Combined Large Cell Neuroendocrine Carcinomas of the Lung: Integrative Molecular Analysis Identifies Subtypes with Potential Therapeutic Implications
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cases
2.2. Immunohistochemistry
2.3. Mutational and Copy Number Variation Status of 409 Cancer Genes
2.4. Tumor Mutational Load and Mutational Signatures
2.5. Fusion Gene Detection
2.6. Gene Expression Analysis by Next-Generation Sequencing
2.7. Gene Set Enrichment Analysis
2.8. Statistical Analysis
3. Results
3.1. Clinico-Pathological Features
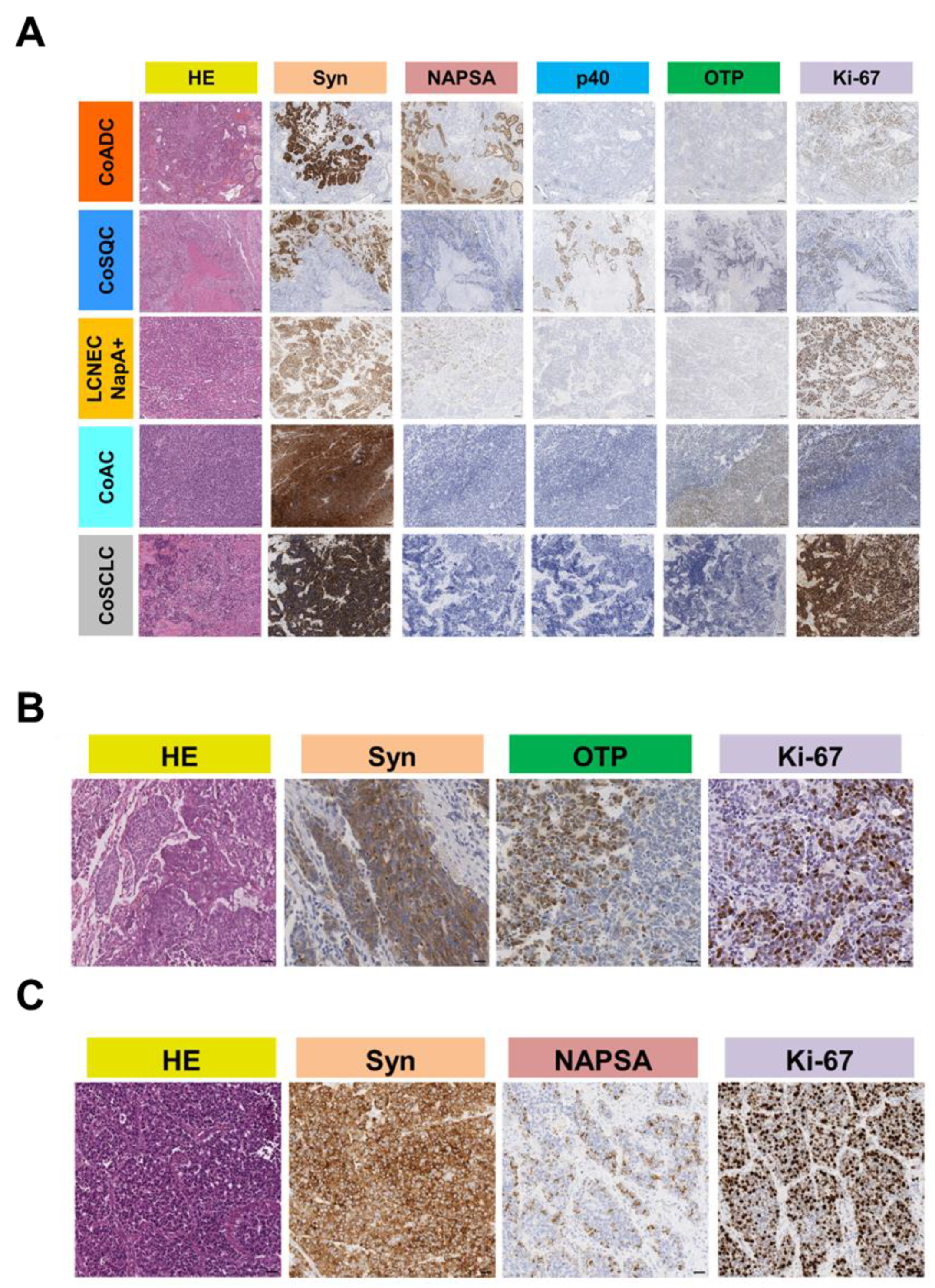
3.2. Immunohistochemical Features
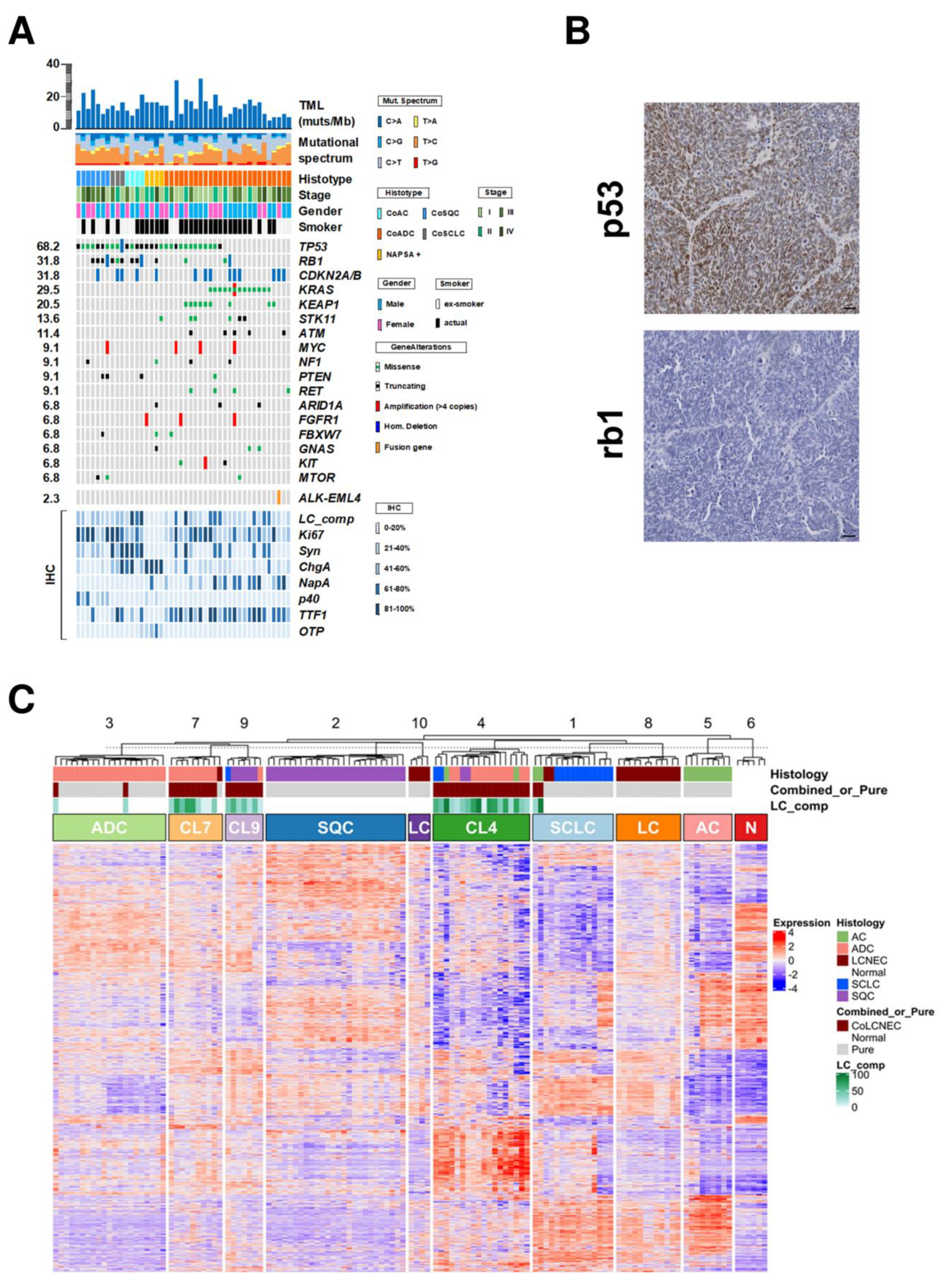
3.3. Mutational and Copy Number Status of 409 Genes
3.4. Fusion Genes
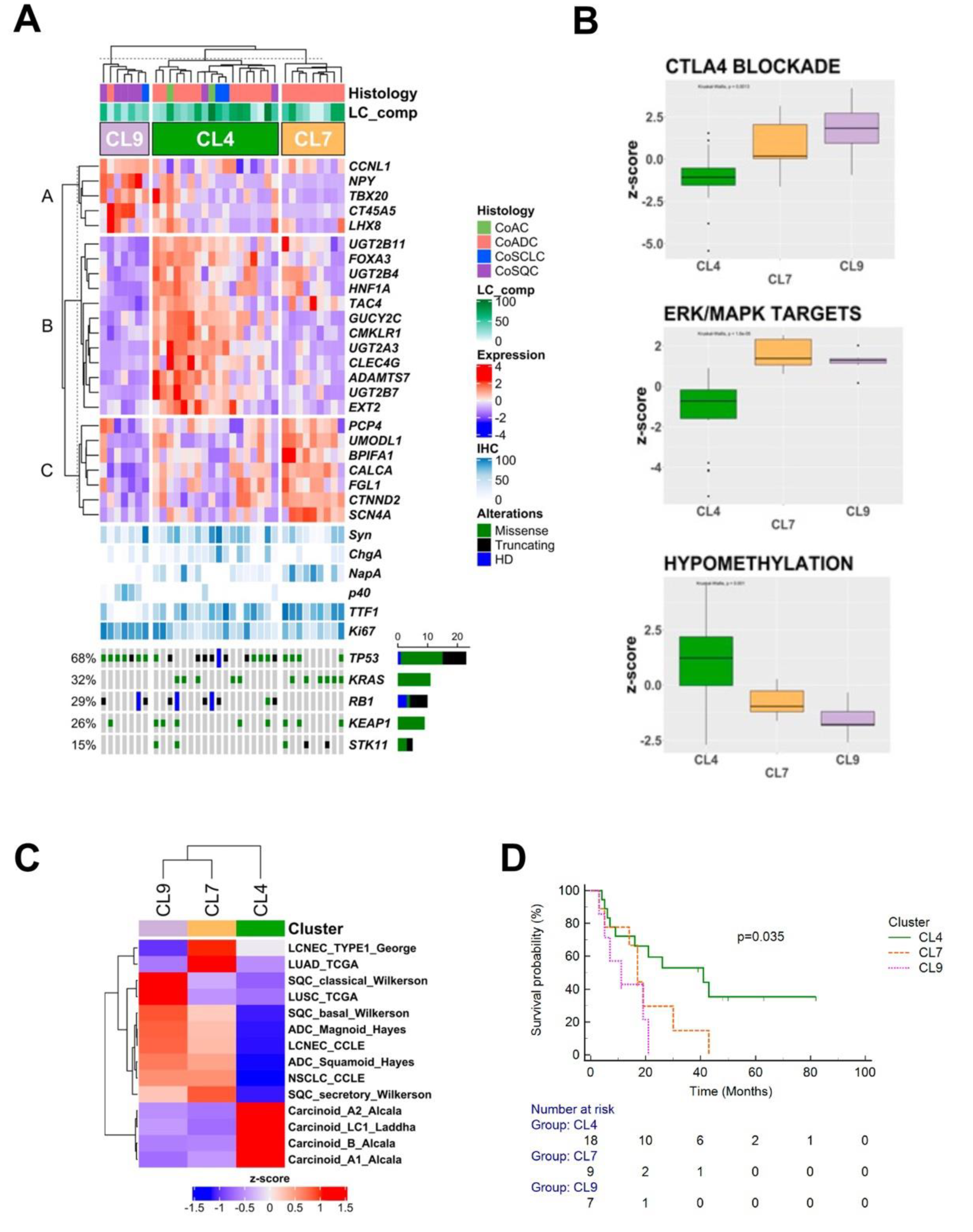
3.5. Gene Expression Profiles
3.6. Survival Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Travis, W.D.; Brambilla, E.; Burke, A.; Marx, A.; Nicholson, A.G. WHO Classification of Tumours. Thoracic Tumours; International Agency for Research on Cancer: Lyon, France, 2021; p. 412. [Google Scholar]
- Righi, L.; Gatti, G.; Volante, M.; Papotti, M. Lung neuroendocrine tumors: Pathological characteristics. J Thorac. Dis. 2017, 9, S1442–S1447. [Google Scholar] [CrossRef] [PubMed]
- Rekhtman, N.; Pietanza, C.M.; Sabari, J.; Montecalvo, J.; Wang, H.; Habeeb, O.; Kadota, K.; Adusumilli, P.; Rudin, C.M.; Ladanyi, M.; et al. Pulmonary large cell neuroendocrine carcinoma with adenocarcinoma-like features: Napsin A expression and genomic alterations. Mod. Pathol. 2018, 31, 111–121. [Google Scholar] [CrossRef]
- Travis, W.; Brambilla, E.; Burke, A.; Marx, A.; Nicholson, A. WHO Classification of Tumours of the Lung, Pleura, Thymus and Heart; International Agency for Research on Cancer: Lyon, France, 2015. [Google Scholar]
