Identification and Validation of a Novel Ferroptotic Prognostic Genes-Based Signature of Clear Cell Renal Cell Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
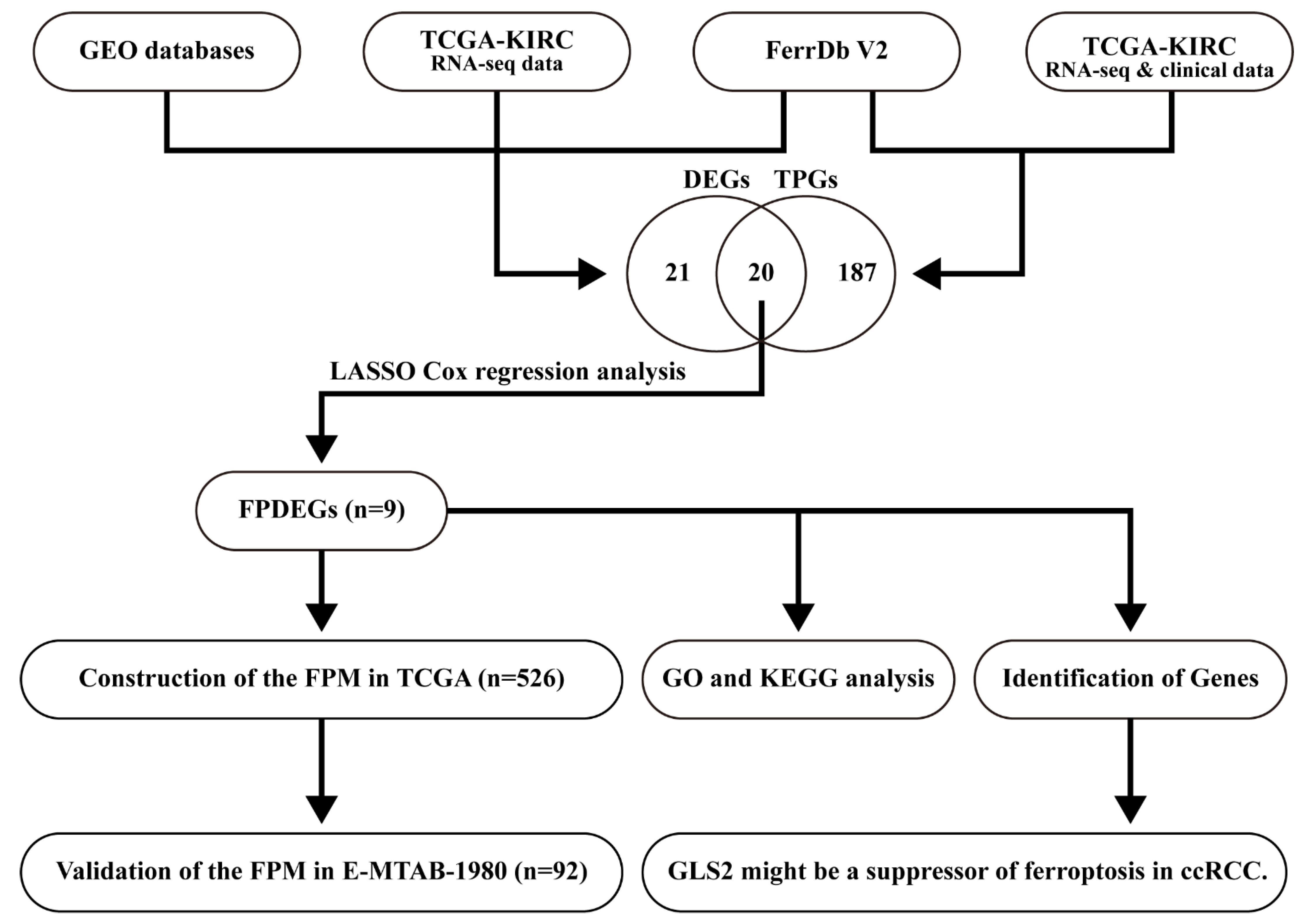
2.1. Collection and Processing of Data
2.2. Establishment and Validation of the FPM
2.3. Independent Prognostic Value of FPM
2.4. Establishment and Validation of the Nomogram
2.5. Molecular Functional Analysis
2.6. Regents and Assay Kits
2.7. Cell lines and Cell Culture
2.8. Cell Viability Assay
2.9. Real Time-PCR Assay
2.10. Lentiviral Infection
2.11. Western Blot
2.12. Detection of MDA and GSH Level
2.13. Lipid Peroxidation Assay
2.14. MBB Staining
2.15. Statistical Analysis
3. Results
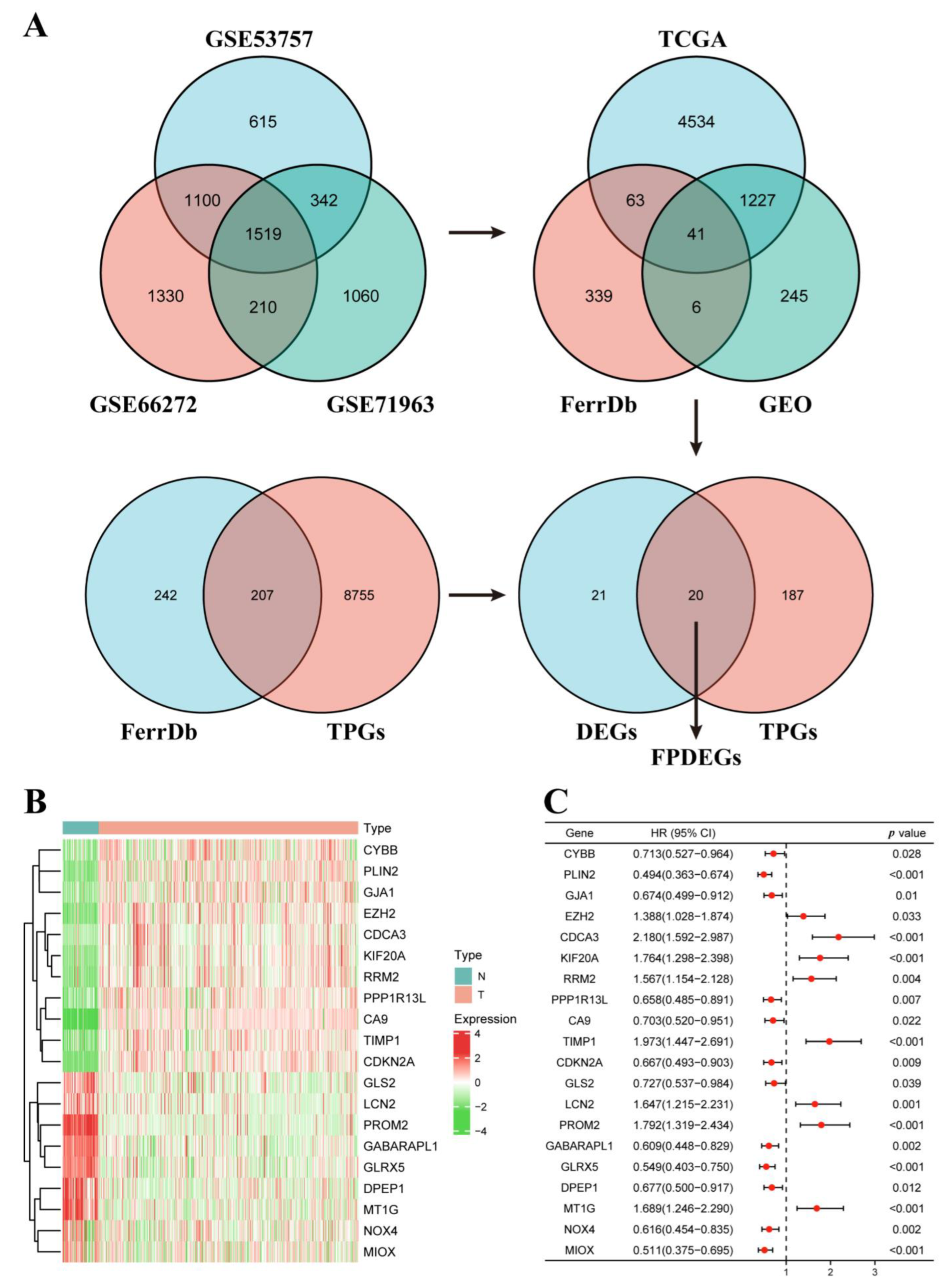
3.1. Identification of the FPDEGs
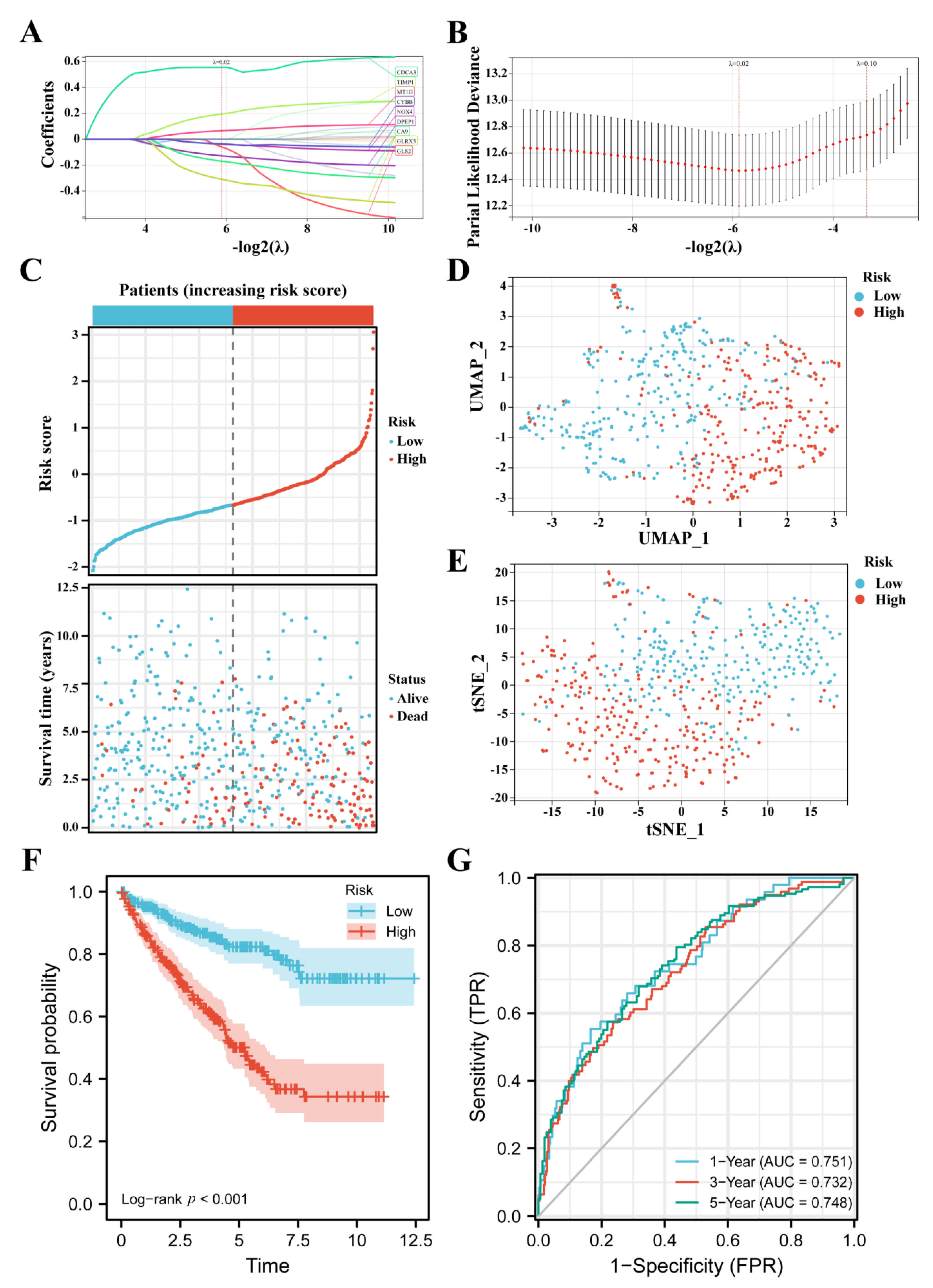
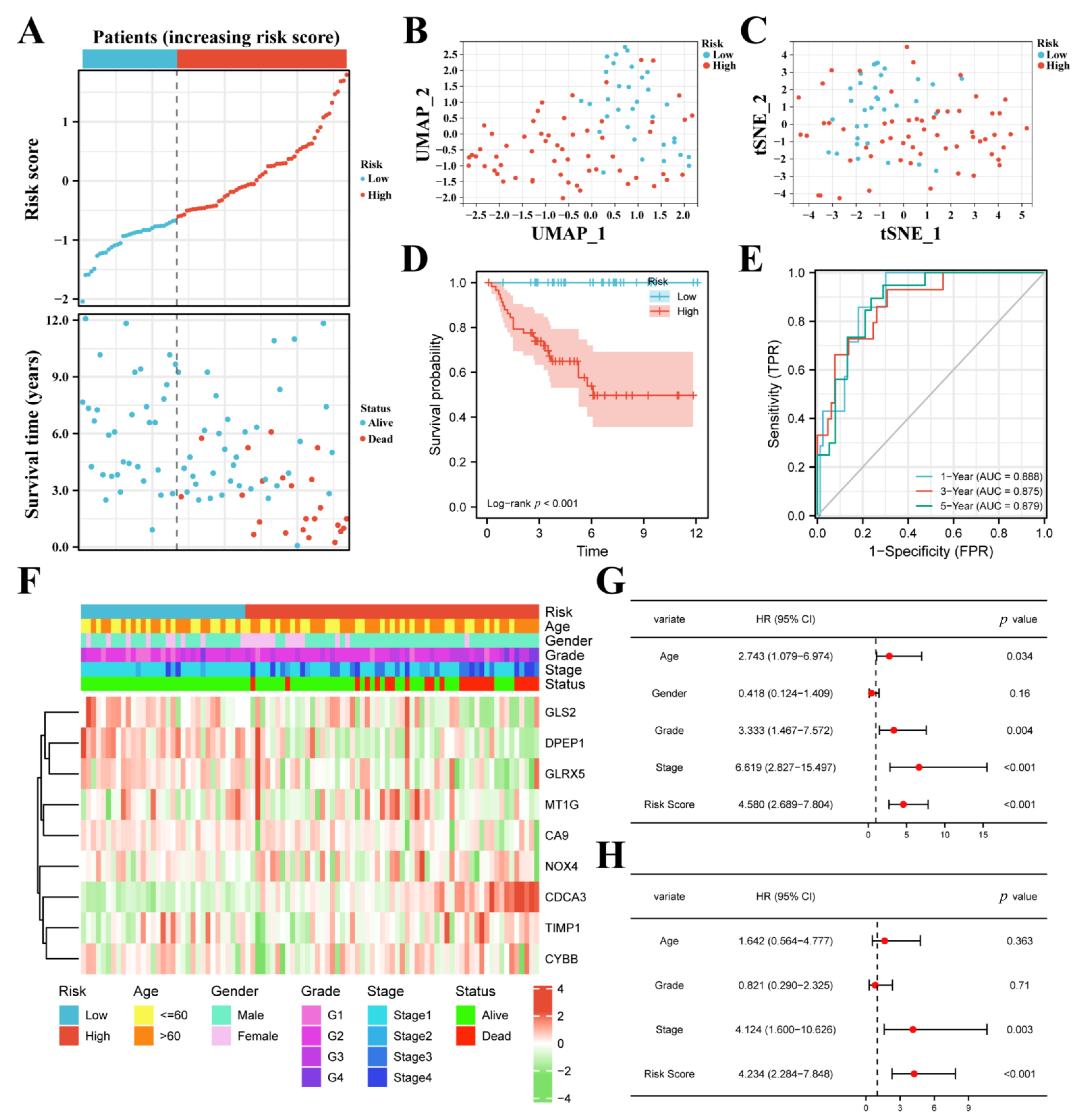
3.2. Construction of the FPM in TCGA
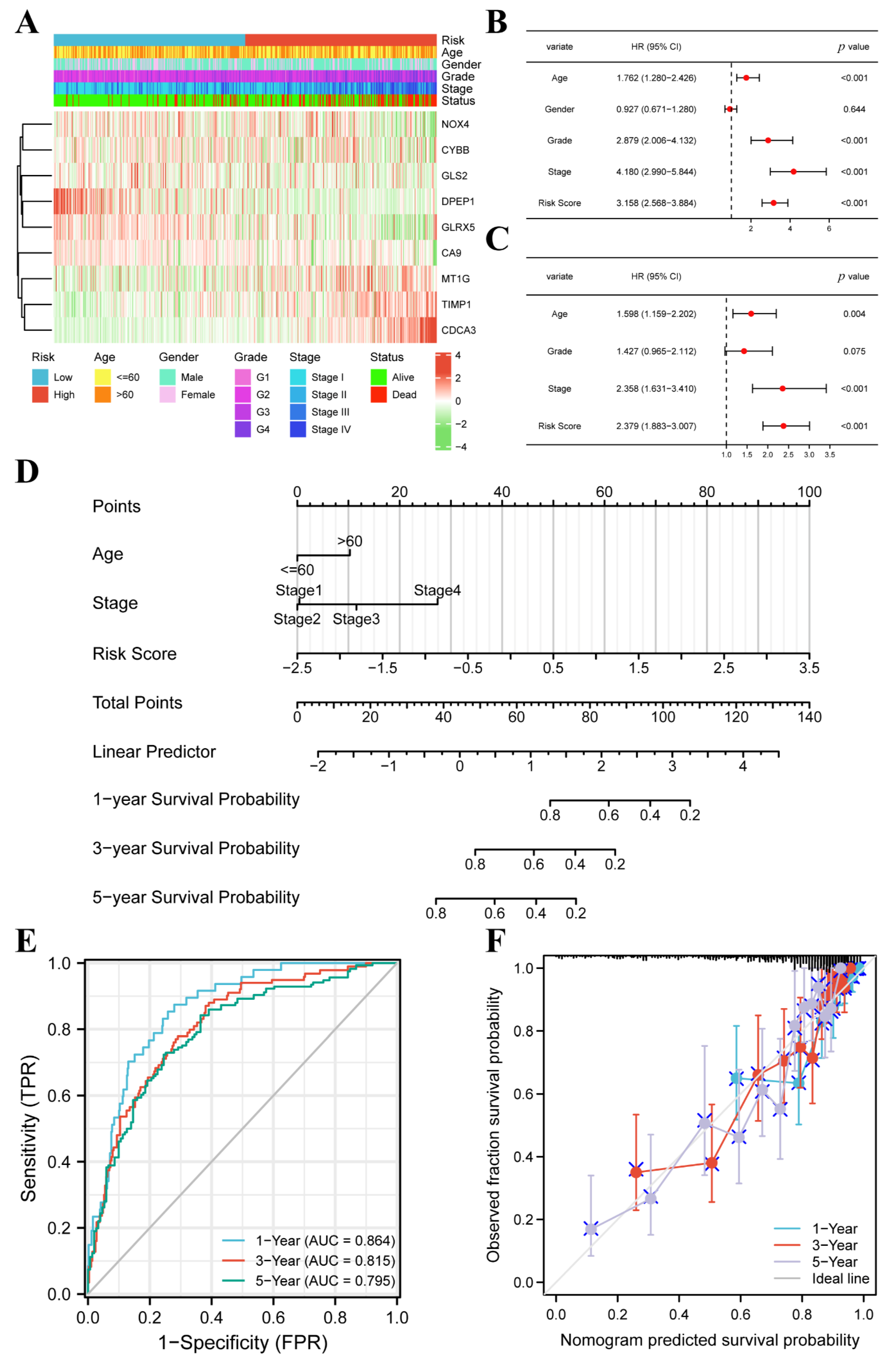
3.3. FPM Could Be a Well Independent Prognostic Factor of ccRCC
3.4. The FPM Could Be a Convincing Independent Prognostic Predictor in E-MTAB-1980 Cohort
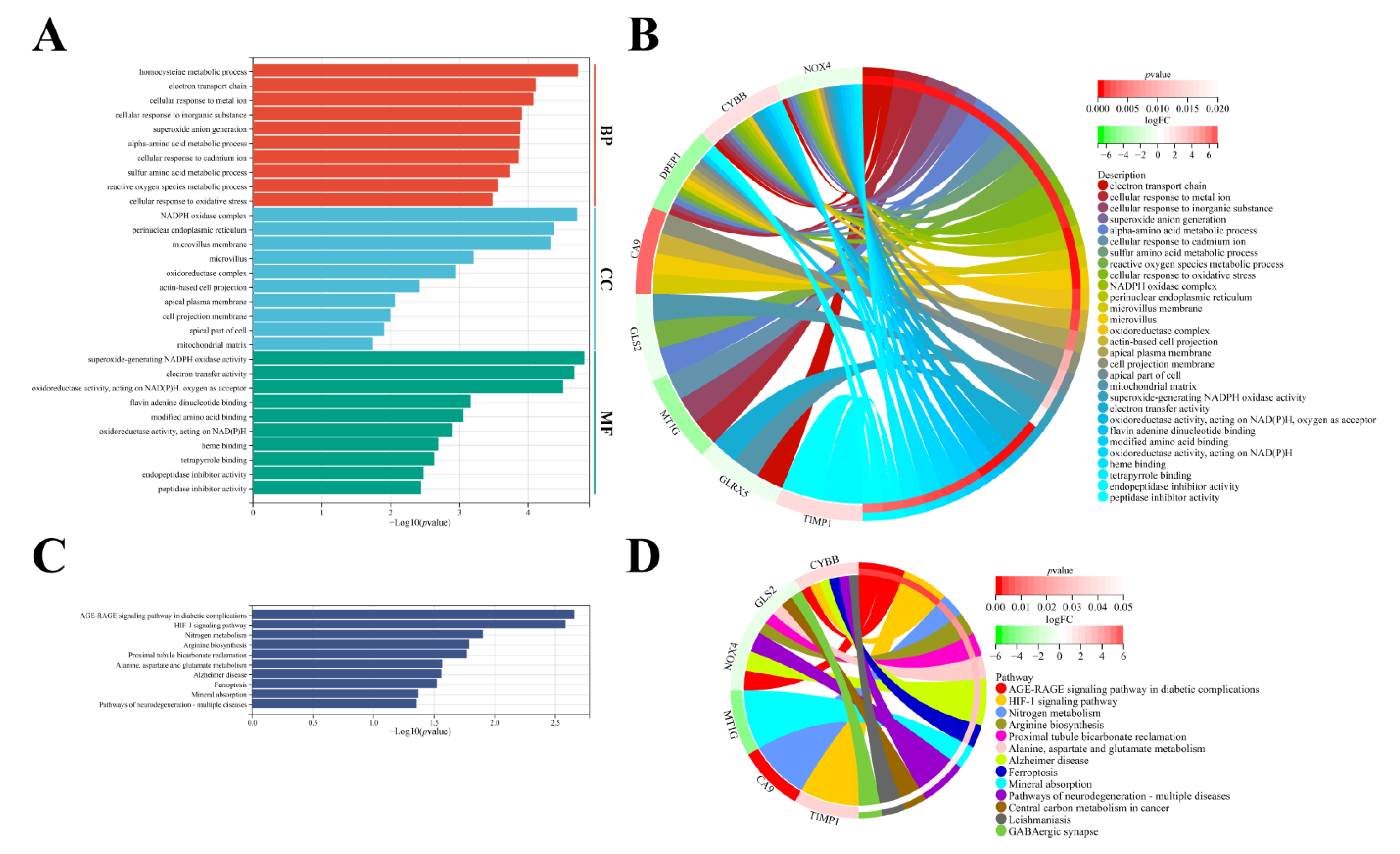
3.5. Molecular Functional Analysis
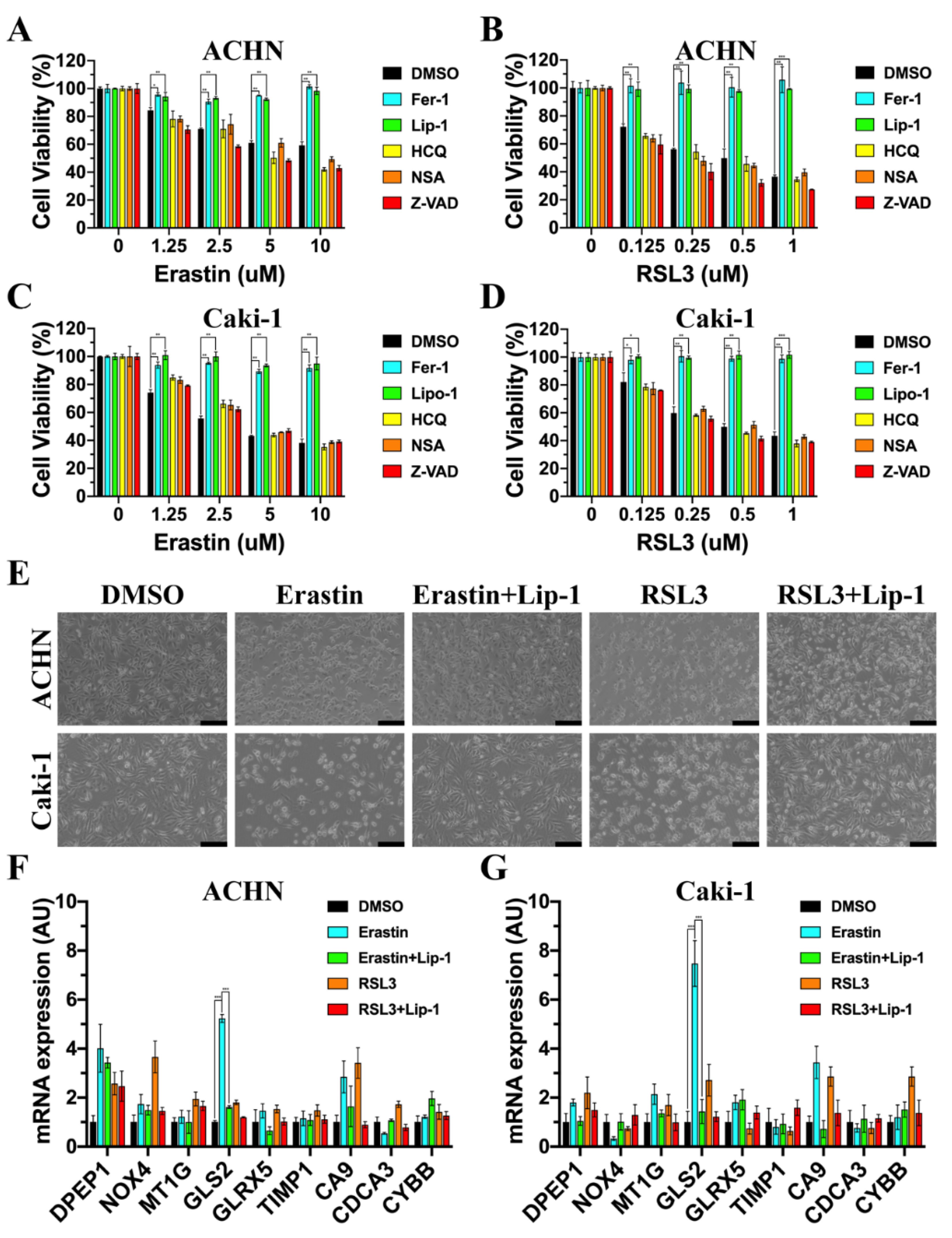
3.6. GLS2 Was Upregulated during Ferroptosis
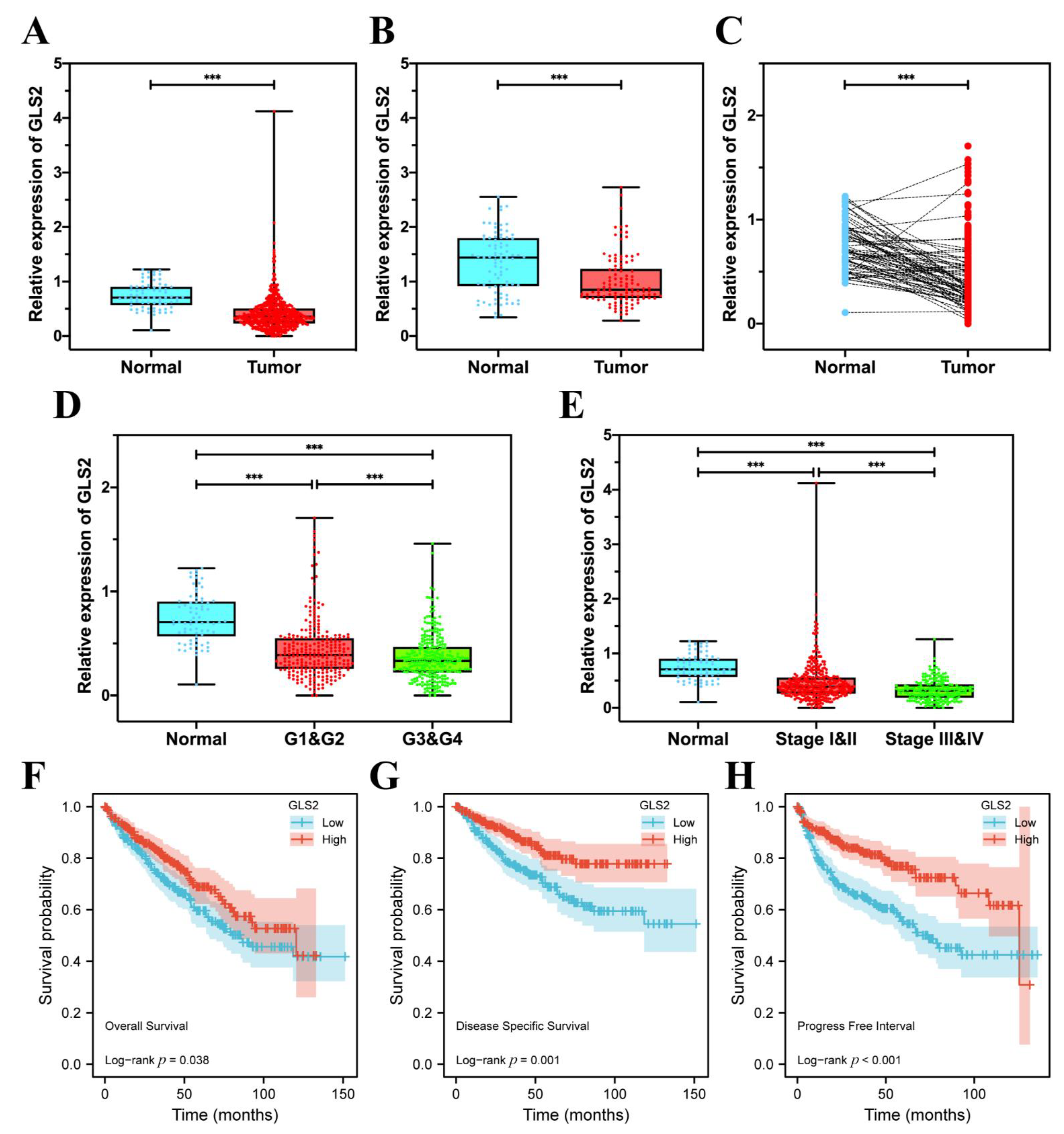
3.7. GLS2 Was Low-Expressed in ccRCC Tissues and Closely Related with Prognosis
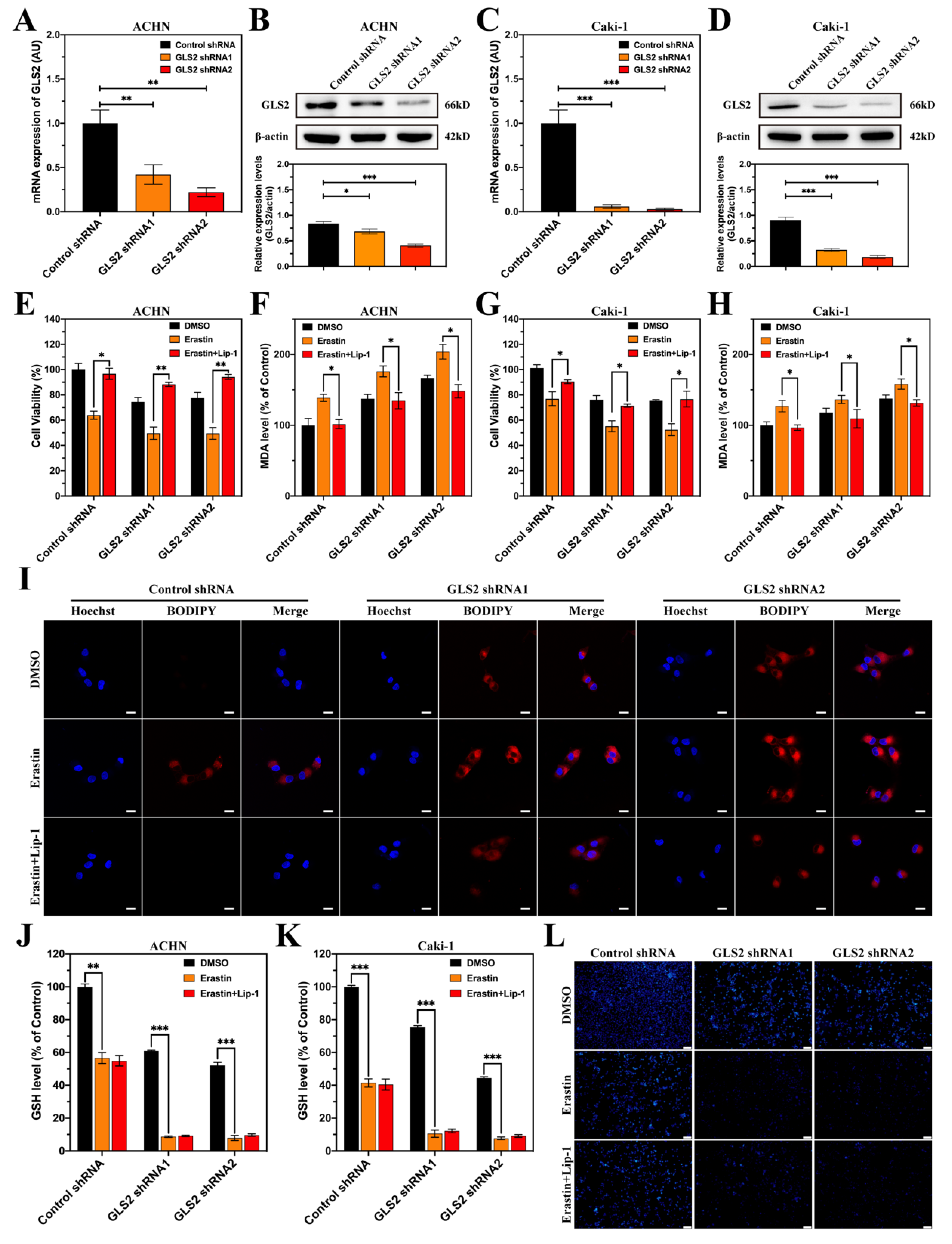
3.8. GLS2 Might Be a Suppressor of Ferroptosis in ccRCC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zheng, R.; Zhang, S.; Zeng, H.; Wang, S.; Sun, K.; Chen, R.; Li, L.; Wei, W.; He, J. Cancer Incidence and Mortality in China, 2016. J. Natl. Cancer Cent. 2022, 2, 1–9. [Google Scholar] [CrossRef]
- Miller, D.K.; Nogueira, L.; Mariotto, A.B.; Rowland, J.H.; Yabroff, K.R.; Alfano, C.M.; Jemal, A.; Kramer, J.L.; Siegel, R.L. Cancer Treatment and Survivorship Statistics, 2019. CA Cancer J. Clin. 2019, 69, 363–385. [Google Scholar] [CrossRef] [PubMed]
- Gray, E.R.; Harris, G.T. Renal Cell Carcinoma: Diagnosis and Management. Am. Fam. Physician 2019, 99, 179–184. [Google Scholar]
- Hsieh, J.J.; Purdue, M.P.; Signoretti, S.; Swanton, C.; Albiges, L.; Schmidinger, M.; Heng, D.Y.; Larkin, J.; Ficarra, V. Renal Cell Carcinoma. Nat. Rev. Dis Primers 2017, 3, 17009. [Google Scholar] [CrossRef]
