Regulating the p53 Tumor Suppressor Network at PML Biomolecular Condensates
Abstract
Simple Summary
Abstract
1. Introduction
2. The PML Nuclear Body and Its Functions
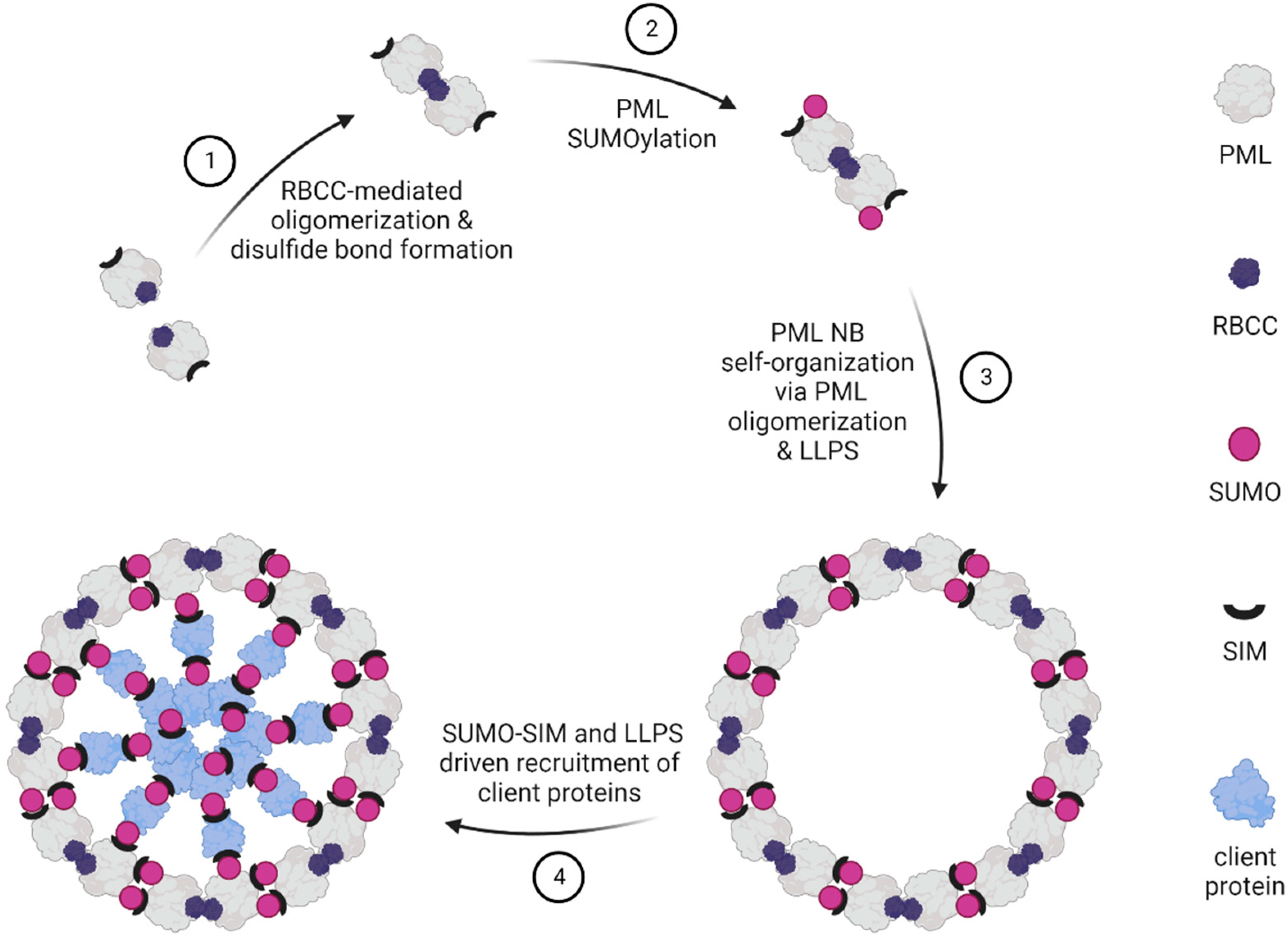
3. Biogenesis of PML Biomolecular Condensates
3.1. The Phases of PML NB Formation
3.2. LLPS Contributes to PML NB Biogenesis
4. PML Biocondensates and the p53 Response
4.1. PML Biocondensates and p53 Post-Translational Modifications
4.2. PML Biocondensates, p53 Downstream Responses, and Cancer
4.3. PML Isoforms and p53
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhang, L.; Dai, T.; Qin, Z.; Lu, H.; Zhang, L.; Zhou, F. Liquid-liquid phase separation in human health and diseases. Signal Transduct. Target Ther. 2021, 6, 290. [Google Scholar] [CrossRef] [PubMed]
- Alberti, S. Phase separation in biology. Curr. Biol. 2017, 27, R1097–R1102. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.; Zhang, J. Liquid-liquid phase separation drives cellular function and dysfunction in cancer. Nat. Rev. Cancer 2022, 22, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Boeynaems, S.; Alberti, S.; Fawzi, N.L.; Mittag, T.; Polymenidou, M.; Rousseau, F.; Schymkowitz, J.; Shorter, J.; Wolozin, B.; Van Den Bosch, L.; et al. Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol. 2018, 28, 420–435. [Google Scholar] [CrossRef]
- Alberti, S.; Saha, S.; Woodruff, J.B.; Franzmann, T.M.; Wang, J.; Hyman, A.A. A User’s Guide for Phase Separation Assays with Purified Proteins. J. Mol. Biol. 2018, 430, 4806–4820. [Google Scholar] [CrossRef]
- Shin, Y.; Brangwynne, C.P. Liquid phase condensation in cell physiology and disease. Science 2017, 357, eaaf4382. [Google Scholar] [CrossRef]
- Bieging, K.T.; Mello, S.S.; Attardi, L.D. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer 2014, 14, 359–370. [Google Scholar] [CrossRef]
- Kastenhuber, E.R.; Lowe, S.W. Putting p53 in Context. Cell 2017, 170, 1062–1078. [Google Scholar] [CrossRef]
- Dietrich, C.; Hofmann, T.G. Ferroptosis Meets Cell-Cell Contacts. Cells 2021, 10, 2462. [Google Scholar] [CrossRef]
- Hafner, A.; Bulyk, M.L.; Jambhekar, A.; Lahav, G. The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell Biol. 2019, 20, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tavana, O.; Gu, W. p53 modifications: Exquisite decorations of the powerful guardian. J. Mol. Cell Biol. 2019, 11, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Guan, D.; Dong, M.; Yang, J.; Wei, H.; Liang, Q.; Song, L.; Xu, L.; Bai, J.; Liu, C.; et al. UFMylation maintains tumour suppressor p53 stability by antagonizing its ubiquitination. Nat. Cell Biol. 2020, 22, 1056–1063. [Google Scholar] [CrossRef] [PubMed]
- Matt, S.; Hofmann, T.G. Crosstalk between p53 modifiers at PML bodies. Mol. Cell Oncol. 2018, 5, e1074335. [Google Scholar] [CrossRef] [PubMed]
