Cross-Domain Text Mining to Predict Adverse Events from Tyrosine Kinase Inhibitors for Chronic Myeloid Leukemia
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
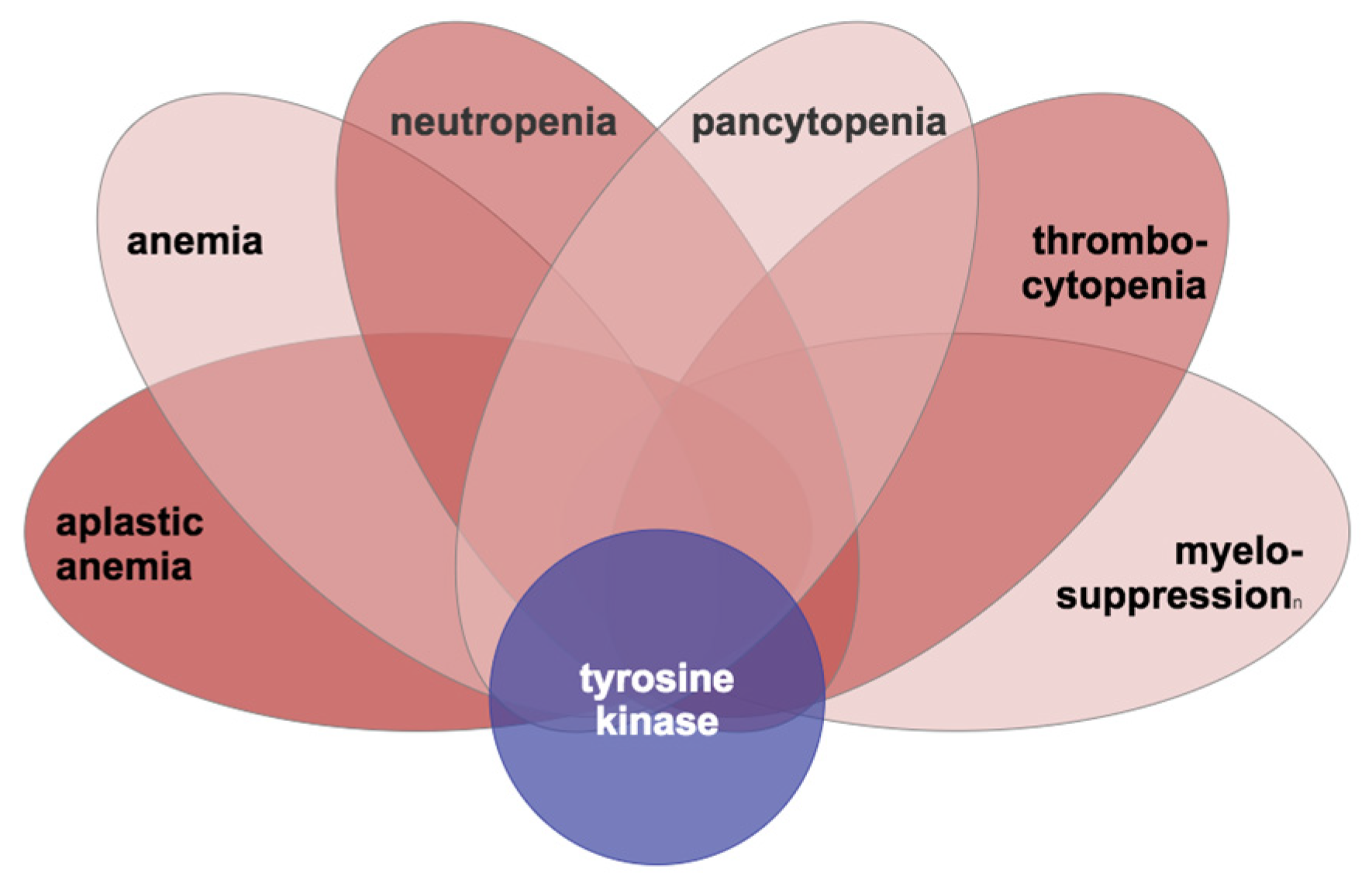
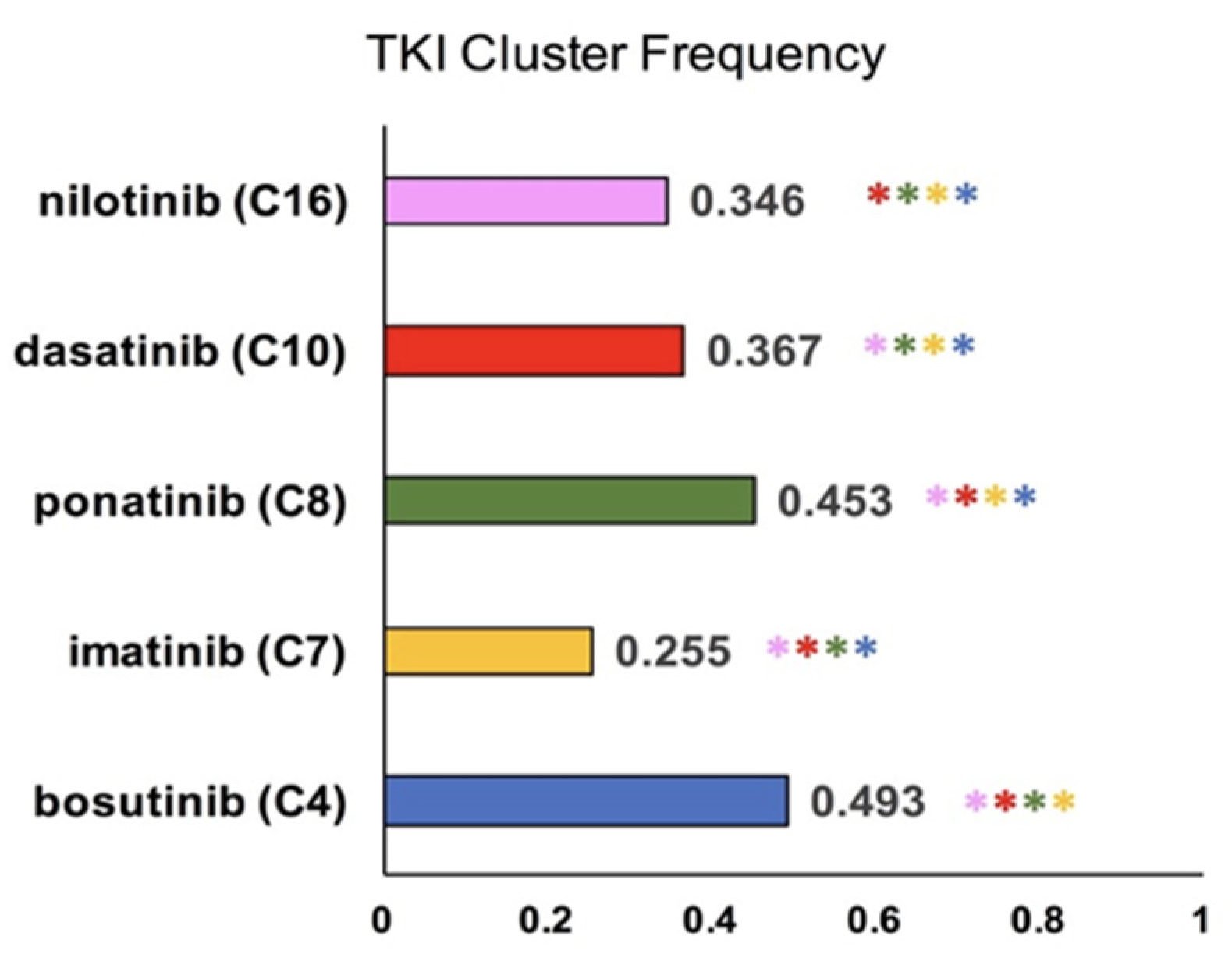
2.1. Cluster Analysis of TKI-Specific Adverse Events
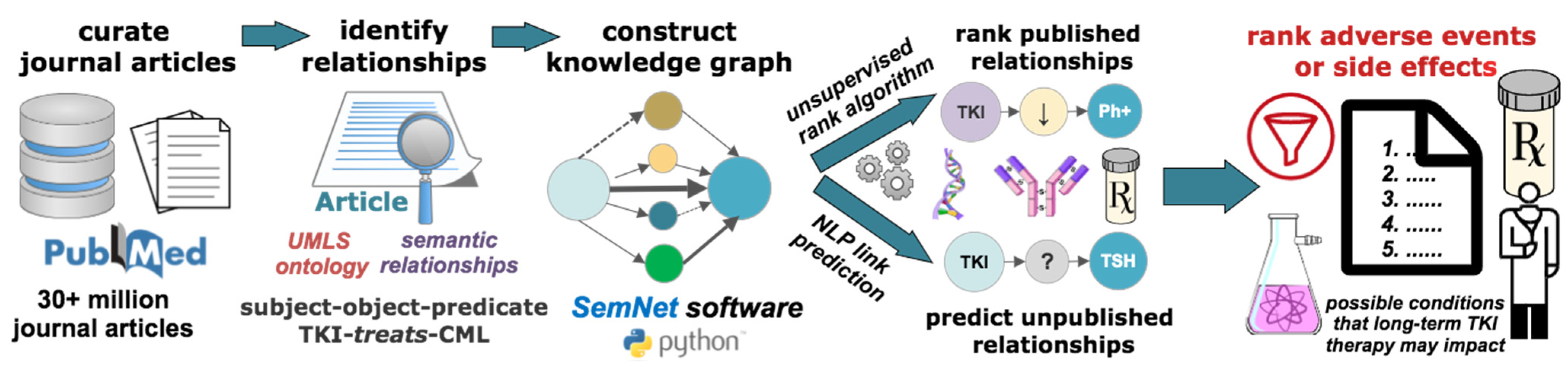
2.2. Overview of Cross-Domain Text Mining of Conditions Linked to Tyrosine Kinase
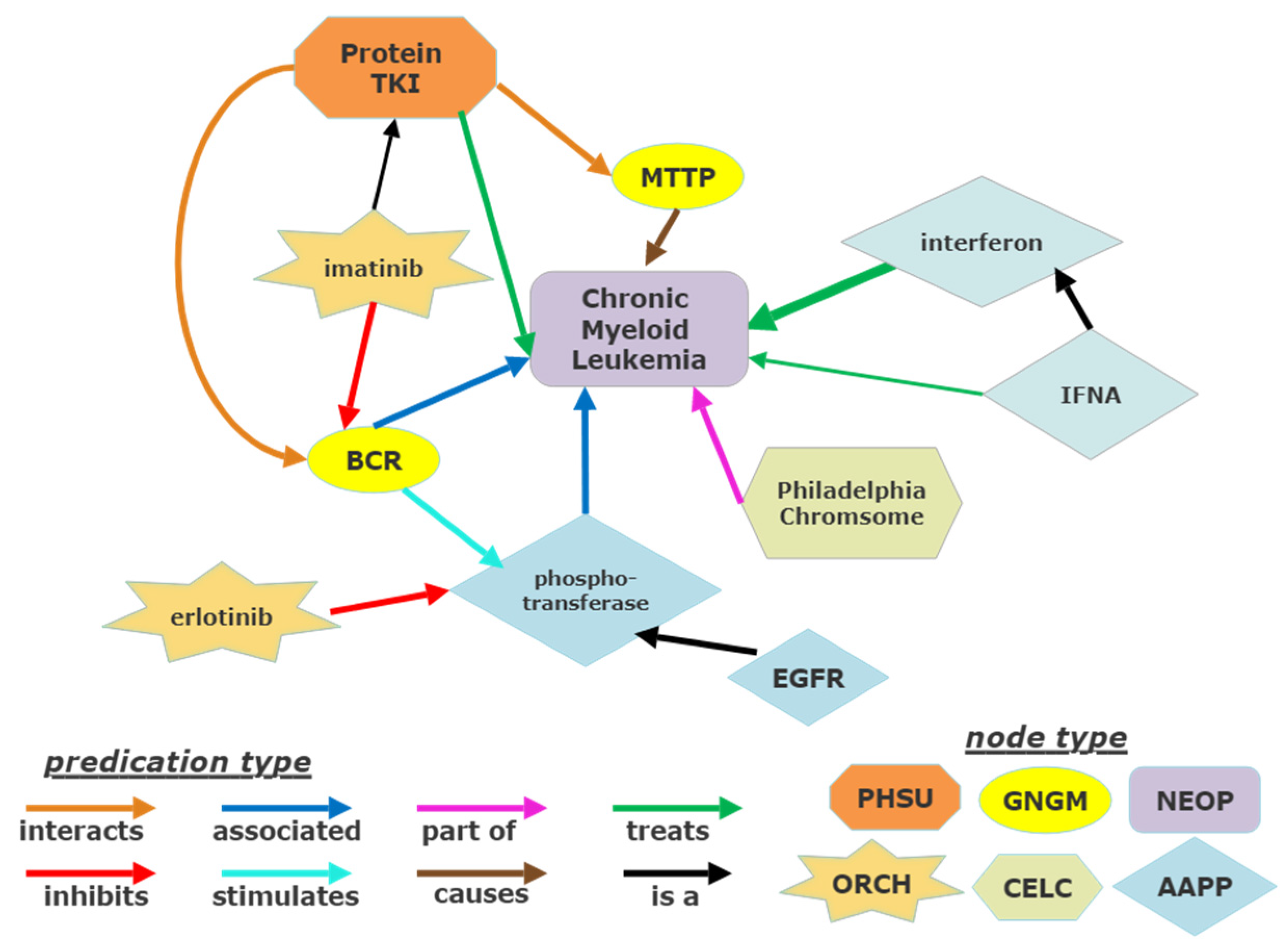
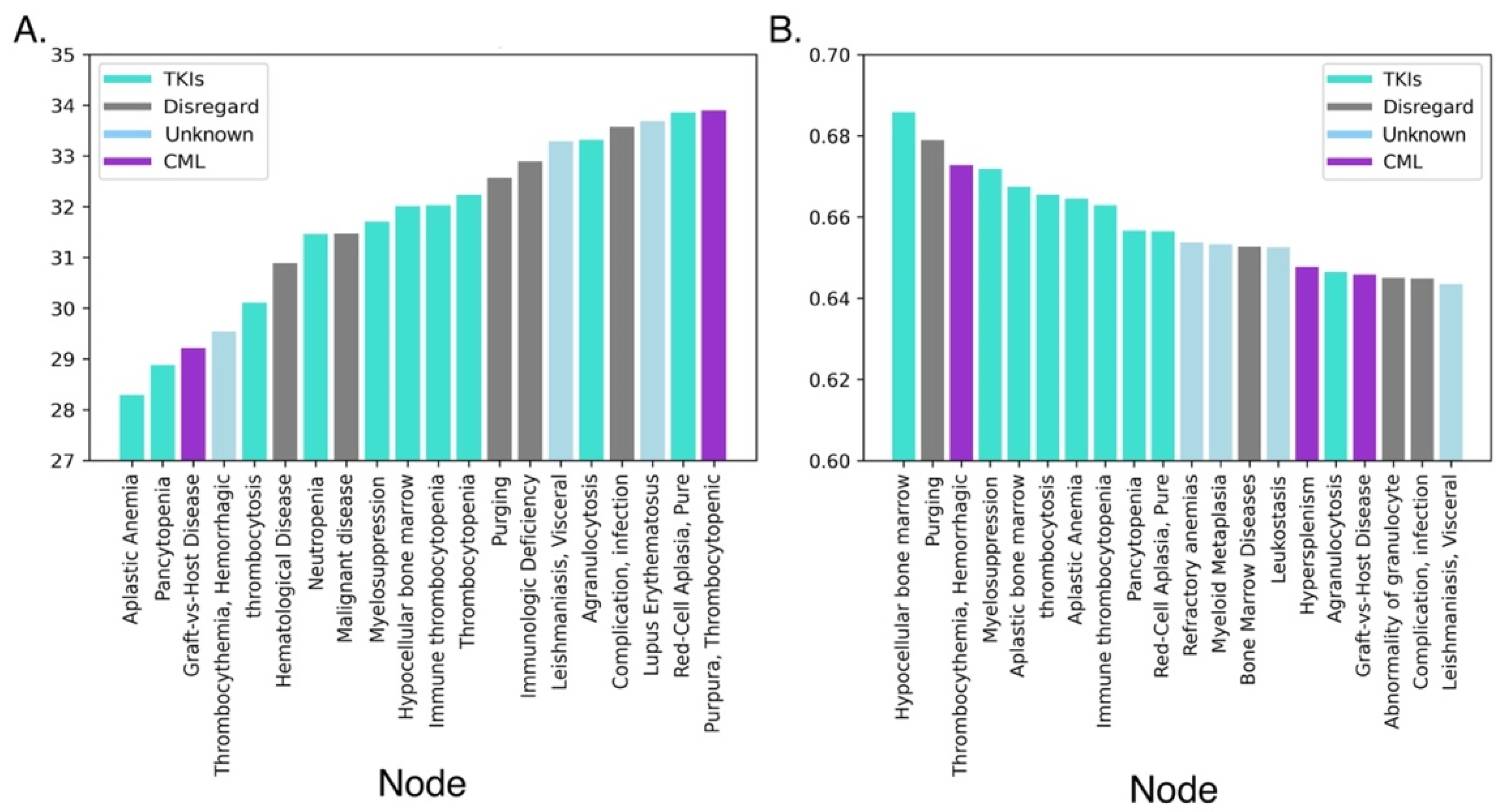
2.3. Examination of Existing Published Nodes to CML and Tyrosine Kinase with SemNet
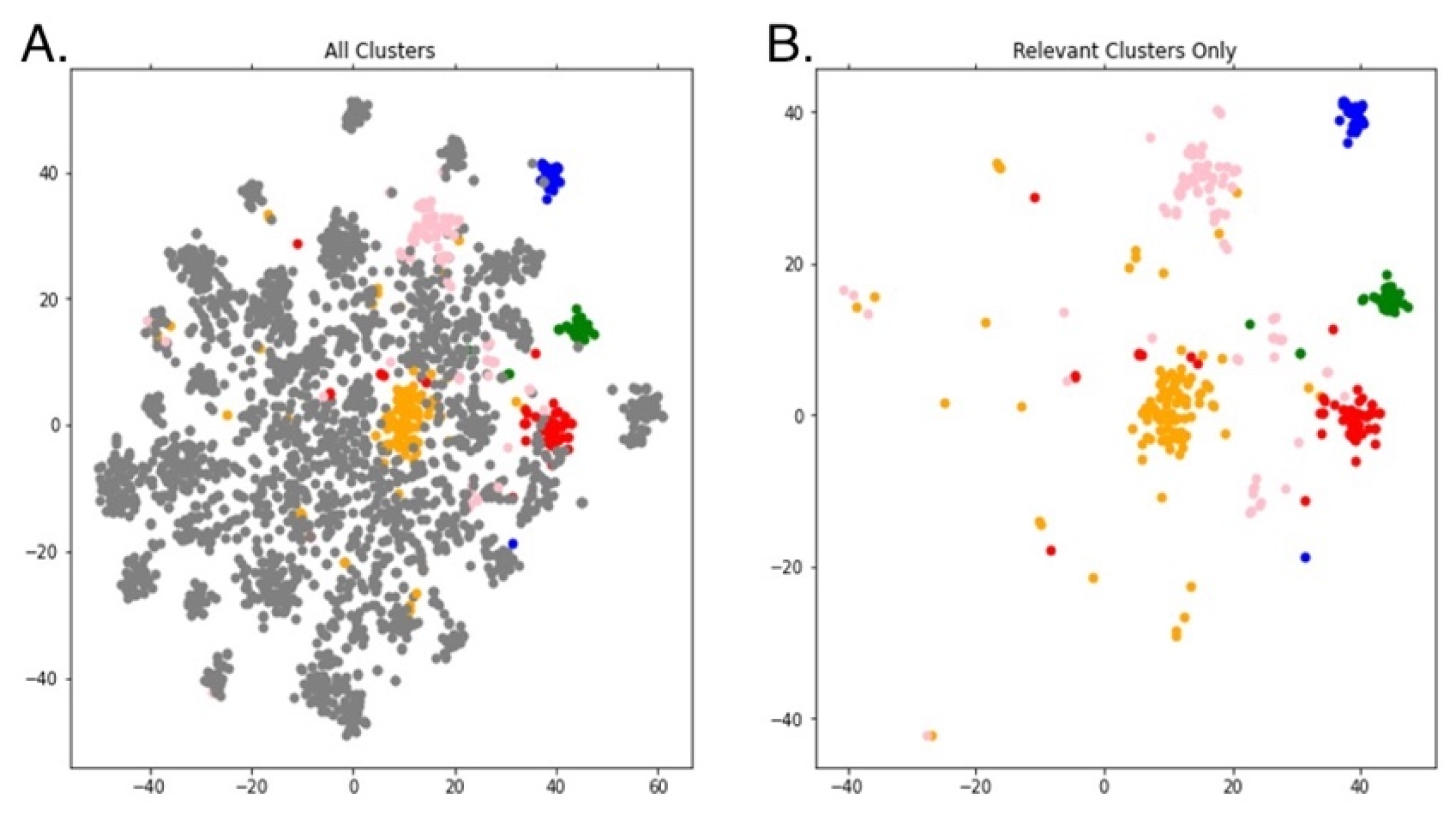
2.4. Link Prediction with Hub Node Network Analysis to Predict Cross-Domain Relationships
2.4.1. Motivation for Hub Node Network Analysis Adapted to Knowledge Graphs
2.4.2. Implementation of Hub Node Network Analysis with Link Prediction
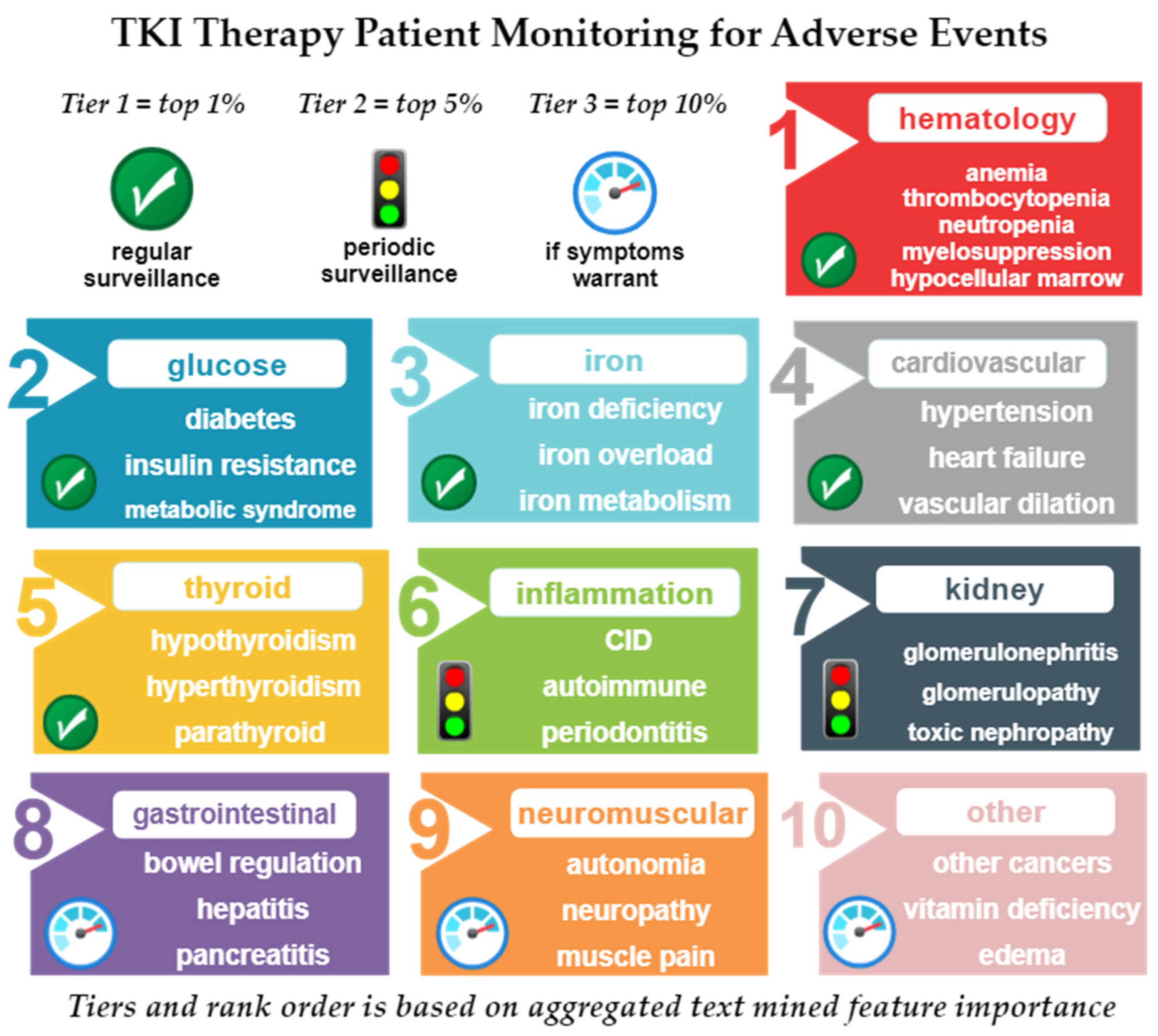
2.4.3. Selection and Validation of Most Relevant Nodes and Aggregation into Tiers and Foci
3. Results
3.1. K-Means Clustering of Adverse Events in the CML TKI Literature
3.2. Initial Cross-Domain Text Mining of Published Relationships to CML
3.3. Prediction of Distant or Novel Predicted Connections to Tyrosine Kinase
4. Discussion
4.1. Predicted Tier 1 Adverse Events Likely Tied to TKI Therapy
4.1.1. Hematological Conditions as Adverse Events from TKI Therapy
4.1.2. Glucose-Related Adverse Events from TKI Therapy
4.1.3. Iron Homeostasis Adverse Events with TKI Therapy
4.1.4. Cardiovascular Adverse Events with TKI Therapy
4.1.5. Thyroid Disorders as Adverse Events with TKI Therapy
4.2. Predicted Tier 2 Adverse Events Likely Tied to TKI Therapy
4.2.1. Kidney Adverse Events with TKI Therapy
4.2.2. Inflammation Adverse Events with TKI Therapy
4.3. Predicted Tier 3 AEs Likely Tied to TKI Therapy
4.3.1. Gastrointestinal Adverse Events with TKI Therapy
4.3.2. Neuromuscular Adverse Events with TKI Therapy
4.3.3. Other Adverse Events with TKI Therapy
4.4. Limitations and Future Work
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| TKI/Cluster | 4 | 7 | 8 | 10 | 16 |
|---|---|---|---|---|---|
| bosutinib | 0.493 | 0.003 | 0.034 | 0.007 | 0.004 |
| imatinib | 0.053 | 0.255 | 0.024 | 0.033 | 0.078 |
| ponatinib | 0.003 | 0.002 | 0.453 | 0.000 | 0.004 |
| dasatinib | 0.026 | 0.009 | 0.023 | 0.367 | 0.062 |
| nilotinib | 0.027 | 0.008 | 0.037 | 0.011 | 0.346 |
References
- SEER. Chronic Myeloid Leukemia—Cancer Stat Facts. Available online: https://seer.cancer.gov/statfacts/html/cmyl.html (accessed on 23 March 2022).
- Athale, U.; Hijiya, N.; Patterson, B.C.; Bergsagel, J.; Andolina, J.R.; Bittencourt, H.; Schultz, K.R.; Burke, M.J.; Redell, M.S.; Kolb, E.A.; et al. Management of chronic myeloid leukemia in children and adolescents: Recommendations from the Children’s Oncology Group CML Working Group. Pediatr. Blood Cancer 2019, 66, e27827. [Google Scholar] [CrossRef] [PubMed]
- Massimino, M.; Stella, S.; Tirro, E.; Pennisi, M.S.; Vitale, S.R.; Puma, A.; Romano, C.; di Gregorio, S.; Tomarchio, C.; Di Raimondo, F.; et al. ABL1-Directed Inhibitors for CML: Efficacy, Resistance and Future Perspectives. Anticancer Res. 2020, 40, 2457–2465. [Google Scholar] [CrossRef] [PubMed]
- Rea, D.; Cayuela, J.M. Treatment-free remission in patients with chronic myeloid leukemia. Int. J. Hematol. 2018, 108, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Atallah, E.; Sweet, K. Treatment-Free Remission: The New Goal in CML Therapy. Curr. Hematol. Malig. Rep. 2021, 16, 433–439. [Google Scholar] [CrossRef]