- Campbell, S.; Uzzo, R.G.; Allaf, M.E.; Bass, E.B.; Cadeddu, J.A.; Chang, A.; Clark, P.E.; Davis, B.J.; Derweesh, I.H.; Giambarresi, L.; et al. Renal Mass and Localized Renal Cancer: Aua Guideline. J. Urol. 2017, 198, 520–529. [Google Scholar] [CrossRef]
- Capitanio, U.; Montorsi, F. Renal Cancer. Lancet 2016, 387, 894–906. [Google Scholar] [CrossRef]
- Rao, A.; Wiggins, C.; Lauer, R.C. Survival Outcomes for Advanced Kidney Cancer Patients in the Era of Targeted Therapies. Ann. Transl. Med. 2018, 6, 165. [Google Scholar] [CrossRef]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef]
- Shi, Z.; Zheng, J.; Tang, W.; Bai, Y.; Zhang, L.; Xuan, Z.; Sun, H.; Shao, C. Multifunctional Nanomaterials for Ferroptotic Cancer Therapy. Front. Chem. 2022, 10, 868630. [Google Scholar] [CrossRef]
- Shi, Z.; Zhang, L.; Zheng, J.; Sun, H.; Shao, C. Ferroptosis: Biochemistry and Biology in Cancers. Front. Oncol. 2021, 11, 579286. [Google Scholar] [CrossRef]
- Yagoda, N.; Von Rechenberg, M.; Zaganjor, E.; Bauer, A.J.; Yang, W.S.; Fridman, D.J.; Wolpaw, A.J.; Smukste, I.; Peltier, J.M.; Boniface, J.J.; et al. Ras-Raf-Mek-Dependent Oxidative Cell Death Involving Voltage-Dependent Anion Channels. Nature 2007, 447, 864–868. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.S.; SriRamaratnam, R.; Welsch, M.E.; Shimada, K.; Skouta, R.; Viswanathan, V.S.; Cheah, J.H.; Clemons, P.A.; Shamji, A.F.; Clish, C.B.; et al. Regulation of Ferroptotic Cancer Cell Death by Gpx4. Cell 2014, 156, 317–331. [Google Scholar] [CrossRef] [PubMed]
- Gaschler, M.M.; Stockwell, B.R. Lipid Peroxidation in Cell Death. Biochem. Biophys. Res. Commun. 2017, 482, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Weïwer, M.; Bittker, J.A.; Lewis, T.; Shimada, K.; Yang, W.S.; MacPherson, L.; Dandapani, S.; Palmer, M.; Stockwell, B.R.; Schreiber, S.L.; et al. Development of Small-Molecule Probes That Selectively Kill Cells Induced to Express Mutant Ras. Bioorg Med. Chem. Lett. 2012, 22, 1822–1826. [Google Scholar] [CrossRef]
- Fenton, H.J.H. Oxidation of Tartaric Acid in Presence of Iron. J. Chem. Soc. Trans. 1894, 65, 899–910. [Google Scholar] [CrossRef]
- Wu, Z.G.; Wang, Q.F.; Xu, Y.K.; Li, Q.L.; Cheng, L. A New Survival Model Based on Ferroptosis-Related Genes for Prognostic Prediction in Clear Cell Renal Cell Carcinoma. Aging 2020, 12, 14933–14948. [Google Scholar] [CrossRef]
