Simple Summary
Colorectal cancer remains one of the most significant sources of cancer-related morbidity and mortality worldwide. The liver is the most common site of metastatic spread. Multiple modalities exist to manage and potentially cure patients with metastatic colorectal cancer. However, reliable biomarkers to assist with clinical decision-making are limited. Recent advances in genomic sequencing technology have greatly expanded our knowledge of colorectal cancer carcinogenesis and significantly reduced the cost and timing of the investigation. In this article, we discuss the current utility of biomarkers in the management of colorectal cancer liver metastases.
Abstract
Surgical management combined with improved systemic therapies have extended 5-year overall survival beyond 50% among patients with colorectal liver metastases (CRLM). Furthermore, a multitude of liver-directed therapies has improved local disease control for patients with unresectable CRLM. Unfortunately, a significant portion of patients treated with curative-intent hepatectomy develops disease recurrence. Traditional markers fail to risk-stratify and prognosticate patients with CRLM appropriately. Over the last few decades, advances in molecular sequencing technology have greatly expanded our knowledge of the pathophysiology and tumor microenvironment characteristics of CRLM. These investigations have revealed biomarkers with the potential to better inform management decisions in patients with CRLM. Actionable biomarkers such as RAS and BRAF mutations, microsatellite instability/mismatch repair status, and tumor mutational burden have been incorporated into national and societal guidelines. Other biomarkers, including circulating tumor DNA and radiomic features, are under active investigation to evaluate their clinical utility. Given the plethora of therapeutic modalities and lack of evidence on timing and sequence, reliable biomarkers are needed to assist clinicians with the development of patient-tailored management plans. In this review, we discuss the current evidence regarding biomarkers for patients with CRLM.
1. Introduction
Colorectal cancer (CRC) is a leading cause of cancer-related morbidity and mortality worldwide [1]. While the global incidence continues to rise, disproportionately so in low- and middle-income countries, advances in CRC screening and multidisciplinary therapy have significantly improved mortality rates, especially among patients living in highly developed nations where mortality rates have declined in recent years [2]. Despite these improvements, CRC remains the second leading cause of cancer-related mortality in the United States [3].
Metastatic liver disease develops in approximately 50% of patients with CRC [3]. While most cases of colorectal liver metastases (CRLM) develop metachronously after treatment for locoregional CRC, 20% to 34% of patients with CRC present with synchronous liver disease. Synchronous disease portends a worse prognosis as these patients more frequently have multiple liver lesions and bilobar disease versus patients with metachronous CRLM [4]. Unfortunately, 80% to 90% of patients with CRLM have an unresectable liver disease at presentation [5,6]. Five-year survival is significantly lower among patients not undergoing surgery [7]. Consequently, metastatic liver disease drives mortality in most patients with CRC.
The current standard of care therapy for patients with resectable CRLM includes both curative intent surgery and perioperative systemic therapy [8,9]. Refined surgical techniques combined with more efficient surgical devices have greatly reduced perioperative morbidity and mortality from liver surgery while improving resection rates [10]. Complete surgical resection with negative margins and adequate liver remnant significantly improves long-term outcomes for patients [9,11]. Recent meta-analyses reported a median 5-year survival of 38% to 71% for select patients with solitary liver metastases following resection with appropriate perioperative systemic therapy [12,13].
However, 50% of patients develop recurrent disease within two years after surgery [14]. While some of these patients are candidates for additional resection, many have an unresectable disease. For patients with unresectable CRLM, liver-directed therapies provide local disease control and, in certain situations, may convert patients to resectable disease. Liver-directed therapies include hepatic artery infusion (HAI), arterial-directed embolization (i.e., radioembolization (Y-90)), ablation, or external beam radiation (i.e., conformal radiation therapy (CRT), stereotactic body radiation therapy (SBRT), and intensity-modulated radiation therapy (IMRT)) [8]. While surgery remains the gold standard therapy, questions remain over the choice and sequence of perioperative systemic therapy, as well as the necessity and timing of liver-directed therapies. In fact, even among expert liver surgeons, significant debate exists regarding the appropriate therapeutic strategy in more than half of the clinical cases proposed in a recent survey [15].
Advances in molecular sequencing technology (e.g., next-generation sequencing (NGS)) and computational data analytics have greatly improved our understanding of CRC pathophysiology, revealing genomic variants responsible for CRC carcinogenesis. These molecular biomarkers, combined with the emerging fields of radiomics and artificial intelligence, provide clinicians with potentially actionable information. Current treatment algorithms for patients with CRLM incorporate certain biomarkers, including RAS, BRAF, and mismatch repair (MMR) status. Other biomarkers require validation on their clinical utility and are actively being investigated in clinical trials. We herein review the current utility of biomarkers in the management of patients with CRLM.
2. Tumor Morphology
Tumor morphology provides significant prognostic information and guides clinical decision-making. The size, number of lesions, and proximity to vascular structures determine resectability, and margin-negative resection is an important factor associated with long-term survival. Furthermore, lesion size, depth from the periphery, and vessel proximity inform combined surgical/ablation approaches to provide definitive management and spare liver parenchyma in the case that future hepatectomy may be needed to clear recurrent disease [16,17]. Second, morphology-based scoring systems demonstrate a prognostic discriminatory ability for long-term survival outcomes (Table 1) [18,19,20,21]. An earlier scoring system developed by Fong et al. included the number of lesions (>1) and size of the largest lesion (>5 cm) among five clinical or morphologic factors and successfully prognosticated survival after hepatectomy [18]. A recent study by Sasaki et al. developed a Tumor Burden Score (TBS) using only the number of lesions and maximum tumor size based on similar studies that included patients with hepatocellular carcinoma [22]. External validation on two cohorts demonstrated accurate stratification of 5-year survival based on the TBS, with higher scores correlating with lower survival among patients undergoing hepatectomy for CRLM. Peng et al. applied the TBS to patients with unresectable CRLM and reported that the TBS successfully predicted conversion to resectable disease [23].

Table 1.
Clinical risk scores.
Radiographic response rates of CRLM while on systemic or liver-directed therapy have been correlated with improved disease-free and overall survival among patients with resectable and unresectable diseases [25]. Patients with a complete radiographic response (i.e., disappearing liver metastases) after therapy that remains undetectable after intraoperative ultrasound have a pathologic complete response (pCR) in 24% to 96% (median 77.5%) of cases [26]. Furthermore, patients with the unresectable disease who converted to resectable disease after therapy demonstrated improved survival compared with patients who did not demonstrate a radiographic response and remained unresectable [27]. Histological growth patterns of CRLM evaluated after resection also provide a prognostic value regarding overall survival [11,28].
In response to the growing amount of clinical information generated from the digitalization of healthcare, investigators have explored artificial intelligence (AI)-based techniques, such as machine learning. While not a biomarker itself, AI optimizes data utility by considering all available data in an unbiased and unsupervised manner with the ability to continuously improve predictions [29]. A novel machine learning approach was used to predict long-term outcomes in 1406 patients with CRLM that underwent hepatic resection [30]. The machine-learning generated model based on clinical and morphological tumor characteristics significantly outperformed traditional clinical risk scores for predicting 1-, 3-, and 5-year recurrence.
Tumor morphology has some significant limitations as a “biomarker”. Tumor morphology provides incomplete information on resectability. For example, specific criteria and thresholds to define resectability vary among institutions, sometimes based more on surgeon expertise than morphologic characteristics. In addition, tumor morphology offers little information to identify the 10% to 15% of patients with a resectable disease who undergo a resection yet develop early recurrence and cancer-related death [31]. While changes in tumor morphology based on serial imaging provide valuable information about the response to therapy during treatment, it poorly predicts response to systemic therapy prior to the initiation of treatment, let alone determine which systemic therapy may provide the best outcome for a particular patient.
3. Molecular Biomarkers
In 1988, Vogelstein et al. published a seminal report characterizing mutations related to KRAS, APC, and TP53 at various stages of CRC carcinogenesis, hypothesizing that invasive carcinoma develops from adenomatous polyps via the sequential acquisition of somatic mutations in multiple genes [32]. Since this foundational work, the development/invention of NGS technology has dramatically advanced the field of oncogenesis with techniques such as whole-genome sequencing, whole-exome sequencing, and targeted sequencing that have revealed a host of genomic alterations [33]. Figure 1 illustrates the interconnected relationships between multiple signaling cascades critical in CRC oncogenesis, highlighting two frequent mutation genes, the tumor suppressor gene TP53 (pathway A) and proto-oncogene PI3K (pathway B).
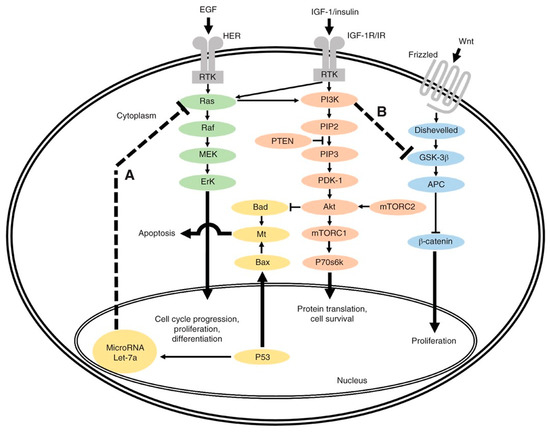
Figure 1.