- Mahon, F.X.; Etienne, G. Deep molecular response in chronic myeloid leukemia: The new goal of therapy? Clin. Cancer Res. 2014, 20, 310–322. [Google Scholar] [CrossRef]
- Cortes, J.; Rea, D.; Lipton, J.H. Treatment-free remission with first- and second-generation tyrosine kinase inhibitors. Am. J. Hematol. 2019, 94, 346–357. [Google Scholar] [CrossRef]
- Rousselot, P.; Huguet, F.; Rea, D.; Legros, L.; Cayuela, J.M.; Maarek, O.; Blanchet, O.; Marit, G.; Gluckman, E.; Reiffers, J.; et al. Imatinib mesylate discontinuation in patients with chronic myelogenous leukemia in complete molecular remission for more than 2 years. Blood 2007, 109, 58–60. [Google Scholar] [CrossRef]
- Ross, D.M.; Branford, S.; Seymour, J.F.; Schwarer, A.P.; Arthur, C.; Yeung, D.T.; Dang, P.; Goyne, J.M.; Slader, C.; Filshie, R.J.; et al. Safety and efficacy of imatinib cessation for CML patients with stable undetectable minimal residual disease: Results from the TWISTER study. Blood 2013, 122, 515–522. [Google Scholar] [CrossRef]
- Saussele, S.; Richter, J.; Guilhot, J.; Gruber, F.X.; Hjorth-Hansen, H.; Almeida, A.; Janssen, J.; Mayer, J.; Koskenvesa, P.; Panayiotidis, P.; et al. Discontinuation of tyrosine kinase inhibitor therapy in chronic myeloid leukaemia (EURO-SKI): A prespecified interim analysis of a prospective, multicentre, non-randomised, trial. Lancet Oncol. 2018, 19, 747–757. [Google Scholar] [CrossRef]
- Ross, D.M.; Masszi, T.; Gomez Casares, M.T.; Hellmann, A.; Stentoft, J.; Conneally, E.; Garcia-Gutierrez, V.; Gattermann, N.; le Coutre, P.D.; Martino, B.; et al. Durable treatment-free remission in patients with chronic myeloid leukemia in chronic phase following frontline nilotinib: 96-week update of the ENESTfreedom study. J. Cancer Res. Clin. Oncol. 2018, 144, 945–954. [Google Scholar] [CrossRef]
- Hughes, T.P.; Clementino, N.C.D.; Fominykh, M.; Lipton, J.H.; Turkina, A.G.; Moiraghi, E.B.; Nicolini, F.E.; Takahashi, N.; Sacha, T.; Kim, D.W.; et al. Long-term treatment-free remission in patients with chronic myeloid leukemia after second-line nilotinib: ENESTop 5-year update. Leukemia 2021, 35, 1631–1642. [Google Scholar] [CrossRef]
- Shah, N.P.; Garcia-Gutierrez, V.; Jimenez-Velasco, A.; Larson, S.; Saussele, S.; Rea, D.; Mahon, F.X.; Levy, M.Y.; Gomez-Casares, M.T.; Pane, F.; et al. Dasatinib discontinuation in patients with chronic-phase chronic myeloid leukemia and stable deep molecular response: The DASFREE study. Leuk. Lymphoma 2020, 61, 650–659. [Google Scholar] [CrossRef]
- Shih, Y.T.; Cortes, J.E.; Kantarjian, H.M. Treatment value of second-generation BCR-ABL1 tyrosine kinase inhibitors compared with imatinib to achieve treatment-free remission in patients with chronic myeloid leukaemia: A modelling study. Lancet Haematol. 2019, 6, e398–e408. [Google Scholar] [CrossRef]
- Reff, M.J.; Shillingburg, A.; Shah, B.; Elder, C.; Prescott, H.; Kennerly-Shah, J. Front-line use of tyrosine kinase inhibitors in chronic phase chronic myeloid leukemia: Practice considerations. J. Oncol. Pharm. Pract. 2020, 26, 156–174. [Google Scholar] [CrossRef]
- Mohanavelu, P.; Mutnick, M.; Mehra, N.; White, B.; Kudrimoti, S.; Hernandez Kluesner, K.; Chen, X.; Nguyen, T.; Horlander, E.; Thenot, H.; et al. Meta-Analysis of Gastrointestinal Adverse Events from Tyrosine Kinase Inhibitors for Chronic Myeloid Leukemia. Cancers 2021, 13, 1643. [Google Scholar] [CrossRef]