- Ishov, A.M.; Sotnikov, A.G.; Negorev, D.; Vladimirova, O.V.; Neff, N.; Kamitani, T.; Yeh, E.T.; Strauss, J.F., 3rd; Maul, G.G. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 1999, 147, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Salomoni, P.; Ronchetti, S.; Guo, A.; Ruggero, D.; Pandolfi, P.P. Promyelocytic leukemia protein (PML) and Daxx participate in a novel nuclear pathway for apoptosis. J. Exp. Med. 2000, 191, 631–640. [Google Scholar] [CrossRef]
- Jensen, K.; Shiels, C.; Freemont, P.S. PML protein isoforms and the RBCC/TRIM motif. Oncogene 2001, 20, 7223–7233. [Google Scholar] [CrossRef]
- Li, Y.; Ma, X.; Wu, W.; Chen, Z.; Meng, G. PML Nuclear Body Biogenesis, Carcinogenesis, and Targeted Therapy. Trends Cancer 2020, 6, 889–906. [Google Scholar] [CrossRef] [PubMed]
- Lallemand-Breitenbach, V.; de The, H. PML nuclear bodies. Cold Spring Harb. Perspect. Biol. 2010, 2, a000661. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, G.A.; Kats, L.; Pandolfi, P.P. Synergy against PML-RARa: Targeting transcription, proteolysis, differentiation, and self-renewal in acute promyelocytic leukemia. J. Exp. Med. 2013, 210, 2793–2802. [Google Scholar] [CrossRef]
- de The, H.; Pandolfi, P.P.; Chen, Z. Acute Promyelocytic Leukemia: A Paradigm for Oncoprotein-Targeted Cure. Cancer Cell 2017, 32, 552–560. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, T.G.; Will, H. Body language: The function of PML nuclear bodies in apoptosis regulation. Cell Death Differ. 2003, 10, 1290–1299. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bernardi, R.; Pandolfi, P.P. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 2007, 8, 1006–1016. [Google Scholar] [CrossRef] [PubMed]
- LaMorte, V.J.; Dyck, J.A.; Ochs, R.L.; Evans, R.M. Localization of nascent RNA and CREB binding protein with the PML-containing nuclear body. Proc. Natl. Acad. Sci. USA 1998, 95, 4991–4996. [Google Scholar] [CrossRef] [PubMed]
- Fuchsova, B.; Novak, P.; Kafkova, J.; Hozak, P. Nuclear DNA helicase II is recruited to IFN-alpha-activated transcription sites at PML nuclear bodies. J. Cell Biol. 2002, 158, 463–473. [Google Scholar] [CrossRef]
- Kiesslich, A.; von Mikecz, A.; Hemmerich, P. Cell cycle-dependent association of PML bodies with sites of active transcription in nuclei of mammalian cells. J. Struct. Biol. 2002, 140, 167–179. [Google Scholar] [CrossRef]
- Arora, R.; Lee, Y.; Wischnewski, H.; Brun, C.M.; Schwarz, T.; Azzalin, C.M. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 2014, 5, 5220. [Google Scholar] [CrossRef]
- Grande, M.A.; van der Kraan, I.; van Steensel, B.; Schul, W.; de The, H.; van der Voort, H.T.; de Jong, L.; van Driel, R. PML-containing nuclear bodies: Their spatial distribution in relation to other nuclear components. J. Cell Biochem. 1996, 63, 280–291. [Google Scholar] [CrossRef]