Overview of interlinked cellular signaling pathways involved in the proliferation and progression of colorectal cancer. In pathway A, TP53 normally inhibits activated RAS through lethal (Let) 7a23. However, Let-7a is not able to regulate activated RAS if TP53 is mutated. In pathway B, overactive phosphoinositide 3-kinase (PI3K), an oncogene, inhibits glycogen synthase kinase (GSK) 3β24, leading to β-catenin accumulation. EGF, epidermal growth factor; HER, human epidermal growth factor receptor; RTK, receptor tyrosine kinase; MEK, mitogen-activated protein kinase; ErK, extracellular signal-regulated kinase; Mt, mitochondria; IGF-1, insulin-like growth factor 1; IGF-1R/IR, IGF-1 receptor/insulin receptor; PIP2, phosphatidylinositol 4,5-bisphosphate; PTEN, phosphatase and tensin homologue; PIP3, phosphatidylinositol (3,4,5)-trisphosphate; PDK-1, phosphoinositide-dependent protein kinase 1; mTORC1/2, mammalian target of rapamycin complex 1/2; P70s6k, P70s6 kinase; APC, adenomatous polyposis coli. Used with permission [34].
The CRC-liver metastatic cascade is a complex process wherein a subset of CRC cells acquires the capacity to evade the primary tumor, migrate through the extracellular matrix and neighboring tissue, intravasate, survive transit through the circulation, extravasate, and ultimately colonize the liver [35]. The molecular alterations required to complete this process create a biologically aggressive and phenotypically distinct disease entity.
Genomic alterations do not always correspond to predictable changes in biological activity [36]. Proteomics provides clarity to the molecular processes occurring in the gap between gene expression and disease phenotype. Recent proteomic profiling studies revealed unique protein and post-translational modifications present in metastatic CRC tumors, especially in proteins associated with the extracellular matrix, energy metabolism, and immune-cell-related migration [37,38,39]. In the following section, we discuss significant genomic, proteomic, and post-translational mutations in CRLM development.
3.1. Genomic Biomarkers
Genome-wide sequencing studies have demonstrated actionable mutations in one-third of patients with CRC metastasis (Figure 2) [40,41]. The current National Comprehensive Cancer Network (NCCN) guidelines recommend testing for RAS (KRAS and NRAS) and BRAF mutations and HER2 amplifications, individually or as part of an NGS panel, for all patients with metastatic CRC and universal MMR or microsatellite instability (MSI) testing in all newly diagnosed patients with CRC [8].
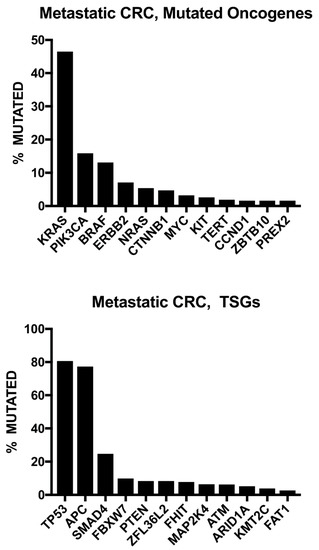
Figure 2.
Frequent gene alterations based on wide-genome sequencing of 372 metastatic colorectal cancer (CRC) patients. Note: TSGs: tumor suppressor genes. Open Access Citation: Testa U, Castelli G, Pelosi E. Genetic Alterations of Metastatic Colorectal Cancer. Biomedicines. 2020; 8(10):414. https://doi.org/10.3390/biomedicines8100414 [42].
Alterations in the RAS proto-oncogene family, most notably KRAS, NRAS, and HRAS, result in unregulated cell proliferation via gain-of-function activity in the MAPK pathway. RAS mutations are found in up to 52% of patients with CRLM and have a high concordance between the primary tumor and CRLM [43,44]. Patients with RAS-mutated CRLM have significantly worse recurrence-free and overall survival versus individuals with wild-type RAS [45,46]. Due to the upregulation of the MAPK pathway, patients with RAS mutations quickly develop resistance to epidermal growth factor receptor (EGFR) antibody therapy [47,48]. As a result, anti-EGFR antibody agents, cetuximab and panitumumab, are only recommended in the treatment of KRAS and NRAS wild-type tumors [8]. Furthermore, RAS-mutated CRLM demonstrate more migratory or invasive biology, causing local tumor progression and a higher incidence of micrometastasis compared with wild-type RAS CRLM [49,50,51]. Consequently, these tumors have a narrower median negative margin (4 mm vs. 7 mm) and double the rate of positive surgical margins after hepatectomy [52]. Similar results are seen for smaller RAS-mutated CRLM treated with ablation therapy [34]. The appropriate negative margin distance for patients with CRLM remains a controversial topic [53]. Where patients with wild-type RAS CRLM benefited from negative surgical margins, patients with RAS-mutated CRLM had similarly poor outcomes between the R0 and R1 resection groups [54,55]. A recent analysis of 1843 patients with CRLM who underwent curative-intent surgery used AI-based analytics to determine the optimal surgical margin in KRAS-variant CRLM [56]. The AI model suggested an optimal margin width of 7 mm for KRAS-variant CRLM. Most of the associated prolongation of survival was seen with a 1 mm margin, with the extension from 1 mm to 7 mm contributing a smaller proportion of the improvement in survival. Ultimately, patients with RAS-mutated tumors should demonstrate disease stability on systemic therapy, with the absence of other poor prognostic factors, prior to attempting curative-intent surgical resection.
BRAF, another protein in the MAPK pathway, has emerged as a very poor prognostic indicator [57]. BRAF mutations occur in 5% of patients with CRLM. BRAFV600E contains a substitution of valine for glutamic acid at codon 600 and is responsible for over 90% of these mutations [58]. Patients with BRAF-mutated CRC rarely present with isolated liver metastases. Furthermore, even in the small proportion of patients who present with resectable disease, median recurrence-free survival and overall survival after curative-intent hepatectomy is half compared to wild-type BRAF patients [59]. Similar to RAS-mutated CRC, patients with a BRAF mutation do not respond to anti-EGFR therapy unless administered with a BRAF inhibitor (i.e., encorafenib) [60]. Although data are limited to small studies, patients with non- BRAFV600E mutations may have better outcomes compared with even wild-type BRAF patients [61].
HER2 amplification is a targetable variant in the MAPK pathway found in 2–3% of metastatic CRCs [62]. While well-studied in breast cancer, the low prevalence of HER2 amplification in CRC cases limits confident statements about the prognostic effect of HER2 amplification. Regarding targeted therapies, phase 2 trials support the use of a dual HER2 blockade in heavily pretreated, HER2-amplified metastatic CRC [63,64]. However, anti-HER2 therapy is only indicated in HER2-amplified tumors that are also RAS and BRAF wild-type.
Approximately 5% of patients with metastatic CRC harbor deficient DNA mismatch repair (dMMR), leading to MSI [34]. Alterations in the MMR system arise through germline mutations (i.e., Lynch syndrome) or sporadically from promoter hypermethylation and silencing of the MMR gene MLH1 [65]. Sporadic mutations are highly associated with BRAFV600E mutations, present in one-third of dMMR patients, and have a worse prognosis versus patients with a germline dMMR [66]. In early-stage CRC, dMMR is associated with reduced metastatic potential and a favorable prognosis; however, dMMR status in metastatic CRC results in a worse prognosis compared with proficient MMR tumors [67]. Metastatic dMMR/MSI-H CRC responds well to immune checkpoint inhibitor (ICI) therapy, doubling the progression-free survival time compared with traditional chemotherapy (16.5 months vs. 8.2 months; HR 0.60, p = 0.0002) [68]. In addition, patients with metastatic CRC can have a high tumor mutation burden leading to MSI independent of the dMMR status. Patients with a high tumor mutational burden may also benefit from ICI therapy [69].
Unfortunately, most of the genomic variants identified in metastatic CRC lack a targeted therapy, including four of the five most mutated genes: APC, TP53, PIK3CA, and SMAD4. However, knowing the status of these mutations does provide prognostic information. Concomitant TP53 and RAS mutations have negative prognostic effects worse than either mutation alone. Specifically, patients with CRLM containing both mutations have significantly worse recurrence-free survival and overall survival [70]. A similar pattern emerges for patients with tumors containing both an APC and PIK3CA mutation. Together, these mutations have a synergistic effect on chemoresistance and, as a result, a worse prognosis [71,72]. Not surprisingly, tumors containing both a tumor suppressor mutation (TP53, APC) and an oncogene (KRAS, PIK3CA) are more biologically aggressive, leading to earlier disease recurrence and mortality.