- Chan, O.; Talati, C.; Isenalumhe, L.; Shams, S.; Nodzon, L.; Fradley, M.; Sweet, K.; Pinilla-Ibarz, J. Side-effects profile and outcomes of ponatinib in the treatment of chronic myeloid leukemia. Blood Adv. 2020, 4, 530–538. [Google Scholar] [CrossRef]
- Abruzzese, E.; Mauro, M.; Apperley, J.; Chelysheva, E. Tyrosine kinase inhibitors and pregnancy in chronic myeloid leukemia: Opinion, evidence, and recommendations. Ther. Adv. Hematol. 2020, 11, 2040620720966120. [Google Scholar] [CrossRef]
- Hughes, T.; White, D. Which TKI? An embarrassment of riches for chronic myeloid leukemia patients. Hematol. Am. Soc. Hematol. Educ. Program 2013, 2013, 168–175. [Google Scholar] [CrossRef]
- Steegmann, J.L.; Baccarani, M.; Breccia, M.; Casado, L.F.; Garcia-Gutierrez, V.; Hochhaus, A.; Kim, D.W.; Kim, T.D.; Khoury, H.J.; Le Coutre, P.; et al. European LeukemiaNet recommendations for the management and avoidance of adverse events of treatment in chronic myeloid leukaemia. Leukemia 2016, 30, 1648–1671. [Google Scholar] [CrossRef]
- Kantarjian, H.; O’Brien, S.; Jabbour, E.; Garcia-Manero, G.; Quintas-Cardama, A.; Shan, J.; Rios, M.B.; Ravandi, F.; Faderl, S.; Kadia, T.; et al. Improved survival in chronic myeloid leukemia since the introduction of imatinib therapy: A single-institution historical experience. Blood 2012, 119, 1981–1987. [Google Scholar] [CrossRef]
- Kirkpatrick, A.O.C.; Kartchner, D.; Allegri, S.; Nakajima An, D.; McCoy, K.; Davalbhakta, E.; Mitchell, C.S. Optimizations for Computing Relatedness in Biomedical Heterogeneous Information Networks: SemNet 2.0. Big Data Cogn. Comput. 2022, 6, 27. [Google Scholar] [CrossRef] [PubMed]
- Sedler, A.R.; Mitchell, C.S. SemNet: Using Local Features to Navigate the Biomedical Concept Graph. Front. Bioeng. Biotechnol. 2019, 7, 156. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Zhang, B.; Cloyd, J.M.; Alaimo, L.; Xu, G.; Du, S.; Mao, Y.; Pawlik, T.M. Novel Drug Candidate Prediction for Intrahepatic Cholangiocarcinoma via Hub Gene Network Analysis and Connectivity Mapping. Cancers 2022, 14, 3284. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.W.; Gao, Z.R.; Liang, H.; Fang, T.; Wang, Y.; Du, Z.Q.; Yang, C.X. Network analysis reveals different hub genes and molecular pathways for pig in vitro fertilized early embryos and parthenogenotes. Reprod. Domest. Anim. 2022, 1–10. [Google Scholar] [CrossRef]
- Nemati, M.; Zare, N.; Hedayat-Evrigh, N.; Asghari, R. Identification of Key Gene Network Modules and Hub Genes Associated with Wheat Response to Biotic Stress Using Combined Microarray Meta-analysis and WGCN Analysis. Mol. Biotechnol. 2022. Online ahead of print. [Google Scholar] [CrossRef]
- Zhang, Y.; Jin, R.; Zhou, Z.-H. Understanding bag-of-words model: A statistical framework. Int. J. Mach. Learn. Cybern. 2010, 1, 43–52. [Google Scholar] [CrossRef]
- MacQueen, J. Some methods for classification and analysis of multivariate observations. In Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability, Berkeley, CA, USA, 21 June–18 July 1965; pp. 281–297. [Google Scholar]
- Pedregosa, F.V.G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; Vanderplas, J.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- McCoy, K.; Gudapati, S.; He, L.; Horlander, E.; Kartchner, D.; Kulkarni, S.; Mehra, N.; Prakash, J.; Thenot, H.; Vanga, S.V.; et al. Biomedical Text Link Prediction for Drug Discovery: A Case Study with COVID-19. Pharmaceutics 2021, 13, 794. [Google Scholar] [CrossRef]