- Simbolo, M.; Barbi, S.; Fassan, M.; Mafficini, A.; Ali, G.; Vicentini, C.; Sperandio, N.; Corbo, V.; Rusev, B.; Mastracci, L.; et al. Gene Expression Profiling of Lung Atypical Carcinoids and Large Cell Neuroendocrine Carcinomas Identifies Three Transcriptomic Subtypes with Specific Genomic Alterations. J. Thorac. Oncol. 2019, 14, 1651–1661. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, G.; Bianchi, F.; Dama, E.; Simbolo, M.; Mafficini, A.; Sonzogni, A.; Pilotto, S.; Harari, S.; Papotti, M.; Volante, M.; et al. Most high-grade neuroendocrine tumours of the lung are likely to secondarily develop from pre-existing carcinoids: Innovative findings skipping the current pathogenesis paradigm. Virchows Arch. 2018, 472, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Milione, M.; Maisonneuve, P.; Grillo, F.; Mangogna, A.; Centonze, G.; Prinzi, N.; Pusceddu, S.; Garzone, G.; Cattaneo, L.; Busico, A.; et al. Ki-67 Index of 55% Distinguishes Two Groups of Bronchopulmonary Pure and Composite Large Cell Neuroendocrine Carcinomas with Distinct Prognosis. Neuroendocrinology 2021, 111, 475–489. [Google Scholar] [CrossRef]
- George, J.; Walter, V.; Peifer, M.; Alexandrov, L.B.; Seidel, D.; Leenders, F.; Maas, L.; Muller, C.; Dahmen, I.; Delhomme, T.M.; et al. Integrative genomic profiling of large-cell neuroendocrine carcinomas reveals distinct subtypes of high-grade neuroendocrine lung tumors. Nat. Commun. 2018, 9, 1048. [Google Scholar] [CrossRef]
- Miyoshi, T.; Umemura, S.; Matsumura, Y.; Mimaki, S.; Tada, S.; Makinoshima, H.; Ishii, G.; Udagawa, H.; Matsumoto, S.; Yoh, K.; et al. Genomic Profiling of Large-Cell Neuroendocrine Carcinoma of the Lung. Clin. Cancer Res. 2017, 23, 757–765. [Google Scholar] [CrossRef]
- Tang, M.; Abbas, H.A.; Negrao, M.V.; Ramineni, M.; Hu, X.; Hubert, S.M.; Fujimoto, J.; Reuben, A.; Varghese, S.; Zhang, J.; et al. The histologic phenotype of lung cancers is associated with transcriptomic features rather than genomic characteristics. Nat. Commun. 2021, 12, 7081. [Google Scholar] [CrossRef]