3.2. Proteomic Biomarkers
Early proteomic studies on CRLM revealed diverse proteomic profiles demonstrating significant metastatic CRC tumor heterogeneity [73]. Current investigations remain in the preclinical exploratory phase, but these studies have identified promising targets for future research. In particular, studies comparing the proteome biology of primary CRC and CRLM have revealed distinct protein dysregulation profiles [37,38,74]. Fahrner et al. compared the matched proteomes of seven patients with CRLM and noted upregulated proteins in liver metastasis involving metabolic processes such as gluconeogenesis (pyruvate carboxylase) and fructose metabolism (fructose-bisphosphate aldolase B (ALDOB), fructose-1,6-bisphosphatase (1), as well as proteins linked to the complement cascade, indicating an active immune response) [37]. ALDOB upregulation indicates a poor prognosis and has been demonstrated to promote tumor progression and CRLM by facilitating the epithelial-mesenchymal transition [75]. Conversely, comparatively downregulated proteins are involved with cellular structural integrity/cell junction assembly (desmin, synemin, and filamin-C). In a separate study of eight patients with CRLM, several extracellular matrix components were significantly upregulated in CRLM compared with the primary tumor, most notably THBS1, which facilitates CRLM by promoting epithelial–mesenchymal transition [38]. Furthermore, THBS1 upregulation was associated with shorter overall survival. CD11b and ITGA2 are two additional proteins with a role in promoting the epithelial–mesenchymal transition that are overexpressed in CRLM [76]. In addition, Ku et al. demonstrated that the filamin A-interacting protein 1-like (FILIP1L) and plasminogen were dysregulated in CRLM compared with primary CRC [74]. FILIP1L overexpression inhibits WNT signaling and decreases metastatic behavior in CRC [77]. Ku et al. reported the downregulation of FILIP1L in CRLM samples. In contrast, plasminogen has been noted to be overexpressed in CRLM samples. Overexpression of the plasminogen activating system in CRLM has been associated with worse overall and cancer-specific survival [78].
The extracellular matrix (ECM) provides physical scaffolding for cells and plays a vital role in biochemical signaling [79]. Alterations in the composition of the ECM are linked to pathological conditions, including carcinogenesis and metastasis. The ECM in cancer patients is often disorganized and characterized by the upregulation of many components (e.g., collagen) compared with normal tissue [80]. Naba et al. performed the first proteomic study of the ECM composition in patients with CRLM, demonstrating that the ECM composition of CRLM resembled the ECM of primary CRC more so than a normal liver [81]. Furthermore, seven proteins were uniquely associated with metastases, including TIMP1 (Tissue Inhibitor of Metalloproteinase-1), a protein that induces pro-invasive ECM remodeling when upregulated and has been associated with poor progression-free survival in patients with CRLM [82]. In addition, TIMP1 may have utility as a non-invasive biomarker for preoperative risk stratification in patients with CRLM [82].
Protein post-translational modifications (PTMs) such as crosslinking, acetylation, ubiquitination, methylation, glycosylation, and citrullination can occur on histone and nonhistone proteins. These modifications often contribute to protein degradation to maintain normal physiological homeostasis; however, the deregulation of PTMs supports carcinogenesis and the development of metastasis. Shen et al. constructed a complete atlas of differentially expressed acetylated proteins in primary CRC and paired CRLM [83]. The authors identified 71 acetylated sites on 55 proteins in CRLM. These proteins were found primarily in the cytoplasm and associated with a broad range of biological processes, including metabolic pathways and carbon metabolism. Of the acetylated nonhistones, TPM2 and K152Ac were the most upregulated, and ADH1B K331Ac was the most downregulated. In another PTM study, Yuzhalin et al. demonstrated that peptidylarginine deiminase 4 (PAD4)-dependent protein citrullination plays an integral role in the development of CRLM [84]. Citrullination is characterized by a post-translational conversion of arginine residues to citrulline. In the study, CRLM exhibited significantly higher levels of PAD4 and citrullination compared to both healthy livers and the primary CRC. Furthermore, citrullination may facilitate CRLM by promoting the epithelial–mesenchymal transition since citrullination of collagen type 1 in the ECM promoted greater adhesion, decreased migration of CRC cells, and increased expression of epithelial markers.
Recently, proteomics has played a central role in the multiomic evaluation of CRLM. Multiomics provides a more comprehensive evaluation of biological processes by integrating data generated from multiple omic analyses (e.g., proteomic, transcriptomic, and genomic) [85]. A recent, large multiomic study integrated genomics, proteomics, and phosphoproteomics to provide a global evaluation of 146 patients with CRC, including 43 patients with CRLM [86]. CRLM were genomically similar to primary CRC but exhibited significant heterogeneity at the proteomic and post-translational level. In addition, in vivo drug testing suggested that phosphoproteomic profiling may more accurately predict drug response to kinase inhibitors than the presence of genomic mutations. Multiomic studies have also demonstrated a correlation with survival [87,88]. Two separate studies by Ma et al. reported that the mutated peptide number had prognostic value and that somatic variants demonstrated corresponding dysregulated protein abundance and biologic function. Relevant variants identified in CRLM included UQCR5, FDFT1, MYH9, and CCT6A.
3.3. Liquid Biopsy and ctDNA
A core tumor biopsy represents the current gold standard for obtaining tissue samples for molecular analyses. Core biopsies can be fraught with technical complications and fail to accurately capture complete tumor spatial heterogeneity and tumor evolution. [89,90]. Liquid biopsy collects body fluid (e.g., blood, urine, saliva) for molecular evaluation and may overcome many of the limitations associated with core biopsy [91]. Patients with solid tumors release isolated tumor DNA fragments into the blood after tumor lysis or apoptosis, called circulating tumor DNA (ctDNA). ctDNA is distinct from normal circulating-free DNA (cfDNA) and contains specific pathologic genetic variants [92]. CRCs tend to shed high amounts of ctDNA relative to other solid tumors, making them an ideal candidate for further investigation [93].
Investigation into the clinical utility of ctDNA as a biomarker for patients with CRC is ongoing (Table 2). Surgical resection of CRLM remains the standard of care, even for patients who have a clinical complete response (cCR) to neoadjuvant therapy, as available biomarkers cannot accurately identify patients who have achieved a pCR [26]. Studies specifically evaluating ctDNA in the neoadjuvant setting among patients with CRLM are limited; however, in retrospective studies of patients with stage II-III CRC, the ctDNA status following neoadjuvant therapy was associated with the pCR status [94,95], disease-free survival, and overall survival [96,97]. Furthermore, sequential sampling before, during, and after therapy allows for the real-time monitoring of the treatment response and disease evolution. Detection of ctDNA during neoadjuvant therapy correlated with primary tumor regression and shorter metastasis-free survival [98].

Table 2.
Notable clinical trials of ctDNA in patients with metastatic colorectal cancer.
In retrospective studies, ctDNA demonstrates a prognostic value after hepatectomy for patients with CRLM. In a single institution study of 63 patients with CRLM, 42 (67%) had ctDNA present after hepatectomy. These patients had significantly worse overall survival, especially when multiple gene mutations were detected [100]. Similar results were demonstrated by Tie et al. in a cohort of 49 patients with resectable CRLM who underwent curative-intent hepatectomy [101]. The 11 patients with positive ctDNA after resection had lower recurrence-free survival and overall survival. However, ctDNA clearance was achieved with adjuvant therapy in three patients, two of whom remained disease free. Patients with positive ctDNA at the completion of therapy (surgery +/− adjuvant chemotherapy) had a 5-year recurrence-free survival of 0% versus 75.6% for patients with undetectable ctDNA after therapy.
Among patients with unresectable metastatic CRC, ctDNA-derived mutational analyses were nearly 100% concordant with the solid tissue biopsies for the detection of multiple clinically relevant mutations, such as the BRAFV600E, KRAS, and NRAS mutations [102,103]. Of note, intra-tumor heterogeneity, the treatment effect, or a low disease burden were responsible for the few discordant results. Furthermore, recent phase II clinical trials demonstrate the efficacy of ctDNA-guided rechallenge therapy with anti-EGFR drugs in the RAS wild-type unresectable metastatic CRC. Sartore-Bianchi et al. evaluated 27 patients who were ctDNA negative for the RAS/BRAF/EGFR mutations. Rechallenge therapy with panitumumab was associated with disease control in 59% of patients with a median progression-free survival of 16 weeks [104]. A separate study by Martinelli et al. included 77 patients with unresectable metastatic CRC that failed second-line therapy after acquiring resistance to first-line chemotherapy plus anti-EGFR drugs. Among the 48 patients with RAS/BRAF wild-type ctDNA, rechallenge therapy with cetuximab plus avelumab improved the median overall survival [105].