- Huang, Z.A.; Huang, Y.A.; You, Z.H.; Zhu, Z.; Sun, Y. Novel link prediction for large-scale miRNA-lncRNA interaction network in a bipartite graph. BMC Med. Genom. 2018, 11, 113. [Google Scholar] [CrossRef]
- Guven, C.; Atzmueller, M. Applying Answer Set Programming for Knowledge-Based Link Prediction on Social Interaction Networks. Front. Big Data 2019, 2, 15. [Google Scholar] [CrossRef]
- Crichton, G.; Guo, Y.; Pyysalo, S.; Korhonen, A. Neural networks for link prediction in realistic biomedical graphs: A multi-dimensional evaluation of graph embedding-based approaches. BMC Bioinform. 2018, 19, 176. [Google Scholar] [CrossRef]
- Gitelman, S.E.; Bundy, B.N.; Ferrannini, E.; Lim, N.; Blanchfield, J.L.; DiMeglio, L.A.; Felner, E.I.; Gaglia, J.L.; Gottlieb, P.A.; Long, S.A.; et al. Imatinib therapy for patients with recent-onset type 1 diabetes: A multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Diabetes Endocrinol. 2021, 9, 502–514. [Google Scholar] [CrossRef]
- Fountas, A.; Diamantopoulos, L.N.; Tsatsoulis, A. Tyrosine Kinase Inhibitors and Diabetes: A Novel Treatment Paradigm? Trends Endocrinol. Metab. 2015, 26, 643–656. [Google Scholar] [CrossRef]
- Elsherbiny, N.M.; El-Sherbiny, M.; Said, E. Amelioration of experimentally induced diabetic nephropathy and renal damage by nilotinib. J. Physiol. Biochem. 2015, 71, 635–648. [Google Scholar] [CrossRef]
- Samaha, M.M.; Said, E.; Salem, H.A. Nilotinib enhances beta-islets integrity and secretory functions in a rat model of STZ-induced diabetes mellitus. Eur. J. Pharmacol. 2019, 860, 172569. [Google Scholar] [CrossRef]
- Valent, P.; Gastl, G.; Geissler, K.; Greil, R.; Hantschel, O.; Lang, A.; Linkesch, W.; Lion, T.; Petzer, A.L.; Pittermann, E.; et al. Nilotinib as frontline and second-line therapy in chronic myeloid leukemia: Open questions. Crit. Rev. Oncol. Hematol. 2012, 82, 370–377. [Google Scholar] [CrossRef]
- Roden, M. Diabetes mellitus: Definition, classification and diagnosis. Wien. Klin. Wochenschr. 2016, 128 (Suppl. S2), S37–S40. [Google Scholar] [CrossRef]
- Soliman, A.T.; De Sanctis, V.; Yassin, M.; Wagdy, M.; Soliman, N. Chronic anemia and thyroid function. Acta Bio-Med. Atenei Parm. 2017, 88, 119–127. [Google Scholar] [CrossRef]
- Kim, D.S.; Na, Y.J.; Kang, M.H.; Yoon, S.Y.; Choi, C.W. Use of deferasirox, an iron chelator, to overcome imatinib resistance of chronic myeloid leukemia cells. Korean J. Intern Med. 2016, 31, 357–366. [Google Scholar] [CrossRef]
- Medeiros, B.C.; Possick, J.; Fradley, M. Cardiovascular, pulmonary, and metabolic toxicities complicating tyrosine kinase inhibitor therapy in chronic myeloid leukemia: Strategies for monitoring, detecting, and managing. Blood Rev. 2018, 32, 289–299. [Google Scholar] [CrossRef]
- Jain, P.; Kantarjian, H.; Boddu, P.C.; Nogueras-Gonzalez, G.M.; Verstovsek, S.; Garcia-Manero, G.; Borthakur, G.; Sasaki, K.; Kadia, T.M.; Sam, P.; et al. Analysis of cardiovascular and arteriothrombotic adverse events in chronic-phase CML patients after frontline TKIs. Blood Adv. 2019, 3, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Mauro, M.J. Lifelong TKI therapy: How to manage cardiovascular and other risks. Hematol. Am. Soc. Hematol. Educ. Program 2021, 2021, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Ahmadieh, H.; Salti, I. Tyrosine kinase inhibitors induced thyroid dysfunction: A review of its incidence, pathophysiology, clinical relevance, and treatment. BioMed Res. Int. 2013, 2013, 725410. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Nayernama, A.; Jones, S.C.; de Claro, R.A.; Waldron, P.E. BCR-ABL1 tyrosine kinase inhibitor-associated thyroid dysfunction: A review of cases reported to the FDA Adverse Event Reporting System and published in the literature. Am. J. Hematol. 2020, 95, E332–E335. [Google Scholar] [CrossRef]