- Boisvert, F.M.; Hendzel, M.J.; Bazett-Jones, D.P. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J. Cell Biol. 2000, 148, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, E.; Laukens, K.; Dang, T.H.; Van Ostade, X. A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics. Int. J. Biol. Sci. 2010, 6, 51–67. [Google Scholar] [CrossRef]
- Bernardi, R.; Pandolfi, P.P. Role of PML and the PML-nuclear body in the control of programmed cell death. Oncogene 2003, 22, 9048–9057. [Google Scholar] [CrossRef]
- Kurihara, M.; Kato, K.; Sanbo, C.; Shigenobu, S.; Ohkawa, Y.; Fuchigami, T.; Miyanari, Y. Genomic Profiling by ALaP-Seq Reveals Transcriptional Regulation by PML Bodies through DNMT3A Exclusion. Mol. Cell 2020, 78, 493–505.e498. [Google Scholar] [CrossRef] [PubMed]
- Dellaire, G.; Ching, R.W.; Ahmed, K.; Jalali, F.; Tse, K.C.; Bristow, R.G.; Bazett-Jones, D.P. Promyelocytic leukemia nuclear bodies behave as DNA damage sensors whose response to DNA double-strand breaks is regulated by NBS1 and the kinases ATM, Chk2, and ATR. J. Cell Biol. 2006, 175, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Varadaraj, A.; Dovey, C.L.; Laredj, L.; Ferguson, B.; Alexander, C.E.; Lubben, N.; Wyllie, A.H.; Rich, T. Evidence for the receipt of DNA damage stimuli by PML nuclear domains. J. Pathol. 2007, 211, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Vancurova, M.; Hanzlikova, H.; Knoblochova, L.; Kosla, J.; Majera, D.; Mistrik, M.; Burdova, K.; Hodny, Z.; Bartek, J. PML nuclear bodies are recruited to persistent DNA damage lesions in an RNF168-53BP1 dependent manner and contribute to DNA repair. DNA Repair 2019, 78, 114–127. [Google Scholar] [CrossRef]
- Hornofova, T.; Pokorna, B.; Hubackova, S.S.; Uvizl, A.; Kosla, J.; Bartek, J.; Hodny, Z.; Vasicova, P. Phospho-SIM and exon8b of PML protein regulate formation of doxorubicin-induced rDNA-PML compartment. DNA Repair 2022, 114, 103319. [Google Scholar] [CrossRef] [PubMed]
- Krieghoff-Henning, E.; Hofmann, T.G. Role of nuclear bodies in apoptosis signalling. Biochim. Biophys. Acta 2008, 1783, 2185–2194. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ivanschitz, L.; De The, H.; Le Bras, M. PML, SUMOylation, and Senescence. Front. Oncol. 2013, 3, 171. [Google Scholar] [CrossRef]
- Scherer, M.; Stamminger, T. Emerging Role of PML Nuclear Bodies in Innate Immune Signaling. J. Virol. 2016, 90, 5850–5854. [Google Scholar] [CrossRef]
- Chang, H.R.; Munkhjargal, A.; Kim, M.J.; Park, S.Y.; Jung, E.; Ryu, J.H.; Yang, Y.; Lim, J.S.; Kim, Y. The functional roles of PML nuclear bodies in genome maintenance. Mutat. Res. 2018, 809, 99–107. [Google Scholar] [CrossRef]
- Negorev, D.; Maul, G.G. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 2001, 20, 7234–7242. [Google Scholar] [CrossRef] [PubMed]