- Scardoni, M.; Vittoria, E.; Volante, M.; Rusev, B.; Bersani, S.; Mafficini, A.; Gottardi, M.; Giandomenico, V.; Malleo, G.; Butturini, G.; et al. Mixed adenoneuroendocrine carcinomas of the gastrointestinal tract: Targeted next-generation sequencing suggests a monoclonal origin of the two components. Neuroendocrinology 2014, 100, 310–316. [Google Scholar] [CrossRef]
- Huang, J.; Behrens, C.; Wistuba, I.I.; Gazdar, A.F.; Jagirdar, J. Clonality of combined tumors. Arch. Pathol. Lab. Med. 2002, 126, 437–441. [Google Scholar] [CrossRef]
- Mahul, B.A.; Stephen, E.; Frederick, L.; David, R.; Robert, K.; Mary, K. AJCC Cancer Staging Manual, 8th ed.; Springer Nature: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Milione, M.; Maisonneuve, P.; Pellegrinelli, A.; Grillo, F.; Albarello, L.; Spaggiari, P.; Vanoli, A.; Tagliabue, G.; Pisa, E.; Messerini, L.; et al. Ki67 proliferative index of the neuroendocrine component drives MANEC prognosis. Endocr. Relat. Cancer 2018, 25, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Volante, M.; Brizzi, M.P.; Faggiano, A.; La Rosa, S.; Rapa, I.; Ferrero, A.; Mansueto, G.; Righi, L.; Garancini, S.; Capella, C.; et al. Somatostatin receptor type 2A immunohistochemistry in neuroendocrine tumors: A proposal of scoring system correlated with somatostatin receptor scintigraphy. Mod. Pathol. 2007, 20, 1172–1182. [Google Scholar] [CrossRef] [PubMed]
- Simbolo, M.; Gottardi, M.; Corbo, V.; Fassan, M.; Mafficini, A.; Malpeli, G.; Lawlor, R.T.; Scarpa, A. DNA qualification workflow for next generation sequencing of histopathological samples. PLoS ONE 2013, 8, e62692. [Google Scholar] [CrossRef] [PubMed]
- Law, C.W.; Alhamdoosh, M.; Su, S.; Dong, X.; Tian, L.; Smyth, G.K.; Ritchie, M.E. RNA-seq analysis is easy as 1-2-3 with limma, Glimma and edgeR. F1000Res 2016, 5. [Google Scholar] [CrossRef]