Despite the growing evidence supporting the utility of ctDNA, its widespread incorporation into clinical decision-making algorithms has yet to occur due to some notable limitations [106]. While patients with CRCs tend to shed higher levels of ctDNA compared with other cancer types, the concentration of ctDNA is still low, especially earlier in the disease process and compared with cfDNA. Highly sensitive tests are required to detect ctDNA accurately. Furthermore, the ratio of ctDNA to cfDNA (ctDNA fraction) significantly influences assay sensitivity and specificity. As a result, variation in the ctDNA fraction leads to unclear thresholds for the limit of detection [89,92]. Finally, currently available evidence supporting the use of ctDNA is overwhelmingly retrospective. Results from active randomized controlled trials will hopefully provide clarity on the clinical utility of ctDNA in patients with CRLM.
4. Radiomics
Radiomics refers to the quantitative analysis of textual features in medical images using data-characterization algorithms [107,108]. In theory, by examining the distinct radiomic features (e.g., shape, intensity, sphericity) in a region of interest, radiomic-based approaches extract biological data directly from the medical images, providing timely diagnostic and prognostic information while avoiding the cost and morbidity of an invasive biopsy [109].
Lubner et al. reported one of the first studies evaluating the clinical utility of radiomic features in patients with CRLM [110] The authors examined CT scans in 77 patients with a single untreated CRLM and noted an association between the radiomic features and tumor grade, KRAS mutation, and overall survival. Radiomic-based approaches have also been used to predict the effectiveness of therapy. In two recent studies, radiomic signatures successfully predicted treatment sensitivity to EGFR-targeted therapy [111]. and FOLFIRI + bevacizumab [112], outperforming known biomarkers such as tumor shrinkage as determined by RECIST. Furthermore, radiomic signatures were significantly associated with overall survival in both studies. In another study, a CT-based radiomics model outperformed a clinical model to detect local tumor progression in 31 lesions after thermal ablation [113]. In addition to predicting the therapy response, radiomics may improve the diagnostic accuracy of chemotherapy-associated liver injury (CALI). In a retrospective study of 78 consecutive patients with CRLM that received preoperative chemotherapy followed by liver resection, 66 demonstrated some degree of CALI on the final pathology [114]. Multiple radiomic features were associated with CALI, and, combined with relevant clinical data, radiomic signatures significantly improved the diagnostic accuracy of CALI versus traditional models.
AI techniques may optimize the predictions of radiomic-based analyses given the abundance of data extracted from medical images during the process. In a retrospective study of 91 patients with CRC, machine learning-derived models were used to predict the occurrence of metachronous CRLM [115]. The model, trained on only radiomic features, significantly outperformed the model that used only clinical features (AUC 85% [95%CI 85–87%] vs. 71% [95%CI 69–72%]). In another retrospective study, a deep learning-based radiomics model better-predicted response to chemotherapy in 192 patients with CRLM compared with the traditional classifier-based radiomics model [116]. In another study of 199 patients with CRC and 550 small hypoattenuating hepatic nodules (<1 cm), convolutional neural networks (CNNs) characterized lesions as benign or malignant with similar accuracy as expert radiologists, yet with greater diagnostic confidence (the CNN had a lower rate of nodules reported with low confidence, 19.6 vs. 31.4 (expert average); p < 0.0001).
Although radiomics has shown potential as a non-invasive biomarker for patients with CRLM, several challenges should be mentioned. While radiomic features have been standardized [117], significant heterogeneity exists between the number and order of feature analysis, and nearly all institutions use a different software application [109], Furthermore, when reported, radiomic feature thresholds vary between studies, limiting reproducibility and generalizability. Few radiomic studies evaluating CRLM have a prospective design or include a validation dataset. Despite these limitations, radiomic features demonstrate diagnostic and prognostic value for patients with CRLM in a timely and non-invasive manner.
5. Conclusions
CRLM continues to be a major source of morbidity and mortality for patients, despite a multitude of therapeutic modalities. However, NGS techniques and advances in mass spectrometry have improved our understanding of the genomic, proteomic, and post-translational variants responsible for CRLM development, revealing many potential biomarkers. Furthermore, the application of AI technology and, more specifically, machine learning algorithms provides an unbiased method to integrate the immense amount of molecular and radiomic data being generated to assist clinicians with developing personalized treatment plans. The many active trials evaluating the clinical utility of ctDNA as a less invasive biomarker are expected to provide clarity on the diagnostic and prognostic role of ctDNA in the management of patients with CRLM. While much work remains to optimize treatment plans, recent advances in the field provide hope for improving outcomes for patients with CRLM.
Author Contributions
Conceptualization, D.B.H. and T.M.P.; methodology, D.B.H.; validation, D.B.H. and T.M.P.; investigation, D.B.H. and Z.J.B.; resources, T.M.P.; writing—original draft preparation, D.B.H.; writing—review and editing, D.B.H., Z.J.B. and T.M.P.; supervision, T.M.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Xi, Y.; Xu, P. Global colorectal cancer burden in 2020 and projections to 2040. Transl. Oncol. 2021, 14, 101174. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Tsai, M.S.; Su, Y.H.; Ho, M.C.; Liang, J.T.; Chen, T.P.; Lai, H.S.; Lee, P.H. Clinicopathological features and prognosis in resectable synchronous and metachronous colorectal liver metastasis. Ann. Surg. Oncol. 2007, 14, 786–794. [Google Scholar] [CrossRef]
- Van Cutsem, E.; Nordlinger, B.; Adam, R.; Köhne, C.H.; Pozzo, C.; Poston, G.; Ychou, M.; Rougier, P.; Group, E.C.M.T. Towards a pan-European consensus on the treatment of patients with colorectal liver metastases. Eur. J. Cancer 2006, 42, 2212–2221. [Google Scholar] [CrossRef]
- Yoo, P.S.; Lopez-Soler, R.I.; Longo, W.E.; Cha, C.H. Liver resection for metastatic colorectal cancer in the age of neoadjuvant chemotherapy and bevacizumab. Clin. Colorectal Cancer 2006, 6, 202–207. [Google Scholar] [CrossRef]
- Stangl, R.; Altendorf-Hofmann, A.; Charnley, R.M.; Scheele, J. Factors influencing the natural history of colorectal liver metastases. Lancet 1994, 343, 1405–1410. [Google Scholar] [CrossRef]
- National Comprehensive Cancer Network. NCCN Guildeline: Colon Cancer (Version 1.2022); NCCN: Philadelphia, PA, USA, 2022. [Google Scholar]
- Van Cutsem, E.; Cervantes, A.; Adam, R.; Sobrero, A.; Van Krieken, J.H.; Aderka, D.; Aranda Aguilar, E.; Bardelli, A.; Benson, A.; Bodoky, G.; et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann. Oncol. 2016, 27, 1386–1422. [Google Scholar] [CrossRef]