- Milovic-Holm, K.; Krieghoff, E.; Jensen, K.; Will, H.; Hofmann, T.G. FLASH links the CD95 signaling pathway to the cell nucleus and nuclear bodies. EMBO J. 2007, 26, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Krieghoff, E.; Milovic-Holm, K.; Hofmann, T.G. FLASH meets nuclear bodies: CD95 receptor signals via a nuclear pathway. Cell Cycle 2007, 6, 771–775. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.C.; Oram, M.K.; Bielinsky, A.K. SUMO-Targeted Ubiquitin Ligases and Their Functions in Maintaining Genome Stability. Int. J. Mol. Sci. 2021, 22, 5391. [Google Scholar] [CrossRef]
- Jansen, N.S.; Vertegaal, A.C.O. A Chain of Events: Regulating Target Proteins by SUMO Polymers. Trends Biochem. Sci. 2021, 46, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Mediani, L.; Guillen-Boixet, J.; Vinet, J.; Franzmann, T.M.; Bigi, I.; Mateju, D.; Carra, A.D.; Morelli, F.F.; Tiago, T.; Poser, I.; et al. Defective ribosomal products challenge nuclear function by impairing nuclear condensate dynamics and immobilizing ubiquitin. EMBO J. 2019, 38, e101341. [Google Scholar] [CrossRef] [PubMed]
- Keiten-Schmitz, J.; Wagner, K.; Piller, T.; Kaulich, M.; Alberti, S.; Muller, S. The Nuclear SUMO-Targeted Ubiquitin Quality Control Network Regulates the Dynamics of Cytoplasmic Stress Granules. Mol. Cell 2020, 79, 54–67.e57. [Google Scholar] [CrossRef] [PubMed]
- Lang, M.; Jegou, T.; Chung, I.; Richter, K.; Munch, S.; Udvarhelyi, A.; Cremer, C.; Hemmerich, P.; Engelhardt, J.; Hell, S.W.; et al. Three-dimensional organization of promyelocytic leukemia nuclear bodies. J. Cell Sci. 2010, 123, 392–400. [Google Scholar] [CrossRef]
- Jeanne, M.; Lallemand-Breitenbach, V.; Ferhi, O.; Koken, M.; Le Bras, M.; Duffort, S.; Peres, L.; Berthier, C.; Soilihi, H.; Raught, B.; et al. PML/RARA oxidation and arsenic binding initiate the antileukemia response of As2O3. Cancer Cell 2010, 18, 88–98. [Google Scholar] [CrossRef]
- Kentsis, A.; Gordon, R.E.; Borden, K.L. Control of biochemical reactions through supramolecular RING domain self-assembly. Proc. Natl. Acad. Sci. USA 2002, 99, 15404–15409. [Google Scholar] [CrossRef]
- Wang, P.; Benhenda, S.; Wu, H.; Lallemand-Breitenbach, V.; Zhen, T.; Jollivet, F.; Peres, L.; Li, Y.; Chen, S.J.; Chen, Z.; et al. RING tetramerization is required for nuclear body biogenesis and PML sumoylation. Nat. Commun. 2018, 9, 1277. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ma, X.; Chen, Z.; Wu, H.; Wang, P.; Wu, W.; Cheng, N.; Zeng, L.; Zhang, H.; Cai, X.; et al. B1 oligomerization regulates PML nuclear body biogenesis and leukemogenesis. Nat. Commun. 2019, 10, 3789. [Google Scholar] [CrossRef] [PubMed]
- Brand, P.; Lenser, T.; Hemmerich, P. Assembly dynamics of PML nuclear bodies in living cells. PMC Biophys. 2010, 3, 3. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Ferhi, O.; Jeanne, M.; Benhenda, S.; Berthier, C.; Jollivet, F.; Niwa-Kawakita, M.; Faklaris, O.; Setterblad, N.; de The, H.; et al. Oxidative stress-induced assembly of PML nuclear bodies controls sumoylation of partner proteins. J. Cell Biol. 2014, 204, 931–945. [Google Scholar] [CrossRef] [PubMed]
