Loh, A.; Adiguzel, Z.; Deheragoda, M.; Qureshi, U.; Batra, S.K.; Swallow, D.M.; Pereira, S.P. MUC4 and MUC5AC are highly specific tumour-associated mucins in biliary tract cancer. Br. J. Cancer 2008, 98, 1675–1681. [Google Scholar] [CrossRef]
- Li, B.; Tang, H.; Zhang, A.; Dong, J. Prognostic Role of Mucin Antigen MUC4 for Cholangiocarcinoma: A Meta-Analysis. PLoS ONE 2016, 11, e0157878. [Google Scholar] [CrossRef]
- Boonla, C.; Wongkham, S.; Sheehan, J.K.; Wongkham, C.; Bhudhisawasdi, V.; Tepsiri, N.; Pairojkul, C. Prognostic value of serum MUC5AC mucin in patients with cholangiocarcinoma. Cancer 2003, 98, 1438–1443. [Google Scholar] [CrossRef]
- Ishida, K.; Osakabe, M.; Eizuka, M.; Tai, S.; Sugimoto, R.; Fujita, Y.; Katagiri, H.; Takahara, T.; Uesugi, N.; Nitta, H.; et al. The expression of gastrointestinal differentiation markers in extrahepatic cholangiocarcinoma: Clinicopathological significance based on tumor location. Hum. Pathol. 2019, 92, 91–100. [Google Scholar] [CrossRef]
- Brewer, M.; Angioli, R.; Scambia, G.; Lorusso, D.; Terranova, C.; Panici, P.B.; Raspagliesi, F.; Scollo, P.; Plotti, F.; Ferrandina, G.; et al. Front-line chemo-immunotherapy with carboplatin-paclitaxel using oregovomab indirect immunization in advanced ovarian cancer: A randomized phase II study. Gynecol. Oncol. 2020, 156, 523–529. [Google Scholar] [CrossRef]
- Lakshmanan, I.; Salfity, S.; Seshacharyulu, P.; Rachagani, S.; Thomas, A.; Das, S.; Majhi, P.D.; Nimmakayala, R.K.; Vengoji, R.; Lele, S.M.; et al. MUC16 Regulates TSPYL5 for Lung Cancer Cell Growth and Chemoresistance by Suppressing p53. Clin. Cancer Res. 2017, 23, 3906–3917. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhang, Q.; Zhang, H.; Qiao, X.; Zhang, X.; Zhang, K.; Gu, X.; Wang, L.; Cui, J. MUC16 promotes EOC proliferation by regulating GLUT1 expression. J. Cell. Mol. Med. 2021, 25, 3031–3040. [Google Scholar] [CrossRef] [PubMed]
- Gubbels, J.A.; Felder, M.; Horibata, S.; Belisle, J.A.; Kapur, A.; Holden, H.; Petrie, S.; Migneault, M.; Rancourt, C.; Connor, J.P.; et al. MUC16 provides immune protection by inhibiting synapse formation between NK and ovarian tumor cells. Mol. Cancer 2010, 9, 11. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Guo, M.; Wang, L.; Cui, X. MUC16 facilitates cervical cancer progression via JAK2/STAT3 phosphorylation-mediated cyclooxygenase-2 expression. Genes Genom. 2020, 42, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Mazewski, C.; Perez, R.E.; Fish, E.N.; Platanias, L.C. Type I Interferon (IFN)-Regulated Activation of Canonical and Non-Canonical Signaling Pathways. Front. Immunol. 2020, 11, 606456. [Google Scholar] [CrossRef]
- Zhao, T.; Li, Y.; Zhang, J.; Zhang, B. PD-L1 expression increased by IFN-γ via JAK2-STAT1 signaling and predicts a poor survival in colorectal cancer. Oncol. Lett. 2020, 20, 1127–1134. [Google Scholar] [CrossRef]
- Moon, J.W.; Kong, S.-K.; Kim, B.S.; Kim, H.J.; Lim, H.; Noh, K.; Kim, Y.; Choi, J.-W.; Lee, J.-H.; Kim, Y.-S. IFNγ induces PD-L1 overexpression by JAK2/STAT1/IRF-1 signaling in EBV-positive gastric carcinoma. Sci. Rep. 2017, 7, 17810. [Google Scholar] [CrossRef]