- Jarnagin, W.R.; Gonen, M.; Fong, Y.; DeMatteo, R.P.; Ben-Porat, L.; Little, S.; Corvera, C.; Weber, S.; Blumgart, L.H. Improvement in perioperative outcome after hepatic resection: Analysis of 1,803 consecutive cases over the past decade. Ann. Surg. 2002, 236, 397–406; discussion 397–406. [Google Scholar] [CrossRef] [PubMed]
- Buisman, F.E.; Giardiello, D.; Kemeny, N.E.; Steyerberg, E.W.; Höppener, D.J.; Galjart, B.; Nierop, P.M.H.; Balachandran, V.P.; Cercek, A.; Drebin, J.A.; et al. Predicting 10-year survival after resection of colorectal liver metastases; an international study including biomarkers and perioperative treatment. Eur. J. Cancer 2022, 168, 25–33. [Google Scholar] [CrossRef]
- Kanas, G.P.; Taylor, A.; Primrose, J.N.; Langeberg, W.J.; Kelsh, M.A.; Mowat, F.S.; Alexander, D.D.; Choti, M.A.; Poston, G. Survival after liver resection in metastatic colorectal cancer: Review and meta-analysis of prognostic factors. Clin. Epidemiol. 2012, 4, 283–301. [Google Scholar] [CrossRef] [PubMed]
- Hadden, W.J.; de Reuver, P.R.; Brown, K.; Mittal, A.; Samra, J.S.; Hugh, T.J. Resection of colorectal liver metastases and extra-hepatic disease: A systematic review and proportional meta-analysis of survival outcomes. HPB 2016, 18, 209–220. [Google Scholar] [CrossRef] [PubMed]
- De Jong, M.C.; Pulitano, C.; Ribero, D.; Strub, J.; Mentha, G.; Schulick, R.D.; Choti, M.A.; Aldrighetti, L.; Capussotti, L.; Pawlik, T.M. Rates and patterns of recurrence following curative intent surgery for colorectal liver metastasis: An international multi-institutional analysis of 1669 patients. Ann. Surg. 2009, 250, 440–448. [Google Scholar] [CrossRef]
- Ignatavicius, P.; Oberkofler, C.E.; Chapman, W.C.; DeMatteo, R.P.; Clary, B.M.; D’Angelica, M.I.; Tanabe, K.K.; Hong, J.C.; Aloia, T.A.; Pawlik, T.M.; et al. Choices of Therapeutic Strategies for Colorectal Liver Metastases Among Expert Liver Surgeons: A Throw of the Dice? Ann. Surg. 2020, 272, 715–722. [Google Scholar] [CrossRef] [PubMed]
- Elfrink, A.K.E.; Nieuwenhuizen, S.; van den Tol, M.P.; Burgmans, M.C.; Prevoo, W.; Coolsen, M.M.E.; van den Boezem, P.B.; van Delden, O.M.; Hagendoorn, J.; Patijn, G.A.; et al. Hospital variation in combined liver resection and thermal ablation for colorectal liver metastases and impact on short-term postoperative outcomes: A nationwide population-based study. HPB 2021, 23, 827–839. [Google Scholar] [CrossRef] [PubMed]
- Deng, G.; Li, H.; Jia, G.Q.; Fang, D.; Tang, Y.Y.; Xie, J.; Chen, K.F.; Chen, Z.Y. Parenchymal-sparing versus extended hepatectomy for colorectal liver metastases: A systematic review and meta-analysis. Cancer Med. 2019, 8, 6165–6175. [Google Scholar] [CrossRef]
- Fong, Y.; Fortner, J.; Sun, R.L.; Brennan, M.F.; Blumgart, L.H. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: Analysis of 1001 consecutive cases. Ann. Surg. 1999, 230, 309–318; discussion 318–321. [Google Scholar] [CrossRef]
- Nordlinger, B.; Guiguet, M.; Vaillant, J.C.; Balladur, P.; Boudjema, K.; Bachellier, P.; Jaeck, D. Surgical resection of colorectal carcinoma metastases to the liver. A prognostic scoring system to improve case selection, based on 1568 patients. Association Française de Chirurgie. Cancer 1996, 77, 1254–1262. [Google Scholar] [CrossRef]
- Nagashima, I.; Takada, T.; Adachi, M.; Nagawa, H.; Muto, T.; Okinaga, K. Proposal of criteria to select candidates with colorectal liver metastases for hepatic resection: Comparison of our scoring system to the positive number of risk factors. World J. Gastroenterol. 2006, 12, 6305–6309. [Google Scholar] [CrossRef]
- Konopke, R.; Kersting, S.; Distler, M.; Dietrich, J.; Gastmeier, J.; Heller, A.; Kulisch, E.; Saeger, H.D. Prognostic factors and evaluation of a clinical score for predicting survival after resection of colorectal liver metastases. Liver Int. 2009, 29, 89–102. [Google Scholar] [CrossRef]
- Sasaki, K.; Morioka, D.; Conci, S.; Margonis, G.A.; Sawada, Y.; Ruzzenente, A.; Kumamoto, T.; Iacono, C.; Andreatos, N.; Guglielmi, A.; et al. The Tumor Burden Score: A New “Metro-ticket” Prognostic Tool For Colorectal Liver Metastases Based on Tumor Size and Number of Tumors. Ann. Surg. 2018, 267, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Liu, Y.; Li, W.; Lin, Y.; Sun, H.; Pan, Z.; Wu, X.; Fan, W.; Lin, J. Application of Tumor Burden Score for predicting conversion outcome in patients with initially unresectable colorectal liver metastases after first-line systemic therapy. Therap. Adv. Gastroenterol. 2021, 14, 17562848211066206. [Google Scholar] [CrossRef] [PubMed]
- Wimmer, K.; Schwarz, C.; Szabo, C.; Bodingbauer, M.; Tamandl, D.; Mittlbock, M.; Kaczirek, K. Impact of Neoadjuvant Chemotherapy on Clinical Risk Scores and Survival in Patients with Colorectal Liver Metastases. Ann. Surg. Oncol. 2017, 24, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Shindoh, J.; Loyer, E.M.; Kopetz, S.; Boonsirikamchai, P.; Maru, D.M.; Chun, Y.S.; Zimmitti, G.; Curley, S.A.; Charnsangavej, C.; Aloia, T.A.; et al. Optimal morphologic response to preoperative chemotherapy: An alternate outcome end point before resection of hepatic colorectal metastases. J. Clin. Oncol. 2012, 30, 4566–4572. [Google Scholar] [CrossRef] [PubMed]
- Barimani, D.; Kauppila, J.H.; Sturesson, C.; Sparrelid, E. Imaging in disappearing colorectal liver metastases and their accuracy: A systematic review. World J. Surg. Oncol. 2020, 18, 264. [Google Scholar] [CrossRef] [PubMed]
- Basso, M.; Dadduzio, V.; Ardito, F.; Lombardi, P.; Strippoli, A.; Vellone, M.; Orlandi, A.; Rossi, S.; Cerchiaro, E.; Cassano, A.; et al. Conversion Chemotherapy for Technically Unresectable Colorectal Liver Metastases: A Retrospective, STROBE-Compliant, Single-Center Study Comparing Chemotherapy Alone and Combination Chemotherapy With Cetuximab or Bevacizumab. Medicine 2016, 95, e3722. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, K.; Rolff, H.C.; Eefsen, R.L.; Vainer, B. The morphological growth patterns of colorectal liver metastases are prognostic for overall survival. Mod. Pathol. 2014, 27, 1641–1648. [Google Scholar] [CrossRef]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef]
- Paredes, A.Z.; Hyer, J.M.; Tsilimigras, D.I.; Moro, A.; Bagante, F.; Guglielmi, A.; Ruzzenente, A.; Alexandrescu, S.; Makris, E.A.; Poultsides, G.A.; et al. A Novel Machine-Learning Approach to Predict Recurrence After Resection of Colorectal Liver Metastases. Ann. Surg. Oncol. 2020, 27, 5139–5147. [Google Scholar] [CrossRef]
- Viganò, L.; Gentile, D.; Galvanin, J.; Corleone, P.; Costa, G.; Cimino, M.; Procopio, F.; Torzilli, G. Very Early Recurrence After Liver Resection for Colorectal Metastases: Incidence, Risk Factors, and Prognostic Impact. J. Gastrointest Surg. 2022, 26, 570–582. [Google Scholar] [CrossRef]
- Vogelstein, B.; Fearon, E.R.; Hamilton, S.R.; Kern, S.E.; Preisinger, A.C.; Leppert, M.; Nakamura, Y.; White, R.; Smits, A.M.; Bos, J.L. Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 1988, 319, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Del Vecchio, F.; Mastroiaco, V.; Di Marco, A.; Compagnoni, C.; Capece, D.; Zazzeroni, F.; Capalbo, C.; Alesse, E.; Tessitore, A. Next-generation sequencing: Recent applications to the analysis of colorectal cancer. J. Transl. Med. 2017, 15, 246. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Chun, Y.S.; Kopetz, S.E.; Vauthey, J.N. Biomarkers in colorectal liver metastases. Br. J. Surg. 2018, 105, 618–627. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Liu, Z.; Wang, Y.; Wen, X.; Amador, E.H.; Yuan, L.; Ran, X.; Xiong, L.; Ran, Y.; Chen, W.; et al. Colorectal liver metastasis: Molecular mechanism and interventional therapy. Signal Transduct. Target Ther. 2022, 7, 70. [Google Scholar] [CrossRef]