- Yau, T.Y.; Sander, W.; Eidson, C.; Courey, A.J. SUMO Interacting Motifs: Structure and Function. Cells 2021, 10, 2825. [Google Scholar] [CrossRef] [PubMed]
- Celen, A.B.; Sahin, U. Sumoylation on its 25th anniversary: Mechanisms, pathology, and emerging concepts. FEBS J. 2020, 287, 3110–3140. [Google Scholar] [CrossRef]
- Kunz, K.; Piller, T.; Muller, S. SUMO-specific proteases and isopeptidases of the SENP family at a glance. J. Cell Sci. 2018, 131, jcs211904. [Google Scholar] [CrossRef]
- Kamitani, T.; Kito, K.; Nguyen, H.P.; Wada, H.; Fukuda-Kamitani, T.; Yeh, E.T. Identification of three major sentrinization sites in PML. J. Biol. Chem. 1998, 273, 26675–26682. [Google Scholar] [CrossRef]
- Duprez, E.; Saurin, A.J.; Desterro, J.M.; Lallemand-Breitenbach, V.; Howe, K.; Boddy, M.N.; Solomon, E.; de The, H.; Hay, R.T.; Freemont, P.S. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: Implications for nuclear localisation. J. Cell Sci. 1999, 112 Pt 3, 381–393. [Google Scholar] [CrossRef] [PubMed]
- Cuchet-Lourenco, D.; Boutell, C.; Lukashchuk, V.; Grant, K.; Sykes, A.; Murray, J.; Orr, A.; Everett, R.D. SUMO pathway dependent recruitment of cellular repressors to herpes simplex virus type 1 genomes. PLoS Pathog. 2011, 7, e1002123. [Google Scholar] [CrossRef]
- Galisson, F.; Mahrouche, L.; Courcelles, M.; Bonneil, E.; Meloche, S.; Chelbi-Alix, M.K.; Thibault, P. A novel proteomics approach to identify SUMOylated proteins and their modification sites in human cells. Mol. Cell Proteom. 2011, 10, S1–S15. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, I.A.; D’Souza, R.C.; Yang, B.; Verlaan-de Vries, M.; Mann, M.; Vertegaal, A.C. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol. 2014, 21, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Lumpkin, R.J.; Gu, H.; Zhu, Y.; Leonard, M.; Ahmad, A.S.; Clauser, K.R.; Meyer, J.G.; Bennett, E.J.; Komives, E.A. Site-specific identification and quantitation of endogenous SUMO modifications under native conditions. Nat. Commun. 2017, 8, 1171. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.H.; Lin, H.K.; Scaglioni, P.P.; Yung, T.M.; Pandolfi, P.P. The mechanisms of PML-nuclear body formation. Mol. Cell 2006, 24, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Peng, Q.; Wan, X.; Sun, H.; Tang, J. C-terminal motifs in promyelocytic leukemia protein isoforms critically regulate PML nuclear body formation. J. Cell Sci. 2017, 130, 3496–3506. [Google Scholar] [CrossRef] [PubMed]
- Keiten-Schmitz, J.; Roder, L.; Hornstein, E.; Muller-McNicoll, M.; Muller, S. SUMO: Glue or Solvent for Phase-Separated Ribonucleoprotein Complexes and Molecular Condensates? Front. Mol. Biosci. 2021, 8, 673038. [Google Scholar] [CrossRef] [PubMed]