- Luo, W.; Friedman, M.S.; Shedden, K.; Hankenson, K.D.; Woolf, P.J. GAGE: Generally applicable gene set enrichment for pathway analysis. BMC Bioinform. 2009, 10, 161. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- Hong, D.S.; Fakih, M.G.; Strickler, J.H.; Desai, J.; Durm, G.A.; Shapiro, G.I.; Falchook, G.S.; Price, T.J.; Sacher, A.; Denlinger, C.S.; et al. KRAS(G12C) Inhibition with Sotorasib in Advanced Solid Tumors. N. Engl. J. Med. 2020, 383, 1207–1217. [Google Scholar] [CrossRef]
- Gu, Z.; Schlesner, M.; Hübschmann, D. Cola: An R/Bioconductor package for consensus partitioning through a general framework. Nucleic Acids Res. 2021, 49, e15. [Google Scholar] [CrossRef]
- Alcala, N.; Leblay, N.; Gabriel, A.A.G.; Mangiante, L.; Hervas, D.; Giffon, T.; Sertier, A.S.; Ferrari, A.; Derks, J.; Ghantous, A.; et al. Integrative and comparative genomic analyses identify clinically relevant pulmonary carcinoid groups and unveil the supra-carcinoids. Nat. Commun. 2019, 10, 3407. [Google Scholar] [CrossRef]
- Laddha, S.V.; da Silva, E.M.; Robzyk, K.; Untch, B.R.; Ke, H.; Rekhtman, N.; Poirier, J.T.; Travis, W.D.; Tang, L.H.; Chan, C.S. Integrative Genomic Characterization Identifies Molecular Subtypes of Lung Carcinoids. Cancer Res. 2019, 79, 4339–4347. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research, N. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014, 511, 543–550. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research, N. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012, 489, 519–525. [Google Scholar] [CrossRef]
- Wilkerson, M.D.; Yin, X.; Hoadley, K.A.; Liu, Y.; Hayward, M.C.; Cabanski, C.R.; Muldrew, K.; Miller, C.R.; Randell, S.H.; Socinski, M.A.; et al. Lung squamous cell carcinoma mRNA expression subtypes are reproducible, clinically important, and correspond to normal cell types. Clin. Cancer Res. 2010, 16, 4864–4875. [Google Scholar] [CrossRef]
- Ghandi, M.; Huang, F.W.; Jane-Valbuena, J.; Kryukov, G.V.; Lo, C.C.; McDonald, E.R., 3rd; Barretina, J.; Gelfand, E.T.; Bielski, C.M.; Li, H.; et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019, 569, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Rudin, C.M.; Poirier, J.T.; Byers, L.A.; Dive, C.; Dowlati, A.; George, J.; Heymach, J.V.; Johnson, J.E.; Lehman, J.M.; MacPherson, D.; et