BRAFV600E Expression in Thyrocytes Causes Recruitment of Immunosuppressive STABILIN-1 Macrophages
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Tissue Collection
2.3. Histology and Immunohistochemistry
2.4. Immunohistofluorescence and Image Analysis
2.5. Blood Analysis and ELISA
2.6. mRNA Quantification
2.7. mRNA Sequencing and Bioinformatic Analysis
2.8. Flow Cytometry
2.9. Western Blotting
2.10. Statistical Analysis
3. Results
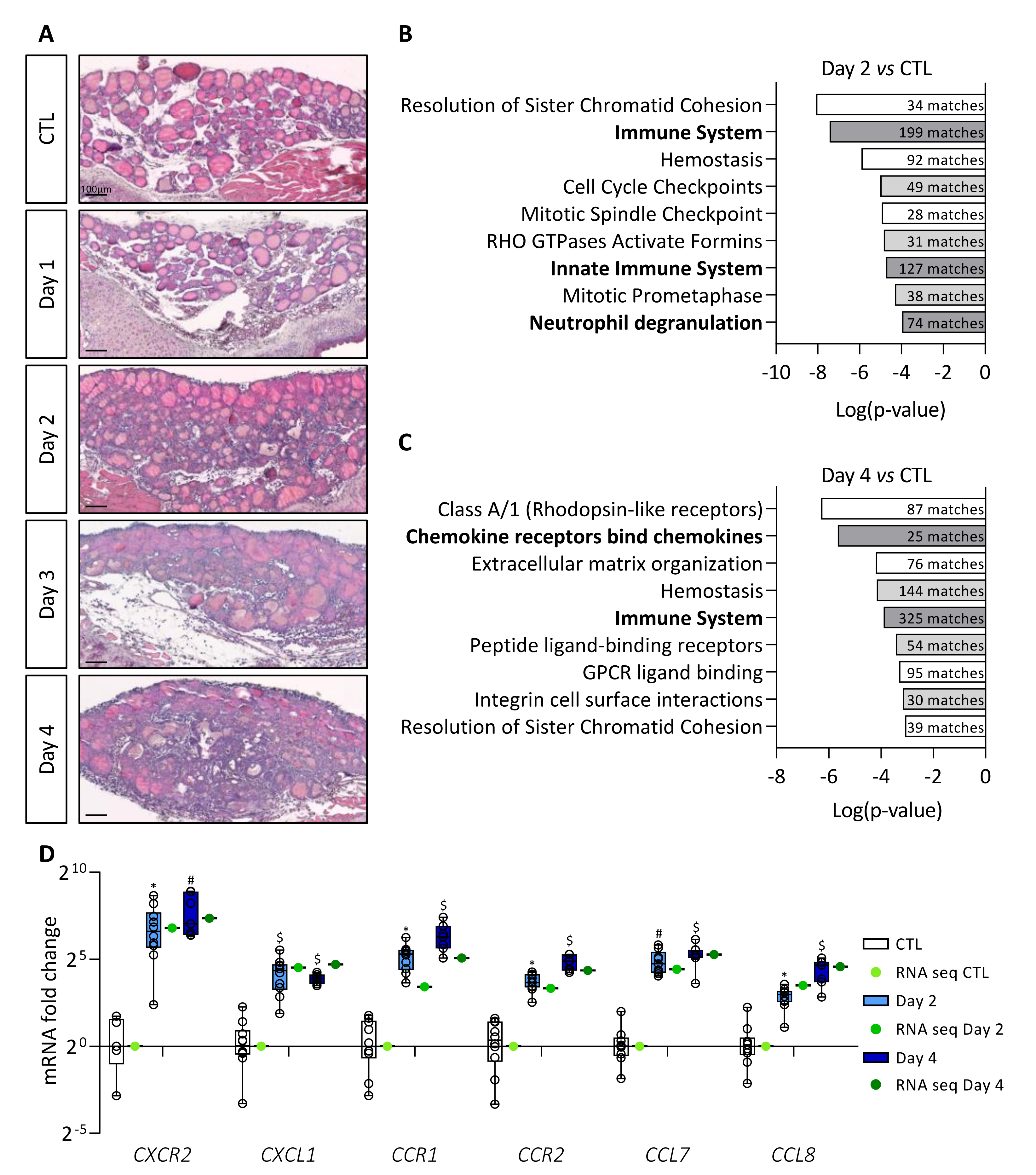
3.1. Induction of BRAFV600E Expression in Thyrocytes Triggers a Rapid Increase in Immune Signaling Pathways
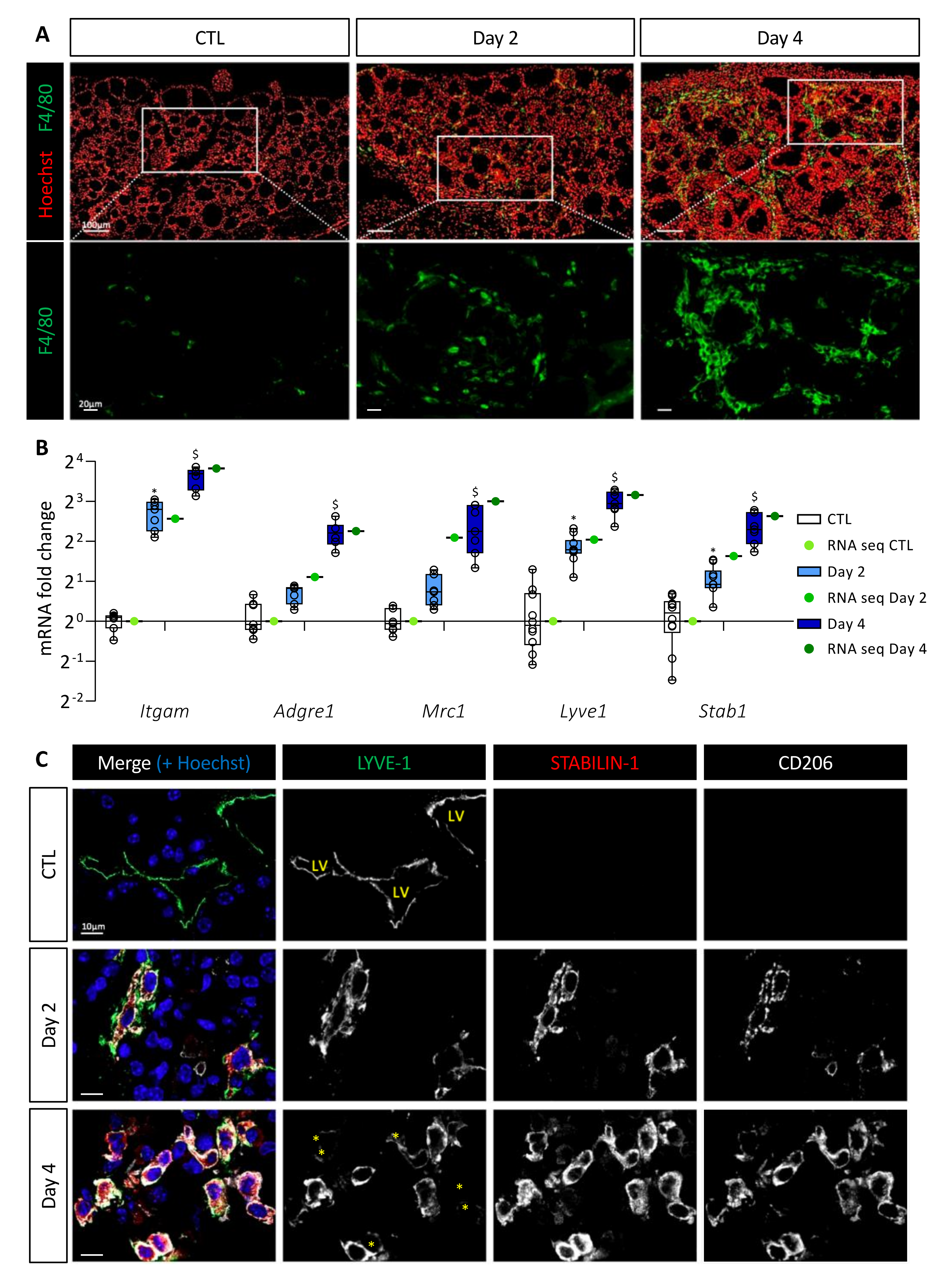
3.2. BRAFV600E Induction Triggers Recruitment of Macrophages, Including a Population of CD11b+/LYVE1+/CD206+/STAB1+ Immunosuppressive Macrophages
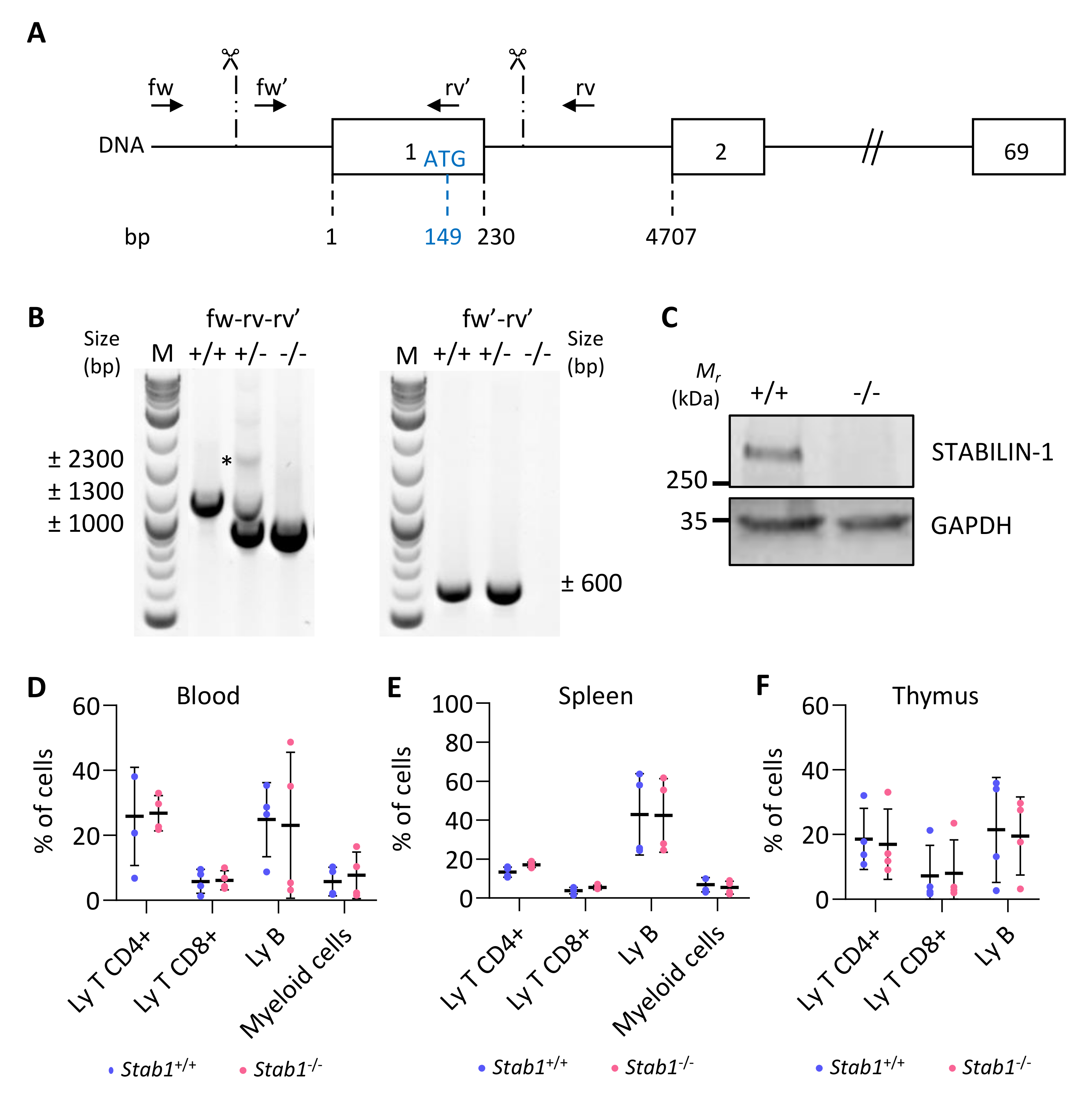
3.3. Generation and Validation of a CRISPR/Cas9 Stabilin-1 Knockout Mouse
3.4. Absence of STABILIN-1 Does Not Affect Epithelial PTC Development in Mice
3.5. Absence of STABILIN-1 Does Not Affect Circulating Cell Populations or Macrophage Recruitment in BRAFV600E-Induced PTC
3.6. Absence of STABILIN-1 Changes the CD4+/CD8+ T-Cell Ratio in BRAFV600E-Dependent PTC
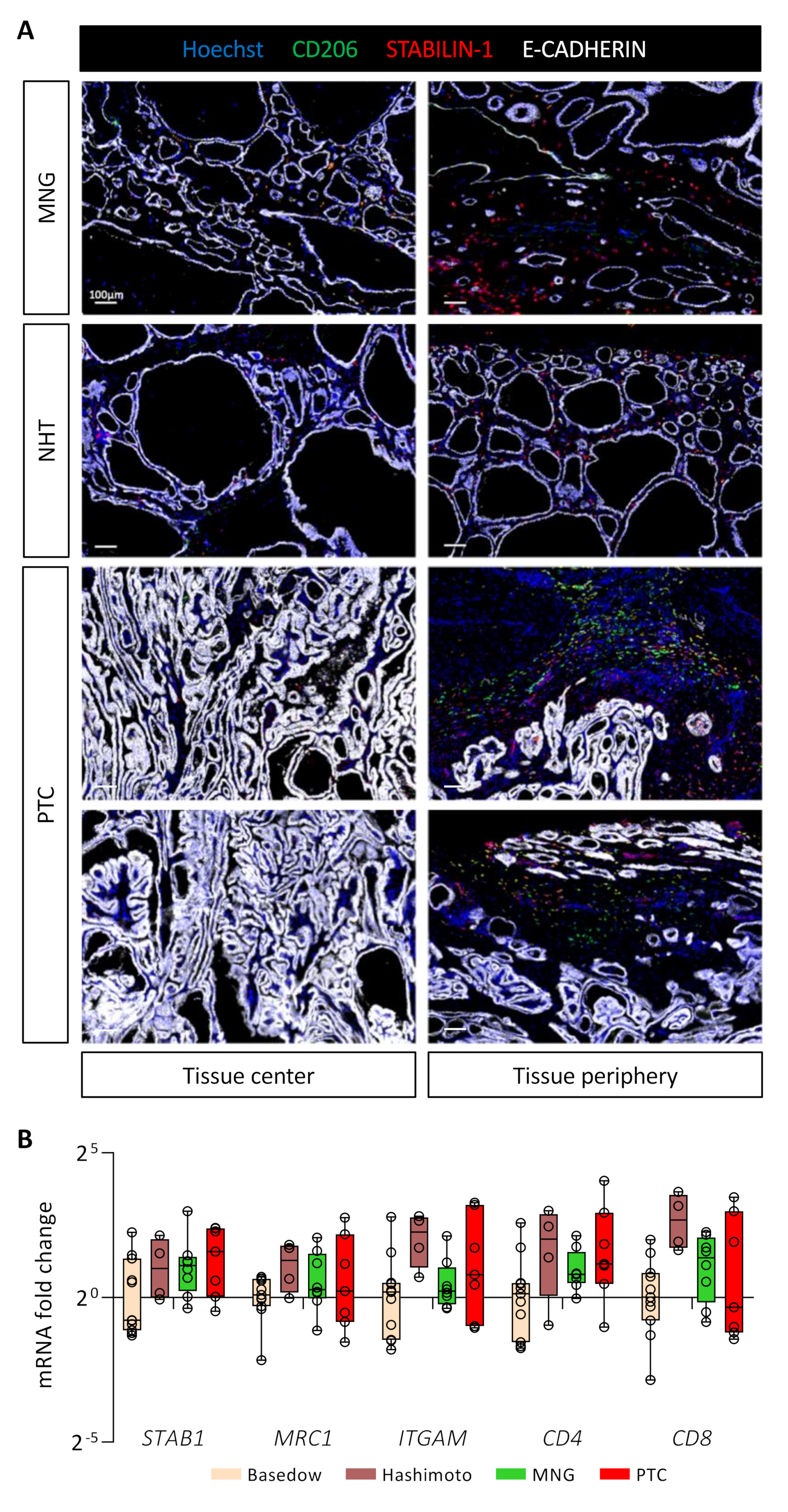
3.7. STABILIN-1+/CD206+ Cells Are Present in Benign and Malignant Human Thyroid Tissues
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cabanillas, M.E.; McFadden, D.G.; Durante, C. Thyroid cancer. Lancet 2016, 388, 2783–2795. [Google Scholar] [CrossRef]
- La Vecchia, C.; Malvezzi, M.; Bosetti, C.; Garavello, W.; Bertuccio, P.; Levi, F.; Negri, E. Thyroid cancer mortality and incidence: A global overview. Int. J. Cancer 2015, 136, 2187–2195. [Google Scholar] [CrossRef] [PubMed]
- Fagin, J.A.; Wells, S.A., Jr. Biologic and Clinical Perspectives on Thyroid Cancer. N. Engl. J. Med. 2016, 375, 1054–1067. [Google Scholar] [CrossRef] [PubMed]
- Nikiforova, M.N.; Wald, A.I.; Roy, S.; Durso, M.B.; Nikiforov, Y.E. Targeted next-generation sequencing panel (ThyroSeq) for detection of mutations in thyroid cancer. J. Clin. Endocrinol. Metab. 2013, 98, E1852–E1860. [Google Scholar] [CrossRef]
- de Biase, D.; Cesari, V.; Visani, M.; Casadei, G.P.; Cremonini, N.; Gandolfi, G.; Sancisi, V.; Ragazzi, M.; Pession, A.; Ciarrocchi, A.; et al. High-sensitivity BRAF mutation analysis: BRAF V600E is acquired early during tumor development but is heterogeneously distributed in a subset of papillary thyroid carcinomas. J. Clin. Endocrinol. Metab. 2014, 99, E1530–E1538. [Google Scholar] [CrossRef]