- Lim, D.J.; Oh, E.J.; Park, C.W.; Kwon, H.S.; Hong, E.J.; Yoon, K.H.; Kang, M.I.; Cha, B.Y.; Lee, K.W.; Son, H.Y.; et al. Pancytopenia and secondary myelofibrosis could be induced by primary hyperparathyroidism. Int. J. Lab. Hematol. 2007, 29, 464–468. [Google Scholar] [CrossRef]
- O’Sullivan, S.; Lin, J.M.; Watson, M.; Callon, K.; Tong, P.C.; Naot, D.; Horne, A.; Aati, O.; Porteous, F.; Gamble, G.; et al. The skeletal effects of the tyrosine kinase inhibitor nilotinib. Bone 2011, 49, 281–289. [Google Scholar] [CrossRef]
- Wang, J.; Zhuang, S. Src family kinases in chronic kidney disease. Am. J. Physiol. Ren. Physiol. 2017, 313, F721–F728. [Google Scholar] [CrossRef]
- Iyoda, M.; Shibata, T.; Hirai, Y.; Kuno, Y.; Akizawa, T. Nilotinib attenuates renal injury and prolongs survival in chronic kidney disease. J. Am. Soc. Nephrol. 2011, 22, 1486–1496. [Google Scholar] [CrossRef]
- Sasaki, K.; Lahoti, A.; Jabbour, E.; Jain, P.; Pierce, S.; Borthakur, G.; Daver, N.; Kadia, T.; Pemmaraju, N.; Ferrajoli, A.; et al. Clinical Safety and Efficacy of Nilotinib or Dasatinib in Patients with Newly Diagnosed Chronic-Phase Chronic Myelogenous Leukemia and Pre-Existing Liver and/or Renal Dysfunction. Clin. Lymphoma Myeloma Leuk. 2016, 16, 152–162. [Google Scholar] [CrossRef]
- Tong, W.G.; Kantarjian, H.; O’Brien, S.; Faderl, S.; Ravandi, F.; Borthakur, G.; Shan, J.; Pierce, S.; Rios, M.B.; Cortes, J. Imatinib front-line therapy is safe and effective in patients with chronic myelogenous leukemia with pre-existing liver and/or renal dysfunction. Cancer 2010, 116, 3152–3159. [Google Scholar] [CrossRef]
- ElShaer, A.; Almasry, M.; Alawar, M.; Masoud, H.; El Kinge, A.R. Dasatinib-Induced Nephrotic Syndrome: A Case Report. Cureus 2021, 13, e20330. [Google Scholar] [CrossRef]
- Masiello, D.; Gorospe, G., 3rd; Yang, A.S. The occurrence and management of fluid retention associated with TKI therapy in CML, with a focus on dasatinib. J. Hematol. Oncol. 2009, 2, 46. [Google Scholar] [CrossRef][Green Version]
- Hailan, Y.M.; Elyas, A.; Abdulla, M.A.; Yassin, M.A. Dasatinib-Induced Pleural and Pericardial Effusions. Cureus 2021, 13, e19024. [Google Scholar] [CrossRef]
- Gunnarsson, N.; Hoglund, M.; Stenke, L.; Wallberg-Jonsson, S.; Sandin, F.; Bjorkholm, M.; Dreimane, A.; Lambe, M.; Markevarn, B.; Olsson-Stromberg, U.; et al. Increased prevalence of prior malignancies and autoimmune diseases in patients diagnosed with chronic myeloid leukemia. Leukemia 2016, 30, 1562–1567. [Google Scholar] [CrossRef]
- Soderlund, S.; Persson, I.; Ilander, M.; Guilhot, J.; Hjorth-Hansen, H.; Koskenvesa, P.; Richter, J.; Saussele, S.; Mustjoki, S.; Olsson-Stromberg, U. Plasma proteomics of biomarkers for inflammation or cancer cannot predict relapse in chronic myeloid leukaemia patients stopping tyrosine kinase inhibitor therapy. Leuk. Res. 2020, 90, 106310. [Google Scholar] [CrossRef]