- Barroso-Gomila, O.; Trulsson, F.; Muratore, V.; Canosa, I.; Merino-Cacho, L.; Cortazar, A.R.; Perez, C.; Azkargorta, M.; Iloro, I.; Carracedo, A.; et al. Identification of proximal SUMO-dependent interactors using SUMO-ID. Nat. Commun. 2021, 12, 6671. [Google Scholar] [CrossRef]
- Banani, S.F.; Rice, A.M.; Peeples, W.B.; Lin, Y.; Jain, S.; Parker, R.; Rosen, M.K. Compositional Control of Phase-Separated Cellular Bodies. Cell 2016, 166, 651–663. [Google Scholar] [CrossRef]
- Cappadocia, L.; Mascle, X.H.; Bourdeau, V.; Tremblay-Belzile, S.; Chaker-Margot, M.; Lussier-Price, M.; Wada, J.; Sakaguchi, K.; Aubry, M.; Ferbeyre, G.; et al. Structural and functional characterization of the phosphorylation-dependent interaction between PML and SUMO1. Structure 2015, 23, 126–138. [Google Scholar] [CrossRef]
- Mascle, X.H.; Gagnon, C.; Wahba, H.M.; Lussier-Price, M.; Cappadocia, L.; Sakaguchi, K.; Omichinski, J.G. Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner. Structure 2020, 28, 157–168.e155. [Google Scholar] [CrossRef]
- Lussier-Price, M.; Wahba, H.M.; Mascle, X.H.; Cappadocia, L.; Bourdeau, V.; Gagnon, C.; Igelmann, S.; Sakaguchi, K.; Ferbeyre, G.; Omichinski, J.G. Zinc controls PML nuclear body formation through regulation of a paralog specific auto-inhibition in SUMO1. Nucleic Acids Res. 2022, 50, 8331–8348. [Google Scholar] [CrossRef] [PubMed]
- Corpet, A.; Kleijwegt, C.; Roubille, S.; Juillard, F.; Jacquet, K.; Texier, P.; Lomonte, P. PML nuclear bodies and chromatin dynamics: Catch me if you can! Nucleic Acids Res. 2020, 48, 11890–11912. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R. A role for macromolecular crowding effects in the assembly and function of compartments in the nucleus. J. Struct. Biol. 2004, 146, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Alberti, S.; Gladfelter, A.; Mittag, T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 2019, 176, 419–434. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 2017, 44, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Fonin, A.V.; Silonov, S.A.; Shpironok, O.G.; Antifeeva, I.A.; Petukhov, A.V.; Romanovich, A.E.; Kuznetsova, I.M.; Uversky, V.N.; Turoverov, K.K. The Role of Non-Specific Interactions in Canonical and ALT-Associated PML-Bodies Formation and Dynamics. Int. J. Mol. Sci. 2021, 22, 5821. [Google Scholar] [CrossRef]
- Lang, A.; Lang, E.; Boe, S.O. PML Bodies in Mitosis. Cells 2019, 8, 893. [Google Scholar] [CrossRef]
- Hoischen, C.; Monajembashi, S.; Weisshart, K.; Hemmerich, P. Multimodal Light Microscopy Approaches to Reveal Structural and Functional Properties of Promyelocytic Leukemia Nuclear Bodies. Front. Oncol. 2018, 8, 125. [Google Scholar] [CrossRef]