- American-Cancer-Society. Cancer Facts & Figures 2022. Available online: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2022/2022-cancer-facts-and-figures.pdf (accessed on 25 June 2022).
- Kitahara, C.M.; Sosa, J.A. The changing incidence of thyroid cancer. Nat. Rev. Endocrinol. 2016, 12, 646–653. [Google Scholar] [CrossRef]
- Jensen, C.B.; Saucke, M.C.; Francis, D.O.; Voils, C.I.; Pitt, S.C. From Overdiagnosis to Overtreatment of Low-Risk Thyroid Cancer: A Thematic Analysis of Attitudes and Beliefs of Endocrinologists, Surgeons, and Patients. Thyroid 2020, 30, 696–703. [Google Scholar] [CrossRef]
- Schiavoni, G.; Gabriele, L.; Mattei, F. The tumor microenvironment: A pitch for multiple players. Front. Oncol. 2013, 3, 90. [Google Scholar] [CrossRef]
- Varol, C.; Mildner, A.; Jung, S. Macrophages: Development and tissue specialization. Annu. Rev. Immunol. 2015, 33, 643–675. [Google Scholar] [CrossRef]
- Hourani, T.; Holden, J.A.; Li, W.; Lenzo, J.C.; Hadjigol, S.; O’Brien-Simpson, N.M. Tumor Associated Macrophages: Origin, Recruitment, Phenotypic Diversity, and Targeting. Front. Oncol. 2021, 11, 788365. [Google Scholar] [CrossRef]
- Pan, Y.; Yu, Y.; Wang, X.; Zhang, T. Tumor-Associated Macrophages in Tumor Immunity. Front. Immunol. 2020, 11, 583084. [Google Scholar] [CrossRef]
- Wang, L.X.; Zhang, S.X.; Wu, H.J.; Rong, X.L.; Guo, J. M2b macrophage polarization and its roles in diseases. J. Leukoc. Biol. 2019, 106, 345–358. [Google Scholar] [CrossRef]
- Menicali, E.; Guzzetti, M.; Morelli, S.; Moretti, S.; Puxeddu, E. Immune Landscape of Thyroid Cancers: New Insights. Front. Endocrinol. 2020, 11, 637826. [Google Scholar] [CrossRef]
- Giannini, R.; Moretti, S.; Ugolini, C.; Macerola, E.; Menicali, E.; Nucci, N.; Morelli, S.; Colella, R.; Mandarano, M.; Sidoni, A.; et al. Immune Profiling of Thyroid Carcinomas Suggests the Existence of Two Major Phenotypes: An ATC-Like and a PDTC-Like. J. Clin. Endocrinol. Metab. 2019, 104, 3557–3575. [Google Scholar] [CrossRef]
- Cunha, L.L.; Morari, E.C.; Guihen, A.C.; Razolli, D.; Gerhard, R.; Nonogaki, S.; Soares, F.A.; Vassallo, J.; Ward, L.S. Infiltration of a mixture of immune cells may be related to good prognosis in patients with differentiated thyroid carcinoma. Clin. Endocrinol. 2012, 77, 918–925. [Google Scholar] [CrossRef]