al. Molecular subtypes of small cell lung cancer: A synthesis of human and mouse model data. Nat. Rev. Cancer 2019, 19, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Patel, V.M.; Coon, M.; Nguyen, T.; Land, S.J.; Ruden, D.M.; Lu, X. Using Drosophila melanogaster as a Model for Genotoxic Chemical Mutational Studies with a New Program, SnpSift. Front. Genet. 2012, 3, 35. [Google Scholar] [CrossRef]
- McLaren, W.; Pritchard, B.; Rios, D.; Chen, Y.; Flicek, P.; Cunningham, F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 2010, 26, 2069–2070. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdottir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Boeva, V.; Popova, T.; Lienard, M.; Toffoli, S.; Kamal, M.; Le Tourneau, C.; Gentien, D.; Servant, N.; Gestraud, P.; Rio Frio, T.; et al. Multi-factor data normalization enables the detection of copy number aberrations in amplicon sequencing data. Bioinformatics 2014, 30, 3443–3450. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Borresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Erson-Omay, E.Z.; Caglayan, A.O.; Schultz, N.; Weinhold, N.; Omay, S.B.; Ozduman, K.; Koksal, Y.; Li, J.; Serin Harmanci, A.; Clark, V.; et al. Somatic POLE mutations cause an ultramutated giant cell high-grade glioma subtype with better prognosis. Neuro Oncol 2015, 17, 1356–1364. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Gay, M.; Vila-Casadesus, M.; Franch-Exposito, S.; Hernandez-Illan, E.; Lozano, J.J.; Castellvi-Bel, S. Mutational Signatures in Cancer (MuSiCa): A web application to implement mutational signatures analysis in cancer samples. BMC Bioinform. 2018, 19, 224. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome. Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed]
- Čuklina, J. Computational Challenges in Biomarker Discovery from High-Throughput Proteomic Data. Ph.D. Thesis, ETH Zurich, Zürich, Switzerland, 2018. [Google Scholar]