- Weidtkamp-Peters, S.; Lenser, T.; Negorev, D.; Gerstner, N.; Hofmann, T.G.; Schwanitz, G.; Hoischen, C.; Maul, G.; Dittrich, P.; Hemmerich, P. Dynamics of component exchange at PML nuclear bodies. J. Cell Sci. 2008, 121, 2731–2743. [Google Scholar] [CrossRef]
- Fonin, A.V.; Silonov, S.A.; Fefilova, A.S.; Stepanenko, O.V.; Gavrilova, A.A.; Petukhov, A.V.; Romanovich, A.E.; Modina, A.L.; Zueva, T.S.; Nedelyaev, E.M.; et al. New Evidence of the Importance of Weak Interactions in the Formation of PML-Bodies. Int. J. Mol. Sci. 2022, 23, 1613. [Google Scholar] [CrossRef]
- Shao, X.; Chen, Y.; Xu, A.; Xiang, D.; Wang, W.; Du, W.; Huang, Y.; Zhang, X.; Cai, M.; Xia, Z.; et al. Deneddylation of PML/RARalpha reconstructs functional PML nuclear bodies via orchestrating phase separation to eradicate APL. Cell Death Differ. 2022, 29, 1654–1668. [Google Scholar] [CrossRef]
- Feric, M.; Vaidya, N.; Harmon, T.S.; Mitrea, D.M.; Zhu, L.; Richardson, T.M.; Kriwacki, R.W.; Pappu, R.V.; Brangwynne, C.P. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 2016, 165, 1686–1697. [Google Scholar] [CrossRef] [PubMed]
- Yoshizawa, T.; Nozawa, R.S.; Jia, T.Z.; Saio, T.; Mori, E. Biological phase separation: Cell biology meets biophysics. Biophys. Rev. 2020, 12, 519–539. [Google Scholar] [CrossRef]
- Lafontaine, D.L.J.; Riback, J.A.; Bascetin, R.; Brangwynne, C.P. The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol. 2021, 22, 165–182. [Google Scholar] [CrossRef] [PubMed]
- Eskiw, C.H.; Dellaire, G.; Bazett-Jones, D.P. Chromatin contributes to structural integrity of promyelocytic leukemia bodies through a SUMO-1-independent mechanism. J. Biol. Chem. 2004, 279, 9577–9585. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Durrin, L.K.; Krontiris, T.G. Specific interaction of PML bodies with the TP53 locus in Jurkat interphase nuclei. Genomics 2003, 82, 250–252. [Google Scholar] [CrossRef]
- Ching, R.W.; Ahmed, K.; Boutros, P.C.; Penn, L.Z.; Bazett-Jones, D.P. Identifying gene locus associations with promyelocytic leukemia nuclear bodies using immuno-TRAP. J. Cell Biol. 2013, 201, 325–335. [Google Scholar] [CrossRef]
- Fischer, M. Census and evaluation of p53 target genes. Oncogene 2017, 36, 3943–3956. [Google Scholar] [CrossRef]
- Gostissa, M.; Hofmann, T.G.; Will, H.; Del Sal, G. Regulation of p53 functions: Let’s meet at the nuclear bodies. Curr. Opin. Cell Biol. 2003, 15, 351–357. [Google Scholar] [CrossRef]
- Gostissa, M.; Hengstermann, A.; Fogal, V.; Sandy, P.; Schwarz, S.E.; Scheffner, M.; Del Sal, G. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 1999, 18, 6462–6471. [Google Scholar] [CrossRef]
- Rodriguez, M.S.; Desterro, J.M.; Lain, S.; Midgley, C.A.; Lane, D.P.; Hay, R.T. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 1999, 18, 6455–6461. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Santockyte, R.; Shen, R.F.; Tekle, E.; Wang, G.; Yang, D.C.; Chock, P.B. Expression of SUMO-2/3 induced senescence through p53- and pRB-mediated pathways. J. Biol. Chem. 2006, 281, 36221–36227. [Google Scholar] [CrossRef]