- Qing, W.; Fang, W.Y.; Ye, L.; Shen, L.Y.; Zhang, X.F.; Fei, X.C.; Chen, X.; Wang, W.Q.; Li, X.Y.; Xiao, J.C.; et al. Density of tumor-associated macrophages correlates with lymph node metastasis in papillary thyroid carcinoma. Thyroid 2012, 22, 905–910. [Google Scholar] [CrossRef]
- Fang, W.; Ye, L.; Shen, L.; Cai, J.; Huang, F.; Wei, Q.; Fei, X.; Chen, X.; Guan, H.; Wang, W.; et al. Tumor-associated macrophages promote the metastatic potential of thyroid papillary cancer by releasing CXCL8. Carcinogenesis 2014, 35, 1780–1787. [Google Scholar] [CrossRef]
- Ryder, M.; Gild, M.; Hohl, T.M.; Pamer, E.; Knauf, J.; Ghossein, R.; Joyce, J.A.; Fagin, J.A. Genetic and pharmacological targeting of CSF-1/CSF-1R inhibits tumor-associated macrophages and impairs BRAF-induced thyroid cancer progression. PLoS ONE 2013, 8, e54302. [Google Scholar] [CrossRef]
- Solinas, G.; Schiarea, S.; Liguori, M.; Fabbri, M.; Pesce, S.; Zammataro, L.; Pasqualini, F.; Nebuloni, M.; Chiabrando, C.; Mantovani, A.; et al. Tumor-conditioned macrophages secrete migration-stimulating factor: A new marker for M2-polarization, influencing tumor cell motility. J. Immunol. 2010, 185, 642–652. [Google Scholar] [CrossRef]
- Chakravarty, D.; Santos, E.; Ryder, M.; Knauf, J.A.; Liao, X.H.; West, B.L.; Bollag, G.; Kolesnick, R.; Thin, T.H.; Rosen, N.; et al. Small-molecule MAPK inhibitors restore radioiodine incorporation in mouse thyroid cancers with conditional BRAF activation. J. Clin. Investig. 2011, 121, 4700–4711. [Google Scholar] [CrossRef]
- Delcorte, O.; Spourquet, C.; Lemoine, P.; Degosserie, J.; Van Der Smissen, P.; Dauguet, N.; Loriot, A.; Knauf, J.A.; Gatto, L.; Marbaix, E.; et al. BRAF(V600E) Induction in Thyrocytes Triggers Important Changes in the miRNAs Content and the Populations of Extracellular Vesicles Released in Thyroid Tumor Microenvironment. Biomedicines 2022, 10, 755. [Google Scholar] [CrossRef]
- Heymans, C.; Delcorte, O.; Spourquet, C.; Villacorte-Tabelin, M.; Dupasquier, S.; Achouri, Y.; Mahibullah, S.; Lemoine, P.; Balda, M.S.; Matter, K.; et al. Spatio-temporal expression pattern and role of the tight junction protein MarvelD3 in pancreas development and function. Sci. Rep. 2021, 11, 14519. [Google Scholar] [CrossRef]