- Oh, D.-Y.; He, A.R.; Qin, S.; Chen, L.-T.; Okusaka, T.; Vogel, A.; Kim, J.W.; Suksombooncharoen, T.; Lee, M.A.; Kitano, M.; et al. A phase 3 randomized, double-blind, placebo-controlled study of durvalumab in combination with gemcitabine plus cisplatin (GemCis) in patients (pts) with advanced biliary tract cancer (BTC): TOPAZ-1. J. Clin. Oncol. 2022, 40, 378. [Google Scholar] [CrossRef]



| Characteristics | MUC16 (−) (n = 111) No. (%) | MUC16 (+) (n = 57) No. (%) | p-Value |
|---|---|---|---|
| Sex | 0.647 | ||
| Female | 39 (35.1) | 18 (31.6) | |
| Male | 72 (64.9) | 39 (68.4) | |
| Age at initial diagnosis | 0.075 | ||
| Mean, years, (range) | 64.3 (38–86) | 67.3 (41–84) | |
| CCA subtype | <0.001 | ||
| iCCA | 80 (72.1) | 23 (40.4) | |
| pCCA | 19 (17.1) | 16 (28.1) | |
| dCCA | 12 (10.8) | 18 (31.6) | |
| ECOG | 0.621 | ||
| 0 | 76 (68.5) | 37 (64.9) | |
| 1 | 32 (28.8) | 31 (31.6) | |
| 2 | 3 (2.7) | 2 (3.5) | |
| CA-19/9 (ng/mL) | 0.011 | ||
| <37 | 47 (42.3) | 14 (24.6) | |
| ≥37 | 41 (36.9) | 32 (56.1) | |
| n.a. | 23 (20.7) | 11 (19.3) | |
| CA125 (U/mL) | 0.301 | ||
| <35 | 4 (3.6) | 1 (1.8) | |
| ≥35 | 0 (0) | 3 (5.3) | |
| n.a. | 107 (96.4) | 53 (93) | |
| Tumor size (cm) | <0.001 | ||
| ≤5 | 52 (46.8) | 46 (80.7) | |
| >5 | 59 (53.2) | 11 (19.3) | |
| Single Tumor | 0.567 | ||
| Yes | 73 (65.8) | 40 (70.2) | |
| No | 38 (34.2) | 17 (29.8) | |
| Pathological grade | 0.707 | ||
| Grade 1 | 1 (0.9) | 2 (3.5) | |
| Grade 2 | 83 (74.8) | 38 (66.7) | |
| Grade 3 | 27 (24.3) | 17 (29.8) | |
| M status | 0.788 | ||
| M0 | 104 (93.7) | 54 (94.7) | |
| M1 | 7 (6.3) | 3 (5.3) | |
| R status | 0.257 | ||
| R0 | 87 (78.4) | 40 (70.2) | |
| R1 | 21 (18.9) | 15 (26.3) | |
| Rx | 3 (2.7) | 2 (3.5) | |
| L status | 0.439 | ||
| L0 | 52 (46.8) | 26 (45.6) | |
| L1 | 38 (34.2) | 25 (43.9) | |
| Lx | 21 (18.9) | 6 (10.5) | |
| Pn status | 0.034 | ||
| Pn0 | 36 (32.4) | 12 (21.1) | |
| Pn1 | 51 (45.9) | 39 (68.4) | |
| Pnx | 24 (21.6) | 6 (10.5) | |
| Recurrence | 0.766 | ||
| Yes | 46 (41.4) | 25 (43.9) | |
| No | 65 (58.6) | 32 (56.1) | |
| Cholelithiasis | 0.788 | ||
| Yes | 7 (6.3) | 3 (5.3) | |
| No | 104 (93.7) | 54 (94.7) | |
| PSC | 0.086 | ||
| Yes | 2 (1.8) | 4 (7) | |
| No | 109 (98.2) | 53 (93) | |
| Viral hepatitis | 0.884 | ||
| Yes | 9 (8.1) | 5 (8.8) | |
| No | 102 (91.9) | 52 (91.2) | |
| Diabetes | 0.464 | ||
| Yes | 27 (24.3) | 11 (19.3) | |
| No | 84 (75.7) | 46 (80.7) | |
| Liver cirrhosis | 0.053 | ||
| Yes | 7 (6.3) | 0 (0) | |
| No | 104 (93.7) | 57 (100) | |
| Disease survival | 0.385 | ||
| Yes | 45 (40.5) | 12 (21.2) | |
| No | 29 (26.1) | 26 (45.6) | |
| Lost to follow-up | 17 (15.3) | 6 (10.5) | |
| n.a. | 20 (18) | 13 (22.8) | |
| Ki-67 (%) | |||
| Mean, (range) | 7.7 (0–40) | 11.9 (0–80) | 0.027 |
| Univariate Analysis | Multivariate Analysis | |||||
|---|---|---|---|---|---|---|
| Characteristics | HR | 95% CI | p-Value | HR | 95% CI | p-Value |