- Black, D.L. Protein diversity from alternative splicing: A challenge for bioinformatics and post-genome biology. Cell 2000, 103, 367–370. [Google Scholar] [CrossRef]
- Fahrner, M.; Bronsert, P.; Fichtner-Feigl, S.; Jud, A.; Schilling, O. Proteome biology of primary colorectal carcinoma and corresponding liver metastases. Neoplasia. 2021, 23, 1240–1251. [Google Scholar] [CrossRef]
- Liu, X.; Xu, D.; Liu, Z.; Li, Y.; Zhang, C.; Gong, Y.; Jiang, Y.; Xing, B. THBS1 facilitates colorectal liver metastasis through enhancing epithelial-mesenchymal transition. Clin. Transl. Oncol. 2020, 22, 1730–1740. [Google Scholar] [CrossRef]
- Kim, E.K.; Song, M.J.; Jung, Y.; Lee, W.S.; Jang, H.H. Proteomic Analysis of Primary Colon Cancer and Synchronous Solitary Liver Metastasis. Cancer Genom. Proteom. 2019, 16, 583–592. [Google Scholar] [CrossRef]
- Zehir, A.; Benayed, R.; Shah, R.H.; Syed, A.; Middha, S.; Kim, H.R.; Srinivasan, P.; Gao, J.; Chakravarty, D.; Devlin, S.M.; et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017, 23, 703–713. [Google Scholar] [CrossRef]
- Priestley, P.; Baber, J.; Lolkema, M.P.; Steeghs, N.; de Bruijn, E.; Shale, C.; Duyvesteyn, K.; Haidari, S.; van Hoeck, A.; Onstenk, W.; et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature 2019, 575, 210–216. [Google Scholar] [CrossRef]
- Testa, U.; Castelli, G.; Pelosi, E. Genetic Alterations of Metastatic Colorectal Cancer. Biomedicines 2020, 8, 414. [Google Scholar] [CrossRef] [PubMed]
- Schirripa, M.; Bergamo, F.; Cremolini, C.; Casagrande, M.; Lonardi, S.; Aprile, G.; Yang, D.; Marmorino, F.; Pasquini, G.; Sensi, E.; et al. BRAF and RAS mutations as prognostic factors in metastatic colorectal cancer patients undergoing liver resection. Br. J. Cancer 2015, 112, 1921–1928. [Google Scholar] [CrossRef] [PubMed]
- Brannon, A.R.; Vakiani, E.; Sylvester, B.E.; Scott, S.N.; McDermott, G.; Shah, R.H.; Kania, K.; Viale, A.; Oschwald, D.M.; Vacic, V.; et al. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome. Biol. 2014, 15, 454. [Google Scholar] [CrossRef] [PubMed]
- Vauthey, J.N.; Zimmitti, G.; Kopetz, S.E.; Shindoh, J.; Chen, S.S.; Andreou, A.; Curley, S.A.; Aloia, T.A.; Maru, D.M. RAS mutation status predicts survival and patterns of recurrence in patients undergoing hepatectomy for colorectal liver metastases. Ann. Surg. 2013, 258, 619–626; discussion 617–626. [Google Scholar] [CrossRef]
- Connor, J.M.; Sanchez Loria, F.; Ardiles, V.; Grondona, J.; Sanchez, P.; Andriani, O.; Fauda, M.; Brancato, F.; Huertas, E.; Alvarez, F.; et al. Prognostic impact of K-RAS mutational status and primary tumor location in patients undergoing resection for colorectal cancer liver metastases: An update. Future Oncol. 2019, 15, 3149–3157. [Google Scholar] [CrossRef]
- Siena, S.; Sartore-Bianchi, A.; Di Nicolantonio, F.; Balfour, J.; Bardelli, A. Biomarkers predicting clinical outcome of epidermal growth factor receptor-targeted therapy in metastatic colorectal cancer. J. Natl. Cancer Inst. 2009, 101, 1308–1324. [Google Scholar] [CrossRef]
- Misale, S.; Yaeger, R.; Hobor, S.; Scala, E.; Janakiraman, M.; Liska, D.; Valtorta, E.; Schiavo, R.; Buscarino, M.; Siravegna, G.; et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012, 486, 532–536. [Google Scholar] [CrossRef]
- Serova, M.; Astorgues-Xerri, L.; Bieche, I.; Albert, S.; Vidaud, M.; Benhadji, K.A.; Emami, S.; Vidaud, D.; Hammel, P.; Theou-Anton, N.; et al. Epithelial-to-mesenchymal transition and oncogenic Ras expression in resistance to the protein kinase Cbeta inhibitor enzastaurin in colon cancer cells. Mol. Cancer Ther. 2010, 9, 1308–1317. [Google Scholar] [CrossRef]
- Odisio, B.C.; Yamashita, S.; Huang, S.Y.; Harmoush, S.; Kopetz, S.E.; Ahrar, K.; Shin Chun, Y.; Conrad, C.; Aloia, T.A.; Gupta, S.; et al. Local tumour progression after percutaneous ablation of colorectal liver metastases according to RAS mutation status. Br. J. Surg. 2017, 104, 760–768. [Google Scholar] [CrossRef]
- Zhang, Q.; Peng, J.; Ye, M.; Weng, W.; Tan, C.; Ni, S.; Huang, D.; Sheng, W.; Wang, L. KRAS Mutation Predicted More Mirometastases and Closer Resection Margins in Patients with Colorectal Cancer Liver Metastases. Ann. Surg. Oncol. 2020, 27, 1164–1173. [Google Scholar] [CrossRef] [PubMed]
- Brudvik, K.W.; Mise, Y.; Chung, M.H.; Chun, Y.S.; Kopetz, S.E.; Passot, G.; Conrad, C.; Maru, D.M.; Aloia, T.A.; Vauthey, J.N. RAS Mutation Predicts Positive Resection Margins and Narrower Resection Margins in Patients Undergoing Resection of Colorectal Liver Metastases. Ann. Surg. Oncol. 2016, 23, 2635–2643. [Google Scholar] [CrossRef]
- Margonis, G.A.; Sergentanis, T.N.; Ntanasis-Stathopoulos, I.; Andreatos, N.; Tzanninis, I.G.; Sasaki, K.; Psaltopoulou, T.; Wang, J.; Buettner, S.; Papalois, A.; et al. Impact of Surgical Margin Width on Recurrence and Overall Survival Following R0 Hepatic Resection of Colorectal Metastases: A Systematic Review and Meta-analysis. Ann. Surg. 2018, 267, 1047–1055. [Google Scholar] [CrossRef] [PubMed]
- Margonis, G.A.; Sasaki, K.; Andreatos, N.; Kim, Y.; Merath, K.; Wagner, D.; Wilson, A.; Buettner, S.; Amini, N.; Antoniou, E.; et al. KRAS Mutation Status Dictates Optimal Surgical Margin Width in Patients Undergoing Resection of Colorectal Liver Metastases. Ann. Surg. Oncol. 2017, 24, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Hatta, A.A.Z.; Pathanki, A.M.; Hodson, J.; Sutcliffe, R.P.; Marudanayagam, R.; Roberts, K.J.; Chatzizacharias, N.; Isaac, J.; Muiesan, P.; Taniere, P.; et al. The effects of resection margin and KRAS status on outcomes after resection of colorectal liver metastases. HPB 2021, 23, 90–98. [Google Scholar] [CrossRef]
- Bertsimas, D.; Margonis, G.A.; Sujichantararat, S.; Boerner, T.; Ma, Y.; Wang, J.; Kamphues, C.; Sasaki, K.; Tang, S.; Gagniere, J.; et al. Using Artificial Intelligence to Find the Optimal Margin Width in Hepatectomy for Colorectal Cancer Liver Metastases. JAMA Surg. 2022, 157, e221819. [Google Scholar] [CrossRef] [PubMed]
- Pikouli, A.; Papaconstantinou, D.; Wang, J.; Kavezou, F.; Pararas, N.; Nastos, C.; Pikoulis, E.; Margonis, G.A. Reevaluating the prognostic role of BRAF mutation in colorectal cancer liver metastases. Am. J. Surg. 2022, 223, 879–883. [Google Scholar] [CrossRef] [PubMed]
- Margonis, G.A.; Buettner, S.; Andreatos, N.; Kim, Y.; Wagner, D.; Sasaki, K.; Beer, A.; Schwarz, C.; Løes, I.M.; Smolle, M.; et al. Association of BRAF Mutations With Survival and Recurrence in Surgically Treated Patients With Metastatic Colorectal Liver Cancer. JAMA Surg. 2018, 153, e180996. [Google Scholar] [CrossRef]
- Gagnière, J.; Dupré, A.; Gholami, S.S.; Pezet, D.; Boerner, T.; Gönen, M.; Kingham, T.P.; Allen, P.J.; Balachandran, V.P.; De Matteo, R.P.; et al. Is Hepatectomy Justified for BRAF Mutant Colorectal Liver Metastases?: A Multi-institutional Analysis of 1497 Patients. Ann. Surg. 2020, 271, 147–154. [Google Scholar] [CrossRef]
- Pietrantonio, F.; Petrelli, F.; Coinu, A.; Di Bartolomeo, M.; Borgonovo, K.; Maggi, C.; Cabiddu, M.; Iacovelli, R.; Bossi, I.; Lonati, V.; et al. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: A meta-analysis. Eur. J. Cancer 2015, 51, 587–594. [Google Scholar] [CrossRef]
- Jones, J.C.; Renfro, L.A.; Al-Shamsi, H.O.; Schrock, A.B.; Rankin, A.; Zhang, B.Y.; Kasi, P.M.; Voss, J.S.; Leal, A.D.; Sun, J.; et al. BRAF Mutations Define a Clinically Distinct Molecular Subtype of Metastatic Colorectal Cancer. J. Clin. Oncol. 2017, 35, 2624–2630. [Google Scholar] [CrossRef]