- Bertrand, C.; Van Meerbeeck, P.; de Streel, G.; Vaherto-Bleeckx, N.; Benhaddi, F.; Rouaud, L.; Noel, A.; Coulie, P.G.; van Baren, N.; Lucas, S. Combined Blockade of GARP:TGF-beta1 and PD-1 Increases Infiltration of T Cells and Density of Pericyte-Covered GARP(+) Blood Vessels in Mouse MC38 Tumors. Front. Immunol. 2021, 12, 704050. [Google Scholar] [CrossRef]
- Heymans, C.; Degosserie, J.; Spourquet, C.; Pierreux, C.E. Pancreatic acinar differentiation is guided by differential laminin deposition. Sci. Rep. 2019, 9, 2711. [Google Scholar] [CrossRef]
- Delcorte, O.; Craps, J.; Mahibullah, S.; Spourquet, C.; D’Auria, L.; Van Der Smissen, P.; Dessy, C.; Marbaix, E.; Mourad, M.; Pierreux, C.E. Two miRNAs enriched in plasma extracellular vesicles are potential biomarkers for thyroid cancer. Endocr. Relat. Cancer 2022, 29, 389–401. [Google Scholar] [CrossRef]
- Hick, A.C.; Delmarcelle, A.S.; Bouquet, M.; Klotz, S.; Copetti, T.; Forez, C.; Van Der Smissen, P.; Sonveaux, P.; Collet, J.F.; Feron, O.; et al. Reciprocal epithelial:endothelial paracrine interactions during thyroid development govern follicular organization and C-cells differentiation. Dev. Biol. 2013, 381, 227–240. [Google Scholar] [CrossRef]
- Dupasquier, S.; Delmarcelle, A.S.; Marbaix, E.; Cosyns, J.P.; Courtoy, P.J.; Pierreux, C.E. Validation of housekeeping gene and impact on normalized gene expression in clear cell renal cell carcinoma: Critical reassessment of YBX3/ZONAB/CSDA expression. BMC Mol. Biol. 2014, 15, 9. [Google Scholar] [CrossRef]
- Andrews, S. FastQC:A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 20 January 2019).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Degosserie, J.; Heymans, C.; Spourquet, C.; Halbout, M.; D’Auria, L.; Van Der Smissen, P.; Vertommen, D.; Courtoy, P.J.; Tyteca, D.; Pierreux, C.E. Extracellular vesicles from endothelial progenitor cells promote thyroid follicle formation. J. Extracell. Vesicles 2018, 7, 1487250. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Schledzewski, K.; Falkowski, M.; Moldenhauer, G.; Metharom, P.; Kzhyshkowska, J.; Ganss, R.; Demory, A.; Falkowska-Hansen, B.; Kurzen, H.; Ugurel, S.; et al. Lymphatic endothelium-specific hyaluronan receptor LYVE-1 is expressed by stabilin-1+, F4/80+, CD11b+ macrophages in malignant tumours and wound healing tissue in vivo and in bone marrow cultures in vitro: Implications for the assessment of lymphangiogenesis. J. Pathol. 2006, 209, 67–77. [Google Scholar] [CrossRef]