- Kaufman, L.R.P. Finding Groups in Data: An Introduction to Cluster Analysis; Wiley: New York, NY, USA, 1990. [Google Scholar]
- Struyf, A.; Hubert, M.; Rousseeuw, P.J. Integrating Robust Clustering Techniques in S-PLUS. Comput. Stat. Data Anal. 1997, 26, 17–37. [Google Scholar] [CrossRef]
- Gates, A.J.; Ahn, Y.Y. The impact of random models on clustering similarity. arXiv 2017, arXiv:1701.06508. [Google Scholar]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef]
- Tarca, A.L.; Draghici, S.; Khatri, P.; Hassan, S.S.; Mittal, P.; Kim, J.S.; Kim, C.J.; Kusanovic, J.P.; Romero, R. A novel signaling pathway impact analysis. Bioinformatics 2009, 25, 75–82. [Google Scholar] [CrossRef]
| All Patients | CoAC | CoADC | NapA+ | CoSCLC | CoSQC | p-Value * | |
|---|---|---|---|---|---|---|---|
| Total | 44 (100) | 4 (100) | 26 (100) | 4 (100) | 3 (100) | 7 (100) | |
| Gender | |||||||
| Female | 18 (40.9) | 2 (50.0) | 9 (34.6) | 2 (50.0) | 1 (33.3) | 4 (57.1) | |
| Male | 26 (59.1) | 2 (50.0) | 17 (65.4) | 2 (50.0) | 2 (66.7) | 3 (42.9) | 0.79 |
| Age (Years) | |||||||
| Median (range) | 67 (43–82) | 57 (54–69) | 67 (43–77) | 58.5 (48–70) | 74 (43–78) | 71 (47–82) | 0.29 |
| Smoke | |||||||
| Actual | 29 (65.9) | 2 (50.0) | 19 (73.1) | 4 (100.0) | 1 (33.3) | 3 (42.9) | |
| Former | 15 (34.1) | 2 (50.0) | 7 (26.9) | 0 (0.0) | 2 (66.7) | 4 (57.1) | 0.17 |
| Site | |||||||
| Central | 10 (22.7) | 3 (75.0) | 4 (15.4) | 0 (0.0) | 3 (100.0) | 0 (0.0) | |
| Peripheral | 34 (77.3) | 1 (25.0) | 22 (84.6) | 4 (100.0) | 0 (0.0) | 7 (100.0) | 0.0009 |
| Stage | |||||||
| I | 17 (38.6) | 2 (50.0) | 11 (42.3) | 1 (25.0) | 1 (33.3) | 2 (28.6) | |
| II | 9 (20.5) | 1 (25.0) | 5 (19.2) | 2 (50.0) | 1 (33.3) | 0 (0.0) | |
| III | 13 (29.5) | 1 (25.0) | 8 (30.8) | 0 (0.0) | 1 (33.3) | 3 (42.8) | |
| IV | 5 (11.4) | 0 (0.0) | 2 (7.7) | 1 (25.0) | 0 (0.0) | 2 (28.6) | 0.61 |
| Mitoses (2 mm2) | |||||||