- Ross, J.S.; Fakih, M.; Ali, S.M.; Elvin, J.A.; Schrock, A.B.B.; Suh, J.; Vergilio, J.A.; Ramkissoon, S.; Severson, E.; Daniel, S.; et al. Targeting HER2 in colorectal cancer: The landscape of amplification and short variant mutations in ERBB2 and ERBB3. Cancer 2018, 124, 1358–1373. [Google Scholar] [CrossRef] [PubMed]
- Meric-Bernstam, F.; Hurwitz, H.; Raghav, K.P.S.; McWilliams, R.R.; Fakih, M.; VanderWalde, A.; Swanton, C.; Kurzrock, R.; Burris, H.; Sweeney, C.; et al. Pertuzumab plus trastuzumab for HER2-amplified metastatic colorectal cancer (MyPathway): An updated report from a multicentre, open-label, phase 2a, multiple basket study. Lancet. Oncol. 2019, 20, 518–530. [Google Scholar] [CrossRef]
- Sartore-Bianchi, A.; Trusolino, L.; Martino, C.; Bencardino, K.; Lonardi, S.; Bergamo, F.; Zagonel, V.; Leone, F.; Depetris, I.; Martinelli, E.; et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): A proof-of-concept, multicentre, open-label, phase 2 trial. Lancet. Oncol. 2016, 17, 738–746. [Google Scholar] [CrossRef]
- Gelsomino, F.; Barbolini, M.; Spallanzani, A.; Pugliese, G.; Cascinu, S. The evolving role of microsatellite instability in colorectal cancer: A review. Cancer Treat. Rev. 2016, 51, 19–26. [Google Scholar] [CrossRef]
- Cohen, R.; Buhard, O.; Cervera, P.; Hain, E.; Dumont, S.; Bardier, A.; Bachet, J.B.; Gornet, J.M.; Lopez-Trabada, D.; Kaci, R.; et al. Clinical and molecular characterisation of hereditary and sporadic metastatic colorectal cancers harbouring microsatellite instability/DNA mismatch repair deficiency. Eur. J. Cancer 2017, 86, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.G.; Ahn, J.B.; Jung, M.; Beom, S.H.; Kim, C.; Kim, J.H.; Heo, S.J.; Park, H.S.; Kim, N.K.; Min, B.S.; et al. Effects of microsatellite instability on recurrence patterns and outcomes in colorectal cancers. Br. J. Cancer 2016, 115, 25–33. [Google Scholar] [CrossRef]
- André, T.; Shiu, K.K.; Kim, T.W.; Jensen, B.V.; Jensen, L.H.; Punt, C.; Smith, D.; Garcia-Carbonero, R.; Benavides, M.; Gibbs, P.; et al. Pembrolizumab in Microsatellite-Instability-High Advanced Colorectal Cancer. N. Engl. J. Med. 2020, 383, 2207–2218. [Google Scholar] [CrossRef]
- Schrock, A.B.; Ouyang, C.; Sandhu, J.; Sokol, E.; Jin, D.; Ross, J.S.; Miller, V.A.; Lim, D.; Amanam, I.; Chao, J.; et al. Tumor mutational burden is predictive of response to immune checkpoint inhibitors in MSI-high metastatic colorectal cancer. Ann. Oncol. 2019, 30, 1096–1103. [Google Scholar] [CrossRef]
- Chun, Y.S.; Passot, G.; Yamashita, S.; Nusrat, M.; Katsonis, P.; Loree, J.M.; Conrad, C.; Tzeng, C.D.; Xiao, L.; Aloia, T.A.; et al. Deleterious Effect of RAS and Evolutionary High-risk TP53 Double Mutation in Colorectal Liver Metastases. Ann. Surg. 2019, 269, 917–923. [Google Scholar] [CrossRef]
- Yamashita, S.; Chun, Y.S.; Kopetz, S.E.; Maru, D.; Conrad, C.; Aloia, T.A.; Vauthey, J.N. APC and PIK3CA Mutational Cooperativity Predicts Pathologic Response and Survival in Patients Undergoing Resection for Colorectal Liver Metastases. Ann. Surg. 2020, 272, 1080–1085. [Google Scholar] [CrossRef]
- Deming, D.A.; Leystra, A.A.; Nettekoven, L.; Sievers, C.; Miller, D.; Middlebrooks, M.; Clipson, L.; Albrecht, D.; Bacher, J.; Washington, M.K.; et al. PIK3CA and APC mutations are synergistic in the development of intestinal cancers. Oncogene 2014, 33, 2245–2254. [Google Scholar] [CrossRef] [PubMed]
- Wong, G.Y.M.; Diakos, C.; Hugh, T.J.; Molloy, M.P. Proteomic Profiling and Biomarker Discovery in Colorectal Liver Metastases. Int. J. Mol. Sci. 2022, 23, 6091. [Google Scholar] [CrossRef]
- Ku, X.; Xu, Y.; Cai, C.; Yang, Y.; Cui, L.; Yan, W. In-Depth Characterization of Mass Spectrometry-Based Proteomic Profiles Revealed Novel Signature Proteins Associated with Liver Metastatic Colorectal Cancers. Anal. Cell Pathol. 2019, 2019, 7653230. [Google Scholar] [CrossRef]
- Li, Q.; Li, Y.; Xu, J.; Wang, S.; Xu, Y.; Li, X.; Cai, S. Aldolase B Overexpression is Associated with Poor Prognosis and Promotes Tumor Progression by Epithelial-Mesenchymal Transition in Colorectal Adenocarcinoma. Cell Physiol. Biochem. 2017, 42, 397–406. [Google Scholar] [CrossRef]
- Yang, Q.; Bavi, P.; Wang, J.Y.; Roehrl, M.H. Immuno-proteomic discovery of tumor tissue autoantigens identifies olfactomedin 4, CD11b, and integrin alpha-2 as markers of colorectal cancer with liver metastases. J. Proteomics 2017, 168, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.; Lee, S.J.; Wang, Y.; Rybak, Y.; Luna, A.; Reddy, S.; Adem, A.; Beaty, B.T.; Condeelis, J.S.; Libutti, S.K. Filamin A interacting protein 1-like inhibits WNT signaling and MMP expression to suppress cancer cell invasion and metastasis. Int J. Cancer 2014, 135, 48–60. [Google Scholar] [CrossRef]
- Seetoo, D.Q.; Crowe, P.J.; Russell, P.J.; Yang, J.L. Quantitative expression of protein markers of plasminogen activation system in prognosis of colorectal cancer. J. Surg. Oncol. 2003, 82, 184–193. [Google Scholar] [CrossRef]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The extracellular matrix at a glance. J. Cell Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Weaver, V.M.; Werb, Z. The extracellular matrix: A dynamic niche in cancer progression. J. Cell Biol. 2012, 196, 395–406. [Google Scholar] [CrossRef]
- Naba, A.; Clauser, K.R.; Whittaker, C.A.; Carr, S.A.; Tanabe, K.K.; Hynes, R.O. Extracellular matrix signatures of human primary metastatic colon cancers and their metastases to liver. BMC Cancer 2014, 14, 518. [Google Scholar] [CrossRef]
- Rao, V.S.; Gu, Q.; Tzschentke, S.; Lin, K.; Ganig, N.; Thepkaysone, M.L.; Wong, F.C.; Polster, H.; Seifert, L.; Seifert, A.M.; et al. Extravesicular TIMP-1 is a non-invasive independent prognostic marker and potential therapeutic target in colorectal liver metastases. Oncogene 2022, 41, 1809–1820. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Wang, B.; Luo, J.; Jiang, K.; Zhang, H.; Mustonen, H.; Puolakkainen, P.; Zhu, J.; Ye, Y.; Wang, S. Global-scale profiling of differential expressed lysine acetylated proteins in colorectal cancer tumors and paired liver metastases. J. Proteom. 2016, 142, 24–32. [Google Scholar] [CrossRef]
- Yuzhalin, A.E.; Gordon-Weeks, A.N.; Tognoli, M.L.; Jones, K.; Markelc, B.; Konietzny, R.; Fischer, R.; Muth, A.; O’Neill, E.; Thompson, P.R.; et al. Colorectal cancer liver metastatic growth depends on PAD4-driven citrullination of the extracellular matrix. Nat. Commun. 2018, 9, 4783. [Google Scholar] [CrossRef] [PubMed]
- Pettini, F.; Visibelli, A.; Cicaloni, V.; Iovinelli, D.; Spiga, O. Multi-Omics Model Applied to Cancer Genetics. Int. J. Mol. Sci. 2021, 22, 5751. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Sun, Y.D.; Yu, G.Y.; Cui, J.R.; Lou, Z.; Zhang, H.; Huang, Y.; Bai, C.G.; Deng, L.L.; Liu, P.; et al. Integrated Omics of Metastatic Colorectal Cancer. Cancer Cell 2020, 38, 734–747.e739. [Google Scholar] [CrossRef]
- Ma, Y.S.; Huang, T.; Zhong, X.M.; Zhang, H.W.; Cong, X.L.; Xu, H.; Lu, G.X.; Yu, F.; Xue, S.B.; Lv, Z.W.; et al. Proteogenomic characterization and comprehensive integrative genomic analysis of human colorectal cancer liver metastasis. Mol. Cancer 2018, 17, 139. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.S.; Wu, Z.J.; Zhang, H.W.; Cai, B.; Huang, T.; Long, H.D.; Xu, H.; Zhao, Y.Z.; Yin, Y.Z.; Xue, S.B.; et al. Dual Regulatory Mechanisms of Expression and Mutation Involving Metabolism-Related Genes FDFT1 and UQCR5 during CLM. Mol. Ther. Oncolytics 2019, 14, 172–178. [Google Scholar] [CrossRef]