- Lim, H.Y.; Lim, S.Y.; Tan, C.K.; Thiam, C.H.; Goh, C.C.; Carbajo, D.; Chew, S.H.S.; See, P.; Chakarov, S.; Wang, X.N.; et al. Hyaluronan Receptor LYVE-1-Expressing Macrophages Maintain Arterial Tone through Hyaluronan-Mediated Regulation of Smooth Muscle Cell Collagen. Immunity 2018, 49, 326–341 e327. [Google Scholar] [CrossRef]
- Dollt, C.; Becker, K.; Michel, J.; Melchers, S.; Weis, C.A.; Schledzewski, K.; Krewer, A.; Kloss, L.; Gebhardt, C.; Utikal, J.; et al. The shedded ectodomain of Lyve-1 expressed on M2-like tumor-associated macrophages inhibits melanoma cell proliferation. Oncotarget 2017, 8, 103682–103692. [Google Scholar] [CrossRef]
- Goerdt, S.; Walsh, L.J.; Murphy, G.F.; Pober, J.S. Identification of a novel high molecular weight protein preferentially expressed by sinusoidal endothelial cells in normal human tissues. J. Cell Biol. 1991, 113, 1425–1437. [Google Scholar] [CrossRef]
- Palani, S.; Maksimow, M.; Miiluniemi, M.; Auvinen, K.; Jalkanen, S.; Salmi, M. Stabilin-1/CLEVER-1, a type 2 macrophage marker, is an adhesion and scavenging molecule on human placental macrophages. Eur. J. Immunol. 2011, 41, 2052–2063. [Google Scholar] [CrossRef]
- Kzhyshkowska, J.; Gratchev, A.; Goerdt, S. Stabilin-1, a homeostatic scavenger receptor with multiple functions. J. Cell Mol. Med. 2006, 10, 635–649. [Google Scholar] [CrossRef]
- Hollmen, M.; Figueiredo, C.R.; Jalkanen, S. New tools to prevent cancer growth and spread: A ‘Clever’ approach. Br. J. Cancer 2020, 123, 501–509. [Google Scholar] [CrossRef]
- Karikoski, M.; Marttila-Ichihara, F.; Elima, K.; Rantakari, P.; Hollmen, M.; Kelkka, T.; Gerke, H.; Huovinen, V.; Irjala, H.; Holmdahl, R.; et al. Clever-1/stabilin-1 controls cancer growth and metastasis. Clin. Cancer Res. 2014, 20, 6452–6464. [Google Scholar] [CrossRef]
- Viitala, M.; Virtakoivu, R.; Tadayon, S.; Rannikko, J.; Jalkanen, S.; Hollmen, M. Immunotherapeutic Blockade of Macrophage Clever-1 Reactivates the CD8(+) T-cell Response against Immunosuppressive Tumors. Clin. Cancer Res. 2019, 25, 3289–3303. [Google Scholar] [CrossRef]
- Ammar, A.; Mohammed, R.A.; Salmi, M.; Pepper, M.; Paish, E.C.; Ellis, I.O.; Martin, S.G. Lymphatic expression of CLEVER-1 in breast cancer and its relationship with lymph node metastasis. Anal. Cell. Pathol. 2011, 34, 67–78. [Google Scholar] [CrossRef]
- Antonelli, A.; Ferrari, S.M.; Corrado, A.; Di Domenicantonio, A.; Fallahi, P. Autoimmune thyroid disorders. Autoimmun. Rev. 2015, 14, 174–180. [Google Scholar] [CrossRef]