- Gao, S.; Ruan, H.; Liu, J.; Liu, Y.; Liu, D.; Tong, J.; Shi, J.; Yang, H.; Xu, T.; Zhang, X. A Novel Ferroptosis-Related Pathway for Regulating Immune Checkpoints in Clear Cell Renal Cell Carcinoma. Front. Oncol. 2021, 11, 678694. [Google Scholar] [CrossRef]
- Hong, Y.; Lin, M.; Ou, D.; Huang, Z.; Shen, P. A Novel Ferroptosis-Related 12-Gene Signature Predicts Clinical Prognosis and Reveals Immune Relevancy in Clear Cell Renal Cell Carcinoma. BMC Cancer 2021, 21, 831. [Google Scholar] [CrossRef]
- Zhao, J.G.; Wu, Z.; Ge, L.; Yang, F.; Hong, K.; Zhang, S.; Ma, L. Ferroptosis-Related Gene-Based Prognostic Model and Immune Infiltration in Clear Cell Renal Cell Carcinoma. Front. Genet. 2021, 12, 650416. [Google Scholar] [CrossRef]
- Wu, J.; Sun, Z.; Bi, Q.; Wang, W. A Ferroptosis-Related Genes Model Allows for Prognosis and Treatment Stratification of Clear Cell Renal Cell Carcinoma: A Bioinformatics Analysis and Experimental Verification. Front. Oncol. 2022, 12, 815223. [Google Scholar] [CrossRef]
- Cheng, G.; Liu, D.; Liang, H.; Yang, H.; Chen, K.; Zhang, X. A Cluster of Long Non-Coding Rnas Exhibit Diagnostic and Prognostic Values in Renal Cell Carcinoma. Aging 2019, 11, 9597–9615. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Liu, C.; Zhao, G.; Ge, L.; Song, Y.; Chen, Z.; Liu, Z.; Hong, K.; Ma, L. Long Non-Coding Rna Linc01234 Regulates Proliferation, Migration and Invasion Via Hif-2α Pathways in Clear Cell Renal Cell Carcinoma Cells. PeerJ 2020, 8, e10149. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, J.; Kang, R.; Klionsky, D.J.; Tang, D. Ferroptosis: Machinery and Regulation. Autophagy 2021, 17, 2054–2081. [Google Scholar] [CrossRef] [PubMed]
- Elgendy, M.S.; Alyammahi, S.K.; Alhamad, D.W.; Abdin, S.M.; Omar, H.A. Ferroptosis: An Emerging Approach for Targeting Cancer Stem Cells and Drug Resistance. Crit. Rev. Oncol. Hematol. 2020, 155, 03095. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.M.; McCarthy, D.J.; Smyth, G.K. Edger: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, X.; Yu, M.; Zhang, W.; Zhang, D.; Zeng, S.; Wang, X.; Hu, X. A Novel Ferroptosis-Related Gene Model for Overall Survival Predictions of Bladder Urothelial Carcinoma Patients. Front. Oncol. 2021, 11, 698856. [Google Scholar] [CrossRef]
- Liu, J.; Lichtenberg, T.M.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated Tcga Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416. [Google Scholar] [CrossRef]