| Median (range) | 25 (10–69) | 28.5 (13–43) | 24 (10–46) | 29.5 (25–44) | 27 (21–69) | 25 (10–29) | 0.51 |
| Ki-67 | |||||||
| Median (range) | 60 (20–95) | 55 (44–75) | 59 (27–94) | 50 (41–76) | 85 (61–91) | 68 (20–95) | 0.61 |
| % LCNEC Component | |||||||
| Median (range) | 40 (10–90) | 80 (30–90) | 35 (10–90) | 75 (30–80) | 50 (40–60) | 50 (20–60) | 0.12 |
| % Non-LCNEC Component | |||||||
| Median (range) | 60 (10–90) | 20 (10–70) | 65 (10–90) | 25 (20–70) | 50 (40–60) | 50 (40–80) | 0.12 |
| N. of Patients (%) | CoAC | CoADC | NapA+ | CoSCLC | CoSQC | p-Value * | Adjusted p-Value † | |
|---|---|---|---|---|---|---|---|---|
| Total | 44 (100) | 4 (100) | 26 (100) | 4 (100) | 3 (100) | 7 (100) | ||
| TP53 ^ | 30 (68.2) | 4 (100) | 12 (46.1) | 4 (100) | 3 (100) | 7 (100) | 0.005 | 0.04 |
| RB1 ^ | 14 (31.8) | 3 (75.0) | 3 (11.5) | 1 (25.0) | 3 (100) | 4 (57.1) | 0.0006 | 0.01 |
| CDKN2A/B ^ | 14 (31.8) | 1 (25.0) | 9 (34.6) | 1 (25.0) | 2 (66.7) | 1 (14.3) | 0.62 | 0.89 |
| KRAS | 13 (29.5) | 0 (0.0) | 13 (50.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0.01 | 0.056 |
| KEAP1 | 8 (18.2) | 0 (0.0) | 8 (30.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0.26 | 0.74 |
| STK11 | 6 (13.6) | 0 (0.0) | 5 (19.2) | 1 (25.0) | 0 (0.0) | 0 (0.0) | 0.68 | 0.89 |
| ATM | 5 (11.4) | 0 (0.0) | 5 (19.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0.77 | 0.94 |
| MYC ° | 4 (9.1) | 0 (0.0) | 3 (11.5) | 0 (0.0) | 0 (0.0) | 1 (14.3) | 1.00 | 1.00 |
| NF1 | 4 (9.1) | 0 (0.0) | 2 (7.7) | 1 (25.0) | 0 (0.0) | 1 (14.3) | 0.60 | 0.89 |
| PTEN ^ | 4 (9.1) | 1 (25.0) | 1 (3.8) | 0 (0.0) | 0 (0.0) | 2 (28.6) | 0.19 | 0.74 |
| RET | 4 (9.1) | 0 (0.0) | 4 (15.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0.86 | 0.97 |
| ARID1A | 3 (6.8) | 0 (0.0) | 2 (7.7) | 1 (25.0) | 0 (0.0) | 0 (0.0) | 0.63 | 0.89 |
| FGFR1 | 3 (6.8) | 0 (0.0) | 2 (7.7) | 1 (25.0) | 0 (0.0) | 0 (0.0) | 0.63 | 0.89 |
| FBXW7 | 3 (6.8) | 0 (0.0) | 1 (3.8) | 1 (25.0) | 0 (0.0) | 1 (14.3) | 0.36 | 0.87 |