- Oshima, N.; Mishima, Y.; Shibagaki, K.; Kawashima, K.; Ishimura, N.; Ikejiri, F.; Onishi, C.; Okada, T.; Inoue, M.; Moriyama, I.; et al. Differential gene expression analysis of dasatinib-induced colitis in a patient with chronic myeloid leukemia followed for 3 years: A case report. BMC Gastroenterol. 2021, 21, 19. [Google Scholar] [CrossRef]
- Bocchia, M.; Galimberti, S.; Aprile, L.; Sicuranza, A.; Gozzini, A.; Santilli, F.; Abruzzese, E.; Barate, C.; Scappini, B.; Fontanelli, G.; et al. Genetic predisposition and induced pro-inflammatory/pro-oxidative status may play a role in increased atherothrombotic events in nilotinib treated chronic myeloid leukemia patients. Oncotarget 2016, 7, 72311–72321. [Google Scholar] [CrossRef][Green Version]
- Demirsoy, E.T.; Mehtap, O.; Atesoglu, E.B.; Tarkun, P.; Eren, N.; Geduk, A.; Hacihanefioglu, A. Dasatinib-induced immune mediated-thrombotic thrombocytopenic purpura. Transfus. Apher. Sci. 2018, 57, 222–224. [Google Scholar] [CrossRef]
- Ptasiewicz, M.; Maksymiuk, P.; Chalas, R. Oral Hygiene Considerations in Adult Patients with Leukemia during a Cycle of Chemotherapy. Int. J. Environ. Res. Public Health 2022, 19, 479. [Google Scholar] [CrossRef]
- Ashok, L.; Sujatha, G.P.; Hema, G. Estimation of salivary amylase and total proteins in leukemia patients and its correlation with clinical feature and radiographic finding. Indian J. Dent. Res. 2010, 21, 486–490. [Google Scholar] [CrossRef]
- Allareddy, V.; Prakasam, S.; Allareddy, V.; Martinez-Schlurmann, N.I.; Rampa, S.; Nalliah, R.P.; Eswaran, S.V.; Elangovan, S. Poor Oral Health Linked with Increased Risk of Infectious Complications in Adults with Leukemia. J. Mass. Dent. Soc. 2015, 64, 38–42. [Google Scholar]
- Wang, Z.; Wang, X.; Wang, Z.; Feng, Y.; Jia, Y.; Jiang, L.; Xia, Y.; Cao, J.; Liu, Y. Comparison of Hepatotoxicity Associated With New BCR-ABL Tyrosine Kinase Inhibitors vs Imatinib Among Patients with Chronic Myeloid Leukemia: A Systematic Review and Meta-analysis. JAMA Netw. Open 2021, 4, e2120165. [Google Scholar] [CrossRef]
- Harbaum, L.; Marx, A.; Goekkurt, E.; Schafhausen, P.; Atanackovic, D. Treatment with dasatinib for chronic myeloid leukemia following imatinib-induced hepatotoxicity. Int. J. Hematol. 2014, 99, 91–94. [Google Scholar] [CrossRef]
- Kumar, V.; Singh, P.; Gupta, S.K.; Ali, V.; Verma, M. Transport and metabolism of tyrosine kinase inhibitors associated with chronic myeloid leukemia therapy: A review. Mol. Cell Biochem. 2022, 477, 1261–1279. [Google Scholar] [CrossRef]
- Nesr, G.; Claudiani, S.; Khorashad, J.; Apperley, J.; Milojkovic, D. The influence of salivary amylase on total amylase elevation in CML patients treated with TKI therapy: A case series of 3 patients. Leuk. Lymphoma 2019, 60, 3333–3334. [Google Scholar] [CrossRef]
- Shamroe, C.L.; Comeau, J.M. Ponatinib: A new tyrosine kinase inhibitor for the treatment of chronic myeloid leukemia and Philadelphia chromosome-positive acute lymphoblastic leukemia. Ann. Pharmacother. 2013, 47, 1540–1546. [Google Scholar] [CrossRef]
- Sasi, S.; Mohamed, M.; Chitrambika, P.; Yassin, M. Myasthenia Gravis and Myeloproliferative Neoplasms—Mere Association or Paraneoplastic Neurologic Syndrome: A Mini-Review. Acta Bio-Med. Atenei Parm. 2022, 92, e2021437. [Google Scholar] [CrossRef]
- Ishida, T.; Akagawa, N.; Miyata, T.; Tominaga, N.; Iizuka, T.; Higashihara, M.; Suzuki, T.; Miyazaki, K. Dasatinib-associated reversible demyelinating peripheral polyneuropathy in a case of chronic myeloid leukemia. Int. J. Hematol. 2018, 107, 373–377. [Google Scholar] [CrossRef]