- Landa, I.; Knauf, J.A. Mouse Models as a Tool for Understanding Progression in Braf(V600E)-Driven Thyroid Cancers. Endocrinol. Metab. 2019, 34, 11–22. [Google Scholar] [CrossRef]
- Knauf, J.A.; Ma, X.; Smith, E.P.; Zhang, L.; Mitsutake, N.; Liao, X.H.; Refetoff, S.; Nikiforov, Y.E.; Fagin, J.A. Targeted expression of BRAFV600E in thyroid cells of transgenic mice results in papillary thyroid cancers that undergo dedifferentiation. Cancer Res. 2005, 65, 4238–4245. [Google Scholar] [CrossRef]
- Franco, A.T.; Malaguarnera, R.; Refetoff, S.; Liao, X.H.; Lundsmith, E.; Kimura, S.; Pritchard, C.; Marais, R.; Davies, T.F.; Weinstein, L.S.; et al. Thyrotrophin receptor signaling dependence of Braf-induced thyroid tumor initiation in mice. Proc. Natl. Acad. Sci. USA 2011, 108, 1615–1620. [Google Scholar] [CrossRef]
- Schoultz, E.; Johansson, E.; Moccia, C.; Jakubikova, I.; Ravi, N.; Liang, S.; Carlsson, T.; Montelius, M.; Patyra, K.; Kero, J.; et al. Tissue architecture delineates field cancerization in BRAFV600E-induced tumor development. Dis. Models Mech. 2022, 15, dmm048887. [Google Scholar] [CrossRef]
- Ryder, M.; Ghossein, R.A.; Ricarte-Filho, J.C.; Knauf, J.A.; Fagin, J.A. Increased density of tumor-associated macrophages is associated with decreased survival in advanced thyroid cancer. Endocr. Relat. Cancer 2008, 15, 1069–1074. [Google Scholar] [CrossRef]
- Zhao, K.; Yang, H.; Kang, H.; Wu, A. Identification of Key Genes in Thyroid Cancer Microenvironment. Med. Sci. Monit. 2019, 25, 9602–9608. [Google Scholar] [CrossRef]
- Wang, F.; Flanagan, J.; Su, N.; Wang, L.C.; Bui, S.; Nielson, A.; Wu, X.; Vo, H.T.; Ma, X.J.; Luo, Y. RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 2012, 14, 22–29. [Google Scholar] [CrossRef]
- Kersigo, J.; Pan, N.; Lederman, J.D.; Chatterjee, S.; Abel, T.; Pavlinkova, G.; Silos-Santiago, I.; Fritzsch, B. A RNAscope whole mount approach that can be combined with immunofluorescence to quantify differential distribution of mRNA. Cell Tissue Res. 2018, 374, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Karikoski, M.; Irjala, H.; Maksimow, M.; Miiluniemi, M.; Granfors, K.; Hernesniemi, S.; Elima, K.; Moldenhauer, G.; Schledzewski, K.; Kzhyshkowska, J.; et al. Clever-1/Stabilin-1 regulates lymphocyte migration within lymphatics and leukocyte entrance to sites of inflammation. Eur. J. Immunol. 2009, 39, 3477–3487. [Google Scholar] [CrossRef] [PubMed]