- Parikh, A.R.; Leshchiner, I.; Elagina, L.; Goyal, L.; Levovitz, C.; Siravegna, G.; Livitz, D.; Rhrissorrakrai, K.; Martin, E.E.; Van Seventer, E.E.; et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat. Med. 2019, 25, 1415–1421. [Google Scholar] [CrossRef]
- Russo, M.; Siravegna, G.; Blaszkowsky, L.S.; Corti, G.; Crisafulli, G.; Ahronian, L.G.; Mussolin, B.; Kwak, E.L.; Buscarino, M.; Lazzari, L.; et al. Tumor Heterogeneity and Lesion-Specific Response to Targeted Therapy in Colorectal Cancer. Cancer Discov. 2016, 6, 147–153. [Google Scholar] [CrossRef]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef]
- Diaz, L.A.; Bardelli, A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. 2014, 32, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra224. [Google Scholar] [CrossRef] [PubMed]
- Murahashi, S.; Akiyoshi, T.; Sano, T.; Fukunaga, Y.; Noda, T.; Ueno, M.; Zembutsu, H. Serial circulating tumour DNA analysis for locally advanced rectal cancer treated with preoperative therapy: Prediction of pathological response and postoperative recurrence. Br. J. Cancer 2020, 123, 803–810. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, C.; Lin, G.; Xiao, Y.; Jia, W.; Xiao, G.; Liu, Q.; Wu, B.; Wu, A.; Qiu, H.; et al. Serial Circulating Tumor DNA in Predicting and Monitoring the Effect of Neoadjuvant Chemoradiotherapy in Patients with Rectal Cancer: A Prospective Multicenter Study. Clin. Cancer Res. 2021, 27, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Cohen, J.D.; Wang, Y.; Li, L.; Christie, M.; Simons, K.; Elsaleh, H.; Kosmider, S.; Wong, R.; Yip, D.; et al. Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: A prospective biomarker study. Gut 2019, 68, 663–671. [Google Scholar] [CrossRef]
- Vidal, J.; Casadevall, D.; Bellosillo, B.; Pericay, C.; Garcia-Carbonero, R.; Losa, F.; Layos, L.; Alonso, V.; Capdevila, J.; Gallego, J.; et al. Clinical Impact of Presurgery Circulating Tumor DNA after Total Neoadjuvant Treatment in Locally Advanced Rectal Cancer: A Biomarker Study from the GEMCAD 1402 Trial. Clin. Cancer Res. 2021, 27, 2890–2898. [Google Scholar] [CrossRef]
- Khakoo, S.; Carter, P.D.; Brown, G.; Valeri, N.; Picchia, S.; Bali, M.A.; Shaikh, R.; Jones, T.; Begum, R.; Rana, I.; et al. MRI Tumor Regression Grade and Circulating Tumor DNA as Complementary Tools to Assess Response and Guide Therapy Adaptation in Rectal Cancer. Clin. Cancer Res. 2020, 26, 183–192. [Google Scholar] [CrossRef]
- Mauri, G.; Vitiello, P.P.; Sogari, A.; Crisafulli, G.; Sartore-Bianchi, A.; Marsoni, S.; Siena, S.; Bardelli, A. Liquid biopsies to monitor and direct cancer treatment in colorectal cancer. Br. J. Cancer 2022, 127, 394–407. [Google Scholar] [CrossRef]
- Mason, M.C.; Tzeng, C.D.; Tran Cao, H.S.; Aloia, T.A.; Newhook, T.E.; Overman, M.J.; Kopetz, S.E.; Vauthey, J.N.; Chun, Y.S. Preliminary Analysis of Liquid Biopsy after Hepatectomy for Colorectal Liver Metastases. J. Am. Coll. Surg. 2021, 233, 82–89.e81. [Google Scholar] [CrossRef]
- Tie, J.; Wang, Y.; Cohen, J.; Li, L.; Hong, W.; Christie, M.; Wong, H.L.; Kosmider, S.; Wong, R.; Thomson, B.; et al. Circulating tumor DNA dynamics and recurrence risk in patients undergoing curative intent resection of colorectal cancer liver metastases: A prospective cohort study. PLoS. Med. 2021, 18, e1003620. [Google Scholar] [CrossRef]
- Siravegna, G.; Mussolin, B.; Buscarino, M.; Corti, G.; Cassingena, A.; Crisafulli, G.; Ponzetti, A.; Cremolini, C.; Amatu, A.; Lauricella, C.; et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat. Med. 2015, 21, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Thierry, A.R.; Mouliere, F.; El Messaoudi, S.; Mollevi, C.; Lopez-Crapez, E.; Rolet, F.; Gillet, B.; Gongora, C.; Dechelotte, P.; Robert, B.; et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat. Med. 2014, 20, 430–435. [Google Scholar] [CrossRef] [PubMed]
- Sartore-Bianchi, A.; Pietrantonio, F.; Lonardi, S.; Mussolin, B.; Rua, F.; Fenocchio, E.; Amatu, A.; Corallo, S.; Manai, C.; Tosi, F.; et al. Phase II study of anti-EGFR rechallenge therapy with panitumumab driven by circulating tumor DNA molecular selection in metastatic colorectal cancer: The CHRONOS trial. J. Clin. Oncol. 2021, 39, 3506. [Google Scholar] [CrossRef]
- Martinelli, E.; Martini, G.; Famiglietti, V.; Troiani, T.; Napolitano, S.; Pietrantonio, F.; Ciardiello, D.; Terminiello, M.; Borrelli, C.; Vitiello, P.P.; et al. Cetuximab Rechallenge Plus Avelumab in Pretreated Patients With RAS Wild-type Metastatic Colorectal Cancer: The Phase 2 Single-Arm Clinical CAVE Trial. JAMA Oncol. 2021, 7, 1529–1535. [Google Scholar] [CrossRef] [PubMed]
- Herberts, C.; Wyatt, A.W. Technical and biological constraints on ctDNA-based genotyping. Trends Cancer 2021, 7, 995–1009. [Google Scholar] [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.; Granton, P.; Zegers, C.M.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [PubMed]
- van Timmeren, J.E.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baessler, B. Radiomics in medical imaging-”how-to” guide and critical reflection. Insights Imaging 2020, 11, 91. [Google Scholar] [CrossRef]
- Fiz, F.; Viganò, L.; Gennaro, N.; Costa, G.; La Bella, L.; Boichuk, A.; Cavinato, L.; Sollini, M.; Politi, L.S.; Chiti, A.; et al. Radiomics of Liver Metastases: A Systematic Review. Cancers 2020, 12, 2881. [Google Scholar] [CrossRef]
- Lubner, M.G.; Stabo, N.; Lubner, S.J.; del Rio, A.M.; Song, C.; Halberg, R.B.; Pickhardt, P.J. CT textural analysis of hepatic metastatic colorectal cancer: Pre-treatment tumor heterogeneity correlates with pathology and clinical outcomes. Abdom. Imaging 2015, 40, 2331–2337. [Google Scholar] [CrossRef]
- Dercle, L.; Lu, L.; Schwartz, L.H.; Qian, M.; Tejpar, S.; Eggleton, P.; Zhao, B.; Piessevaux, H. Radiomics Response Signature for Identification of Metastatic Colorectal Cancer Sensitive to Therapies Targeting EGFR Pathway. Natl. Cancer Inst. 2020, 112, 902–912. [Google Scholar] [CrossRef]
- Dohan, A.; Gallix, B.; Guiu, B.; Le Malicot, K.; Reinhold, C.; Soyer, P.; Bennouna, J.; Ghiringhelli, F.; Barbier, E.; Boige, V.; et al. Early evaluation using a radiomic signature of unresectable hepatic metastases to predict outcome in patients with colorectal cancer treated with FOLFIRI and bevacizumab. Gut. 2020, 69, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Taghavi, M.; Staal, F.; Gomez Munoz, F.; Imani, F.; Meek, D.B.; Simões, R.; Klompenhouwer, L.G.; van der Heide, U.A.; Beets-Tan, R.G.H.; Maas, M. CT-Based Radiomics Analysis Before Thermal Ablation to Predict Local Tumor Progression for Colorectal Liver Metastases. Cardiovasc. Intervent. Radiol. 2021, 44, 913–920. [Google Scholar] [CrossRef]
- Costa, G.; Cavinato, L.; Masci, C.; Fiz, F.; Sollini, M.; Politi, L.S.; Chiti, A.; Balzarini, L.; Aghemo, A.; di Tommaso, L.; et al. Virtual Biopsy for Diagnosis of Chemotherapy-Associated Liver Injuries and Steatohepatitis: A Combined Radiomic and Clinical Model in Patients with Colorectal Liver Metastases. Cancers 2021, 13, 3077. [Google Scholar] [CrossRef] [PubMed]
- Taghavi, M.; Trebeschi, S.; Simões, R.; Meek, D.B.; Beckers, R.C.J.; Lambregts, D.M.J.; Verhoef, C.; Houwers, J.B.; van der Heide, U.A.; Beets-Tan, R.G.H.; et al. Machine learning-based analysis of CT radiomics model for prediction of colorectal metachronous liver metastases. Abdom. Radiol. 2021, 46, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Cheng, J.; Gu, D.; Chai, F.; Hong, N.; Wang, Y.; Tian, J. Deep learning-based radiomics predicts response to chemotherapy in colorectal liver metastases. Med. Phys. 2021, 48, 513–522. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).