- Kavanagh, S.; Bril, V.; Lipton, J.H. Peripheral neuropathy associated with imatinib therapy for chronic myeloid leukemia. Blood Res. 2018, 53, 172–174. [Google Scholar] [CrossRef]
- Inoue, H.; Taji, H.; Yamada, K.; Iriyama, C.; Saito, T.; Kato, H.; Yanada, M.; Yamamoto, K.; Matsukawa, N. Dasatinib-induced Reversible Demyelinating Peripheral Neuropathy and Successful Conversion to Nilotinib in Chronic Myelogenous Leukemia. Intern. Med. 2020, 59, 2419–2421. [Google Scholar] [CrossRef]
- Rotstein, D.L.; Sawicka, K.; Bharatha, A.; Montalban, X.; Lipton, J.H. CNS demyelination after initiating the tyrosine kinase inhibitor imatinib: A report of two cases. Mult. Scler. 2020, 26, 1121–1124. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.; Shah, P.; Moguel-Cobos, G. Nilotinib-Induced Dystonia and Cognitive Deficits in a Neurologically Normal Patient with Chronic Myeloid Leukemia. Case Rep. Neurol. Med. 2019, 2019, 3679319. [Google Scholar] [CrossRef] [PubMed]
- Chamoun, K.; Rabinovich, E.; Baer, L.; Fastenau, P.; Lima, M. A case of neurocognitive deficit strongly related to dasatinib therapy. Hematol. Transfus. Cell Ther. 2020, 42, 80–82. [Google Scholar] [CrossRef] [PubMed]
- Chow, E.J.; Doody, D.R.; Wilkes, J.J.; Becker, L.K.; Chennupati, S.; Morin, P.E.; Winestone, L.E.; Henk, H.J.; Lyman, G.H. Adverse events among chronic myelogenous leukemia patients treated with tyrosine kinase inhibitors: A real-world analysis of health plan enrollees. Leuk. Lymphoma 2021, 62, 1203–1210. [Google Scholar] [CrossRef]
- Yu, L.; Huang, X.; Gale, R.P.; Wang, H.; Jiang, Q. Variables associated with patient-reported symptoms in persons with chronic phase chronic myeloid leukemia receiving tyrosine kinase inhibitor therapy. Medicine 2019, 98, e18079. [Google Scholar] [CrossRef] [PubMed]
- Breccia, M.; Efficace, F.; Colafigli, G.; Scalzulli, E.; Di Prima, A.; Martelli, M.; Foa, R. Tyrosine kinase inhibitor discontinuation in the management of chronic myeloid leukemia: A critical review of the current practice. Expert Rev. Hematol. 2020, 13, 1311–1318. [Google Scholar] [CrossRef]
- Duman, B.B.; Paydas, S.; Disel, U.; Besen, A.; Gurkan, E. Secondary malignancy after imatinib therapy: Eight cases and review of the literature. Leuk. Lymphoma 2012, 53, 1706–1708. [Google Scholar] [CrossRef]
- Kumar, V.; Garg, M.; Chaudhary, N.; Chandra, A.B. An observational study on risk of secondary cancers in chronic myeloid leukemia patients in the TKI era in the United States. PeerJ 2018, 6, e4342. [Google Scholar] [CrossRef]
- Miranda, M.B.; Lauseker, M.; Kraus, M.P.; Proetel, U.; Hanfstein, B.; Fabarius, A.; Baerlocher, G.M.; Heim, D.; Hossfeld, D.K.; Kolb, H.J.; et al. Secondary malignancies in chronic myeloid leukemia patients after imatinib-based treatment: Long-term observation in CML Study IV. Leukemia 2016, 30, 1255–1262. [Google Scholar] [CrossRef]
- Narra, R.K.; Flynn, K.E.; Atallah, E. Chronic Myeloid Leukemia-the Promise of Tyrosine Kinase Inhibitor Discontinuation. Curr Hematol. Malig. Rep. 2017, 12, 415–423. [Google Scholar] [CrossRef]
- Albayrak, M.; Celebi, H.; Albayrak, A.; Can, E.S.; Aslan, V.; Onec, B.; Coban, I. Serious skin reaction associated with imatinib in a patient with chronic myeloid leukemia. Eurasian J. Med. 2011, 43, 192–195. [Google Scholar] [CrossRef]
- Cortes, J.E.; Jimenez, C.A.; Mauro, M.J.; Geyer, A.; Pinilla-Ibarz, J.; Smith, B.D. Pleural Effusion in Dasatinib-Treated Patients with Chronic Myeloid Leukemia in Chronic Phase: Identification and Management. Clin. Lymphoma Myeloma Leuk. 2017, 17, 78–82. [Google Scholar] [CrossRef]