| Sex | ||||||
| Female | ref | |||||
| Male | 1.293 | 0.889–1.88 | 0.179 | |||
| CCA subtype | ||||||
| iCCA | ref | |||||
| pCCA | 1.18 | 0.769–1.812 | 0.449 | |||
| dCCA | 1.162 | 0.749–1.802 | 0.503 | |||
| ECOG | ||||||
| 0 | ref | ref | ||||
| 1 | 2.674 | 1.813–3.944 | <0.001 | 2.031 | 1.265–3.262 | 0.003 |
| 2 | 1.728 | 0.544–5.488 | 0.354 | 1.148 | 0.35–3.769 | 0.82 |
| CA-19/9 (ng/mL) | ||||||
| <37 | ref | ref | ||||
| ≥37 | 2.136 | 1.422–3.208 | <0.001 | 1.646 | 1.036–2.615 | 0.035 |
| Tumor size (cm) | ||||||
| ≤5 | ref | |||||
| >5 | 1.021 | 0.717–1.456 | 0.907 | |||
| Single Tumor | ||||||
| Yes | ref | ref | ||||
| No | 1.795 | 1.242–2.594 | 0.002 | 1.714 | 1.069–2.747 | 0.025 |
| MUC16 | ||||||
| Negative | ref | ref | ||||
| Positive | 1.937 | 1.337–2.806 | <0.001 | 1.636 | 1.043–2.568 | 0.032 |
| Pathological grade | ||||||
| Grade 1 | ref | ref | ||||
| Grade 2 | 1.521 | 0.374–6.194 | 0.558 | 0.672 | 0.086–5.272 | 0.705 |
| Grade 3 | 4.806 | 1.152–20.052 | 0.031 | 1.779 | 0.219–14.48 | 0.59 |
| M status | ||||||
| M0 | ref | ref | ||||
| M1 | 2.688 | 1.466–4.928 | 0.001 | 1.801 | 0.836–3.88 | 0.133 |
| R status | ||||||
| R0 | ref | ref | ||||
| R1 | 1.608 | 1.087–2.379 | 0.018 | 1.06 | 0.637–1.764 | 0.822 |
| Recurrence | ||||||
| No | ref | |||||
| Yes | 1.16 | 0.818–1.646 | 0.405 | |||
| PSC | ||||||
| No | ref | |||||
| Yes | 1.734 | 0.762–3.946 | 0.19 | |||
| Diabetes | ||||||
| No | ref | |||||
| Yes | 1.141 | 0.759–1.716 | 0.525 | |||
| Viral hepatitis | ||||||
| No | ref | |||||
| Yes | 0.541 | 0.252–1.161 | 0.115 | |||
| Liver cirrhosis | ||||||
| No | ref | |||||
| Yes | 0.744 | 0.274–2.02 | 0.562 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kinzler, M.N.; Schulze, F.; Gretser, S.; Abedin, N.; Trojan, J.; Zeuzem, S.; Schnitzbauer, A.A.; Walter, D.; Wild, P.J.; Bankov, K. Expression of MUC16/CA125 Is Associated with Impaired Survival in Patients with Surgically Resected Cholangiocarcinoma. Cancers 2022, 14, 4703. https://doi.org/10.3390/cancers14194703
Kinzler MN, Schulze F, Gretser S, Abedin N, Trojan J, Zeuzem S, Schnitzbauer AA, Walter D, Wild PJ, Bankov K. Expression of MUC16/CA125 Is Associated with Impaired Survival in Patients with Surgically Resected Cholangiocarcinoma. Cancers. 2022; 14(19):4703. https://doi.org/10.3390/cancers14194703
Chicago/Turabian StyleKinzler, Maximilian N., Falko Schulze, Steffen Gretser, Nada Abedin, Jörg Trojan, Stefan Zeuzem, Andreas A. Schnitzbauer, Dirk Walter, Peter J. Wild, and Katrin Bankov. 2022. "Expression of MUC16/CA125 Is Associated with Impaired Survival in Patients with Surgically Resected Cholangiocarcinoma" Cancers 14, no. 19: 4703. https://doi.org/10.3390/cancers14194703
APA StyleKinzler, M. N., Schulze, F., Gretser, S., Abedin, N., Trojan, J., Zeuzem, S., Schnitzbauer, A. A., Walter, D., Wild, P. J., & Bankov, K. (2022). Expression of MUC16/CA125 Is Associated with Impaired Survival in Patients with Surgically Resected Cholangiocarcinoma. Cancers, 14(19), 4703. https://doi.org/10.3390/cancers14194703