- Simon, N.; Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Cox’s Proportional Hazards Model via Coordinate Descent. J. Stat. Softw. 2011, 39, 1–13. [Google Scholar] [CrossRef]
- Tibshirani, R. The Lasso Method for Variable Selection in the Cox Model. Stat. Med. 1997, 16, 385–395. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. Clusterprofiler: An R Package for Comparing Biological Themes among Gene Clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Kuang, F.; Liu, J.; Xie, Y.; Tang, D.; Kang, R. Mgst1 Is a Redox-Sensitive Repressor of Ferroptosis in Pancreatic Cancer Cells. Cell Chem. Biol. 2021, 28, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zheng, J.; Xiao, J.; Yue, J.; Shi, Z.; Xuan, Z.; Chen, C.; Zhao, Y.; Tang, W.; Ye, S.; et al. Topk/Pbk Is Phosphorylated by Erk2 at Serine 32, Promotes Tumorigenesis and Is Involved in Sorafenib Resistance in Rcc. Cell Death Dis. 2022, 13, 450. [Google Scholar] [CrossRef] [PubMed]
- Deng, F.; Sharma, I.; Dai, Y.; Yang, M.; Kanwar, Y.S. Myo-Inositol Oxygenase Expression Profile Modulates Pathogenic Ferroptosis in the Renal Proximal Tubule. J. Clin. Investig. 2019, 129, 5033–5049. [Google Scholar] [CrossRef] [PubMed]
- Capitanio, U.; Becker, F.; Blute, M.L.; Mulders, P.; Patard, J.J.; Russo, P.; Studer, U.E.; van Poppel, H. Lymph Node Dissection in Renal Cell Carcinoma. Eur. Urol. 2011, 60, 1212–1220. [Google Scholar] [CrossRef]
- Xuan, Z.; Chen, C.; Tang, W.; Ye, S.; Zheng, J.; Zhao, Y.; Shi, Z.; Zhang, L.; Sun, H.; Shao, C. Tki-Resistant Renal Cancer Secretes Low-Level Exosomal Mir-549a to Induce Vascular Permeability and Angiogenesis to Promote Tumor Metastasis. Front. Cell Dev. Biol. 2021, 9, 689947. [Google Scholar] [CrossRef]
- Wu, H.Z.; Tang, Y.; Yu, H.; Li, H.D. The Role of Ferroptosis in Breast Cancer Patients: A Comprehensive Analysis. Cell Death Discov. 2021, 7, 93. [Google Scholar] [CrossRef]
- Li, R.; Zhang, J.; Zhou, Y.; Gao, Q.; Wang, R.; Fu, Y.; Zheng, L.; Yu, H. Transcriptome Investigation and in Vitro Verification of Curcumin-Induced Ho-1 as a Feature of Ferroptosis in Breast Cancer Cells. Oxid Med. Cell Longev. 2020, 11, 3469840. [Google Scholar] [CrossRef]
- Yang, X.; Liu, J.; Wang, C.; Cheng, K.K.-Y.; Xu, H.; Li, Q.; Hua, T.; Jiang, X.; Sheng, L.; Mao, J.; et al. Mir-18a Promotes Glioblastoma Development by Down-Regulating Aloxe3-Mediated Ferroptotic and Anti-Migration Activities. Oncogenesis 2021, 10, 15. [Google Scholar] [CrossRef]