- Tadayon, S.; Dunkel, J.; Takeda, A.; Eichin, D.; Virtakoivu, R.; Elima, K.; Jalkanen, S.; Hollmen, M. Lymphatic Endothelial Cell Activation and Dendritic Cell Transmigration Is Modified by Genetic Deletion of Clever-1. Front. Immunol. 2021, 12, 602122. [Google Scholar] [CrossRef] [PubMed]
- Riabov, V.; Yin, S.; Song, B.; Avdic, A.; Schledzewski, K.; Ovsiy, I.; Gratchev, A.; Llopis Verdiell, M.; Sticht, C.; Schmuttermaier, C.; et al. Stabilin-1 is expressed in human breast cancer and supports tumor growth in mammary adenocarcinoma mouse model. Oncotarget 2016, 7, 31097–31110. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Z.; Yang, K.; Ye, L.; You, Z.; Chen, R.; Liu, Y. Decorin gene upregulation mediated by an adeno-associated virus vector increases intratumoral uptake of nab-paclitaxel in neuroblastoma via inhibition of stabilin-1. Investig. New Drugs 2017, 35, 566–575. [Google Scholar] [CrossRef]
- Yin, S.; Wang, N.; Riabov, V.; Mossel, D.M.; Larionova, I.; Schledzewski, K.; Trofimova, O.; Sevastyanova, T.; Zajakina, A.; Schmuttermaier, C.; et al. SI-CLP inhibits the growth of mouse mammary adenocarcinoma by preventing recruitment of tumor-associated macrophages. Int. J. Cancer 2020, 146, 1396–1408. [Google Scholar] [CrossRef]
- Liang, M.; Jia, J.; Chen, L.; Wei, B.; Guan, Q.; Ding, Z.; Yu, J.; Pang, R.; He, G. LncRNA MCM3AP-AS1 promotes proliferation and invasion through regulating miR-211-5p/SPARC axis in papillary thyroid cancer. Endocrine 2019, 65, 318–326. [Google Scholar] [CrossRef]
- Cheng, S.P.; Lee, J.J.; Chang, Y.C.; Lin, C.H.; Li, Y.S.; Liu, C.L. Overexpression of chitinase-3-like protein 1 is associated with structural recurrence in patients with differentiated thyroid cancer. J. Pathol. 2020, 252, 114–124. [Google Scholar] [CrossRef]
- Palani, S.; Elima, K.; Ekholm, E.; Jalkanen, S.; Salmi, M. Monocyte Stabilin-1 Suppresses the Activation of Th1 Lymphocytes. J. Immunol. 2016, 196, 115–123. [Google Scholar] [CrossRef]
- Irjala, H.; Alanen, K.; Grenman, R.; Heikkila, P.; Joensuu, H.; Jalkanen, S. Mannose receptor (MR) and common lymphatic endothelial and vascular endothelial receptor (CLEVER)-1 direct the binding of cancer cells to the lymph vessel endothelium. Cancer Res. 2003, 63, 4671–4676. [Google Scholar]