| GNAS | 3 (6.8) | 0 (0.0) | 2 (7.7) | 1 (25.0) | 0 (0.0) | 0 (0.0) | 0.63 | 0.89 |
| KIT ^ | 3 (6.8) | 0 (0.0) | 3 (11.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1.00 | 1.00 |
| MTOR | 3 (6.8) | 0 (0.0) | 1 (3.8) | 0 (0.0) | 0 (0.0) | 2 (28.6) | 0.25 | 0.74 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Simbolo, M.; Centonze, G.; Giudice, L.; Grillo, F.; Maisonneuve, P.; Gkountakos, A.; Ciaparrone, C.; Cattaneo, L.; Sabella, G.; Giugno, R.; et al. Combined Large Cell Neuroendocrine Carcinomas of the Lung: Integrative Molecular Analysis Identifies Subtypes with Potential Therapeutic Implications. Cancers 2022, 14, 4653. https://doi.org/10.3390/cancers14194653
Simbolo M, Centonze G, Giudice L, Grillo F, Maisonneuve P, Gkountakos A, Ciaparrone C, Cattaneo L, Sabella G, Giugno R, et al. Combined Large Cell Neuroendocrine Carcinomas of the Lung: Integrative Molecular Analysis Identifies Subtypes with Potential Therapeutic Implications. Cancers. 2022; 14(19):4653. https://doi.org/10.3390/cancers14194653
Chicago/Turabian StyleSimbolo, Michele, Giovanni Centonze, Luca Giudice, Federica Grillo, Patrick Maisonneuve, Anastasios Gkountakos, Chiara Ciaparrone, Laura Cattaneo, Giovanna Sabella, Rosalba Giugno, and et al. 2022. "Combined Large Cell Neuroendocrine Carcinomas of the Lung: Integrative Molecular Analysis Identifies Subtypes with Potential Therapeutic Implications" Cancers 14, no. 19: 4653. https://doi.org/10.3390/cancers14194653
APA StyleSimbolo, M., Centonze, G., Giudice, L., Grillo, F., Maisonneuve, P., Gkountakos, A., Ciaparrone, C., Cattaneo, L., Sabella, G., Giugno, R., Bossi, P., Spaggiari, P., Del Gobbo, A., Ferrero, S., Mastracci, L., Fabbri, A., Filugelli, M., Garzone, G., Prinzi, N., ... Milione, M. (2022). Combined Large Cell Neuroendocrine Carcinomas of the Lung: Integrative Molecular Analysis Identifies Subtypes with Potential Therapeutic Implications. Cancers, 14(19), 4653. https://doi.org/10.3390/cancers14194653