- Xia, L.; Gong, M.; Zou, Y.; Wang, Z.; Wu, B.; Zhang, S.; Li, L.; Jin, K.; Sun, C. Apatinib Induces Ferroptosis of Glioma Cells through Modulation of the Vegfr2/Nrf2 Pathway. Oxid Med. Cell Longev. 2022, 5, 9925919. [Google Scholar] [CrossRef]
- Asperti, M.; Bellini, S.; Grillo, E.; Gryzik, M.; Cantamessa, L.; Ronca, R.; Maccarinelli, F.; Salvi, A.; De Petro, G.; Arosio, P.; et al. H-Ferritin Suppression and Pronounced Mitochondrial Respiration Make Hepatocellular Carcinoma Cells Sensitive to Rsl3-Induced Ferroptosis. Free Radic. Biol. Med. 2021, 169, 294–303. [Google Scholar] [CrossRef]
- Sun, F.X.; Niu, X.H.; Chen, R.C.; He, W.Y.; Chen, D.; Kang, R.; Tang, D.L. Metallothionein-1g Facilitates Sorafenib Resistance through Inhibition of Ferroptosis. Hepatology 2016, 64, 488–500. [Google Scholar] [CrossRef]
- Koppula, P.; Lei, G.; Zhang, Y.; Yan, Y.; Mao, C.; Kondiparthi, L.; Shi, J.; Liu, X.; Horbath, A.; Das, M.; et al. A Targetable Coq-Fsp1 Axis Drives Ferroptosis- and Radiation-Resistance in Keap1 Inactive Lung Cancers. Nat. Commun. 2022, 13, 2206. [Google Scholar] [CrossRef]
- Meng, C.; Zhan, J.; Chen, D.; Shao, G.; Zhang, H.; Gu, W.; Luo, J. The Deubiquitinase Usp11 Regulates Cell Proliferation and Ferroptotic Cell Death Via Stabilization of Nrf2 Usp11 Deubiquitinates and Stabilizes Nrf2. Oncogene 2021, 40, 1706–1720. [Google Scholar] [CrossRef] [PubMed]
- Kremer, D.M.; Nelson, B.S.; Lin, L.; Yarosz, E.L.; Halbrook, C.J.; Kerk, S.A.; Sajjakulnukit, P.; Myers, A.; Thurston, G.; Hou, S.W.; et al. Got1 Inhibition Promotes Pancreatic Cancer Cell Death by Ferroptosis. Nat. Commun. 2021, 12, 4860. [Google Scholar] [CrossRef]
- Zhou, Z.; Yang, Z.; Cui, Y.; Lu, S.; Huang, Y.; Che, X.; Yang, L.; Zhang, Y. Identification and Validation of a Ferroptosis-Related Long Non-Coding Rna (Frlncrna) Signature to Predict Survival Outcomes and the Immune Microenvironment in Patients with Clear Cell Renal Cell Carcinoma. Front. Genet. 2022, 13, 787884. [Google Scholar] [CrossRef]
- Dong, Y.; Liu, D.; Zhou, H.; Gao, Y.; Nueraihemaiti, Y.; Xu, Y. A Prognostic Signature for Clear Cell Renal Cell Carcinoma Based on Ferroptosis-Related Lncrnas and Immune Checkpoints. Front. Genet. 2022, 13, 912190. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Tu, J.; Ma, L.; Huang, Y.; Yang, C.; Yuan, X. Analysis of Ferroptosis-Related Lncrnas Signatures Associated with Tumor Immune Infiltration and Experimental Validation in Clear Cell Renal Cell Carcinoma. Int. J. Gen. Med. 2022, 15, 3215–3235. [Google Scholar] [CrossRef] [PubMed]