- Algars, A.; Irjala, H.; Vaittinen, S.; Huhtinen, H.; Sundstrom, J.; Salmi, M.; Ristamaki, R.; Jalkanen, S. Type and location of tumor-infiltrating macrophages and lymphatic vessels predict survival of colorectal cancer patients. Int. J. Cancer 2012, 131, 864–873. [Google Scholar] [CrossRef]
- Bostrom, M.M.; Irjala, H.; Mirtti, T.; Taimen, P.; Kauko, T.; Algars, A.; Jalkanen, S.; Bostrom, P.J. Tumor-Associated Macrophages Provide Significant Prognostic Information in Urothelial Bladder Cancer. PLoS ONE 2015, 10, e0133552. [Google Scholar] [CrossRef]
- Tervahartiala, M.; Taimen, P.; Mirtti, T.; Koskinen, I.; Ecke, T.; Jalkanen, S.; Bostrom, P.J. Immunological tumor status may predict response to neoadjuvant chemotherapy and outcome after radical cystectomy in bladder cancer. Sci. Rep. 2017, 7, 12682. [Google Scholar] [CrossRef]
- Kwon, M.; Yeo, S.C.; Lee, J.S.; Park, J.J. Not CD68 but stabilin-1 expression is associated with the risk of recurrence in patients with oral cavity squamous cell carcinoma. Head Neck 2019, 41, 2058–2064. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spourquet, C.; Delcorte, O.; Lemoine, P.; Dauguet, N.; Loriot, A.; Achouri, Y.; Hollmén, M.; Jalkanen, S.; Huaux, F.; Lucas, S.; et al. BRAFV600E Expression in Thyrocytes Causes Recruitment of Immunosuppressive STABILIN-1 Macrophages. Cancers 2022, 14, 4687. https://doi.org/10.3390/cancers14194687
Spourquet C, Delcorte O, Lemoine P, Dauguet N, Loriot A, Achouri Y, Hollmén M, Jalkanen S, Huaux F, Lucas S, et al. BRAFV600E Expression in Thyrocytes Causes Recruitment of Immunosuppressive STABILIN-1 Macrophages. Cancers. 2022; 14(19):4687. https://doi.org/10.3390/cancers14194687
Chicago/Turabian StyleSpourquet, Catherine, Ophélie Delcorte, Pascale Lemoine, Nicolas Dauguet, Axelle Loriot, Younes Achouri, Maija Hollmén, Sirpa Jalkanen, François Huaux, Sophie Lucas, and et al. 2022. "BRAFV600E Expression in Thyrocytes Causes Recruitment of Immunosuppressive STABILIN-1 Macrophages" Cancers 14, no. 19: 4687. https://doi.org/10.3390/cancers14194687
APA StyleSpourquet, C., Delcorte, O., Lemoine, P., Dauguet, N., Loriot, A., Achouri, Y., Hollmén, M., Jalkanen, S., Huaux, F., Lucas, S., Meerkeeck, P. V., Knauf, J. A., Fagin, J. A., Dessy, C., Mourad, M., Henriet, P., Tyteca, D., Marbaix, E., & Pierreux, C. E. (2022). BRAFV600E Expression in Thyrocytes Causes Recruitment of Immunosuppressive STABILIN-1 Macrophages. Cancers, 14(19), 4687. https://doi.org/10.3390/cancers14194687