- Gunnarsson, N.; Stenke, L.; Hoglund, M.; Sandin, F.; Bjorkholm, M.; Dreimane, A.; Lambe, M.; Markevarn, B.; Olsson-Stromberg, U.; Richter, J.; et al. Second malignancies following treatment of chronic myeloid leukaemia in the tyrosine kinase inhibitor era. Br. J. Haematol. 2015, 169, 683–688. [Google Scholar] [CrossRef]
- Nye, B.; Jessy Li, J.; Patel, R.; Yang, Y.; Marshall, I.J.; Nenkova, A.; Wallace, B.C. A Corpus with Multi-Level Annotations of Patients, Interventions and Outcomes to Support Language Processing for Medical Literature. Proc. Conf. Assoc. Comput. Linguist Meet. 2018, 2018, 197–207. [Google Scholar]
- Allegri, S.A.; McCoy, K.; Mitchell, C.S. CompositeView: A Network-Based Visualization Tool. Big Data Cogn. Comput. 2022, 6, 66. [Google Scholar] [CrossRef]
| Event | C4 Bosutinib | C7 Imatinib | C8 Ponatinib | C10 Dasatinib | C16 Nilotinib |
|---|---|---|---|---|---|
| diarrhea | 0.045 | 0.005 | 0.000 | 0.010 | 0.001 |
| gastrointestinal | 0.017 | 0.034 | 0.000 | 0.020 | 0.006 |
| nausea | 0.016 | 0.007 | 0.000 | 0.002 | 0.001 |
| vomiting | 0.014 | 0.002 | 0.000 | 0.000 | 0.001 |
| cardiovascular | 0.014 | 0.000 | 0.051 | 0.004 | 0.031 |
| pleural | 0.011 | 0.003 | 0.000 | 0.029 | 0.002 |
| effusion | 0.011 | 0.005 | 0.000 | 0.032 | 0.002 |
| pulmonary | 0.009 | 0.001 | 0.000 | 0.066 | 0.002 |
| edema | 0.004 | 0.020 | 0.000 | 0.001 | 0.006 |
| rash | 0.003 | 0.028 | 0.002 | 0.011 | 0.009 |
| liver | 0.008 | 0.022 | 0.006 | 0.000 | 0.013 |
| platelet | 0.000 | 0.019 | 0.021 | 0.028 | 0.005 |
| myelosuppression | 0.003 | 0.012 | 0.002 | 0.007 | 0.000 |
| hepatitis | 0.000 | 0.012 | 0.000 | 0.003 | 0.000 |
| hematological | 0.000 | 0.011 | 0.002 | 0.004 | 0.006 |
| hypertension | 0.000 | 0.000 | 0.019 | 0.037 | 0.003 |
| thrombocytopenia | 0.005 | 0.007 | 0.004 | 0.004 | 0.001 |
| pneumonia | 0.000 | 0.000 | 0.000 | 0.012 | 0.005 |
| inflammation | 0.000 | 0.007 | 0.000 | 0.012 | 0.002 |
| vascular | 0.004 | 0.002 | 0.035 | 0.022 | 0.025 |
| arterial | 0.000 | 0.000 | 0.021 | 0.029 | 0.024 |
| diabetes | 0.000 | 0.000 | 0.000 | 0.000 | 0.013 |
| lesion | 0.000 | 0.020 | 0.002 | 0.000 | 0.013 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mehra, N.; Varmeziar, A.; Chen, X.; Kronick, O.; Fisher, R.; Kota, V.; Mitchell, C.S. Cross-Domain Text Mining to Predict Adverse Events from Tyrosine Kinase Inhibitors for Chronic Myeloid Leukemia. Cancers 2022, 14, 4686. https://doi.org/10.3390/cancers14194686
Mehra N, Varmeziar A, Chen X, Kronick O, Fisher R, Kota V, Mitchell CS. Cross-Domain Text Mining to Predict Adverse Events from Tyrosine Kinase Inhibitors for Chronic Myeloid Leukemia. Cancers. 2022; 14(19):4686. https://doi.org/10.3390/cancers14194686
Chicago/Turabian StyleMehra, Nidhi, Armon Varmeziar, Xinyu Chen, Olivia Kronick, Rachel Fisher, Vamsi Kota, and Cassie S. Mitchell. 2022. "Cross-Domain Text Mining to Predict Adverse Events from Tyrosine Kinase Inhibitors for Chronic Myeloid Leukemia" Cancers 14, no. 19: 4686. https://doi.org/10.3390/cancers14194686
APA StyleMehra, N., Varmeziar, A., Chen, X., Kronick, O., Fisher, R., Kota, V., & Mitchell, C. S. (2022). Cross-Domain Text Mining to Predict Adverse Events from Tyrosine Kinase Inhibitors for Chronic Myeloid Leukemia. Cancers, 14(19), 4686. https://doi.org/10.3390/cancers14194686