- Cochetti, G.; Cari, L.; Maulà, V.; Cagnani, R.; Paladini, A.; del Zingaro, M.; Nocentini, G.; Mearin, E.I. Validation in an Independent Cohort of Mir-122, Mir-1271, and Mir-15b as Urinary Biomarkers for the Potential Early Diagnosis of Clear Cell Renal Cell Carcinoma. Cancers 2022, 14, 1112. [Google Scholar] [CrossRef]
- Zheng, B.; Niu, Z.; Si, S.; Zhao, G.; Wang, J.; Yao, Z.; Cheng, F.; He, W. Comprehensive Analysis of New Prognostic Signature Based on Ferroptosis-Related Genes in Clear Cell Renal Cell Carcinoma. Aging 2021, 13, 19789–19804. [Google Scholar] [CrossRef]
- Chen, J.; Zhan, Y.; Zhang, R.; Chen, B.; Huang, J.; Li, C.; Zhang, W.; Wang, Y.; Gao, Y.; Zheng, J.; et al. A New Prognostic Risk Signature of Eight Ferroptosis-Related Genes in the Clear Cell Renal Cell Carcinoma. Front. Oncol. 2021, 11, 700084. [Google Scholar] [CrossRef]
- Li, S.; Xu, X.; Zhang, R.; Huang, Y. Identification of Co-Expression Hub Genes for Ferroptosis in Kidney Renal Clear Cell Carcinoma Based on Weighted Gene Co-Expression Network Analysis and the Cancer Genome Atlas Clinical Data. Sci. Rep. 2022, 12, 4821. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Liang, X.; Ma, Z.; Hu, H.; Liu, H.; Miao, Z.; Linkermann, A.; Hellwege, J.N.; Voight, B.F.; Susztak, K. A Single Genetic Locus Controls Both Expression of Dpep1/Chmp1a and Kidney Disease Development Via Ferroptosis. Nat. Commun. 2021, 12, 5078. [Google Scholar] [CrossRef] [PubMed]
- Park, W.M.; Cha, H.W.; Kim, J.; Kim, J.H.; Yang, H.; Yoon, S.; Boonpraman, N.; Yi, S.S.; Yoo, I.D.; Moon, J.S. Nox4 Promotes Ferroptosis of Astrocytes by Oxidative Stress-Induced Lipid Peroxidation Via the Impairment of Mitochondrial Metabolism in Alzheimer's Diseases. Redox Biol. 2021, 41, 101947. [Google Scholar] [CrossRef]
- Wang, Z.; Ding, Y.; Wang, X.; Lu, S.; Wang, C.; He, C.; Wang, L.; Piao, M.; Chi, G.; Luo, Y.; et al. Pseudolaric Acid B Triggers Ferroptosis in Glioma Cells Via Activation of Nox4 and Inhibition of Xct. Cancer Lett. 2018, 428, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.; Li, M.; Song, C.; Qi, H.; Ba, L.; Cao, Y.; Zhang, M.; Xie, Y.; Ren, J.; Wu, J.; et al. Neutrophil-Like Cell Membrane-Coated Sirna of Lncrna Aabr07017145.1 Therapy for Cardiac Hypertrophy Via Inhibiting Ferroptosis of Cmecs. Mol. Ther. Nucleic Acids 2022, 27, 16–36. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cheng, G.; Huang, Z.; Bao, L.; Liu, J.; Wang, C.; Xiong, Z.; Zhou, L.; Xu, T.; Liu, D.; et al. Long Noncoding Rna Snhg12 Promotes Tumour Progression and Sunitinib Resistance by Upregulating Cdca3 in Renal Cell Carcinoma. Cell Death Dis. 2020, 11, 515. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Henry, W.S.; Ricq, E.L.; Graham, E.T.; Phadnis, V.V.; Maretich, P.; Paradkar, S.; Boehnke, N.; Deik, A.A.; Reinhardt, F.; et al. Plasticity of Ether Lipids Promotes Ferroptosis Susceptibility and Evasion. Nature 2020, 585, 603. [Google Scholar] [CrossRef]
- Lee, W.J.; You, J.H.; Shin, D.; Roh, J.L. Inhibition of Glutaredoxin 5 Predisposes Cisplatin-Resistant Head and Neck Cancer Cells to Ferroptosis. Theranostics 2020, 10, 7775–7786. [Google Scholar] [CrossRef]
- Supuran, C.T. Carbonic Anhydrases: Novel Therapeutic Applications for Inhibitors and Activators. Nat. Rev. Drug Discov. 2008, 7, 168–181. [Google Scholar] [CrossRef]
- Wykoff, C.C.; Beasley, N.J.; Watson, P.; Turner, K.J.; Pastorek, J.; Sibtain, A.; Wilson, G.; Turley, H.; Talks, K.L.; Maxwell, P.; et al. Hypoxia-Inducible Expression of Tumor-Associated Carbonic Anhydrases. Cancer Res. 2000, 60, 7075–7083. [Google Scholar]
- Li, Z.; Jiang, L.; Chew, S.H.; Hirayama, T.; Sekido, Y.; Toyokuni, S. Carbonic Anhydrase 9 Confers Resistance to Ferroptosis/Apoptosis in Malignant Mesothelioma under Hypoxia. Redox Biol. 2019, 26, 101297. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Sun, Q.; Zhang, C.; She, J.; Cao, S.; Cao, M.; Zhang, N.; Adiila, A.V.; Zhong, J.; Yao, C.; et al. Identification and Validation of Cybb, Cd86, and C3ar1 as the Key Genes Related to Macrophage Infiltration of Gastric Cancer. Front. Mol. Biosci. 2021, 8, 756085. [Google Scholar] [CrossRef] [PubMed]
- Masisi, K.B.; el Ansari, R.; Alfarsi, L.; Rakha, E.A.; Green, A.R.; Craze, M.L. The Role of Glutaminase in Cancer. Histopathology 2020, 76, 498–508. [Google Scholar] [CrossRef]
- Katt, P.W.; Lukey, M.J.; Cerione, R.A. A Tale of Two Glutaminases: Homologous Enzymes with Distinct Roles in Tumorigenesis. Future Med. Chem. 2017, 9, 223–243. [Google Scholar] [CrossRef] [PubMed]
- Cassago, A.; Ferreira, A.P.; Ferreira, I.M.; Fornezari, C.; Gomes, E.R.; Greene, K.S.; Pereira, H.M.; Garratt, R.C.; Dias, S.M.; Ambrosio, A.L. Mitochondrial Localization and Structure-Based Phosphate Activation Mechanism of Glutaminase C with Implications for Cancer Metabolism. Proc. Natl. Acad. Sci. USA 2012, 109, 1092–1097. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Shi, X.; Meng, G.; Chen, J.; Yan, C.; Jiang, Y.; Wei, J.; Ding, Y. Kidney-Type Glutaminase (Gls1) Is a Biomarker for Pathologic Diagnosis and Prognosis of Hepatocellular Carcinoma. Oncotarget 2015, 6, 7619–7631. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Zhang, J.; Zhao, G.; Liu, M.; Hu, J.; Lin, F.; Wang, J.; Zhao, W.; Ma, H.; Zhang, C.; et al. Filamentous Gls1 Promotes Ros-Induced Apoptosis Upon Glutamine Deprivation Via Insufficient Asparagine Synthesis. Mol. Cell 2022, 82, 1821–1835. [Google Scholar] [CrossRef]
- Hu, W.; Zhang, C.; Wu, R.; Sun, Y.; Levine, A.; Feng, Z. Glutaminase 2, a Novel P53 Target Gene Regulating Energy Metabolism and Antioxidant Function. Proc. Natl. Acad. Sci. USA 2010, 107, 7455–7460. [Google Scholar] [CrossRef]
- Gao, M.; Monian, P.; Quadri, N.; Ramasamy, R.; Jiang, X. Glutaminolysis and Transferrin Regulate Ferroptosis. Mol. Cell 2015, 59, 298–308. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, Z.; Zheng, J.; Liang, Q.; Liu, Y.; Yang, Y.; Wang, R.; Wang, M.; Zhang, Q.; Xuan, Z.; Sun, H.; et al. Identification and Validation of a Novel Ferroptotic Prognostic Genes-Based Signature of Clear Cell Renal Cell Carcinoma. Cancers 2022, 14, 4690. https://doi.org/10.3390/cancers14194690
Shi Z, Zheng J, Liang Q, Liu Y, Yang Y, Wang R, Wang M, Zhang Q, Xuan Z, Sun H, et al. Identification and Validation of a Novel Ferroptotic Prognostic Genes-Based Signature of Clear Cell Renal Cell Carcinoma. Cancers. 2022; 14(19):4690. https://doi.org/10.3390/cancers14194690
Chicago/Turabian StyleShi, Zhiyuan, Jianzhong Zheng, Qing Liang, Yankuo Liu, Yi Yang, Rui Wang, Mingshan Wang, Qian Zhang, Zuodong Xuan, Huimin Sun, and et al. 2022. "Identification and Validation of a Novel Ferroptotic Prognostic Genes-Based Signature of Clear Cell Renal Cell Carcinoma" Cancers 14, no. 19: 4690. https://doi.org/10.3390/cancers14194690
APA StyleShi, Z., Zheng, J., Liang, Q., Liu, Y., Yang, Y., Wang, R., Wang, M., Zhang, Q., Xuan, Z., Sun, H., Wang, K., & Shao, C. (2022). Identification and Validation of a Novel Ferroptotic Prognostic Genes-Based Signature of Clear Cell Renal Cell Carcinoma. Cancers, 14(19), 4690. https://doi.org/10.3390/cancers14194690