- Kwek, S.S.; Derry, J.; Tyner, A.L.; Shen, Z.; Gudkov, A.V. Functional analysis and intracellular localization of p53 modified by SUMO-1. Oncogene 2001, 20, 2587–2599. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, D.; Muller, S. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl. Acad. Sci. USA 2002, 99, 2872–2877. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.Y.; Chiang, C.M. Crosstalk between sumoylation and acetylation regulates p53-dependent chromatin transcription and DNA binding. EMBO J. 2009, 28, 1246–1259. [Google Scholar] [CrossRef] [PubMed]
- Fogal, V.; Gostissa, M.; Sandy, P.; Zacchi, P.; Sternsdorf, T.; Jensen, K.; Pandolfi, P.P.; Will, H.; Schneider, C.; Del Sal, G. Regulation of p53 activity in nuclear bodies by a specific PML isoform. EMBO J. 2000, 19, 6185–6195. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.; Salomoni, P.; Luo, J.; Shih, A.; Zhong, S.; Gu, W.; Pandolfi, P.P. The function of PML in p53-dependent apoptosis. Nat. Cell Biol. 2000, 2, 730–736. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yoshida, N.; Murakami, N.; Kawata, K.; Ishizaki, H.; Tanaka-Okamoto, M.; Miyoshi, J.; Zinn, A.R.; Shime, H.; Inoue, N. Dynamic regulation of p53 subnuclear localization and senescence by MORC3. Mol. Biol. Cell 2007, 18, 1701–1709. [Google Scholar] [CrossRef] [PubMed]
- Mimura, Y.; Takahashi, K.; Kawata, K.; Akazawa, T.; Inoue, N. Two-step colocalization of MORC3 with PML nuclear bodies. J. Cell Sci. 2010, 123, 2014–2024. [Google Scholar] [CrossRef]
- Liebl, M.C.; Hofmann, T.G. The Role of p53 Signaling in Colorectal Cancer. Cancers 2021, 13, 2125. [Google Scholar] [CrossRef]
- Willms, A.; Schupp, H.; Poelker, M.; Adawy, A.; Debus, J.F.; Hartwig, T.; Krichel, T.; Fritsch, J.; Singh, S.; Walczak, H.; et al. TRAIL-receptor 2-a novel negative regulator of p53. Cell Death Dis. 2021, 12, 757. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, R.; Scaglioni, P.P.; Bergmann, S.; Horn, H.F.; Vousden, K.H.; Pandolfi, P.P. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat. Cell Biol. 2004, 6, 665–672. [Google Scholar] [CrossRef]
- Yang, Q.; Liao, L.; Deng, X.; Chen, R.; Gray, N.S.; Yates, J.R., 3rd; Lee, J.D. BMK1 is involved in the regulation of p53 through disrupting the PML-MDM2 interaction. Oncogene 2013, 32, 3156–3164. [Google Scholar] [CrossRef]
- Wan, J.; Block, S.; Scribano, C.M.; Thiry, R.; Esbona, K.; Audhya, A.; Weaver, B.A. Mad1 destabilizes p53 by preventing PML from sequestering MDM2. Nat. Commun. 2019, 10, 1540. [Google Scholar] [CrossRef]
- Louria-Hayon, I.; Grossman, T.; Sionov, R.V.; Alsheich, O.; Pandolfi, P.P.; Haupt, Y. The promyelocytic leukemia protein protects p53 from Mdm2-mediated inhibition and degradation. J. Biol. Chem. 2003, 278, 33134–33141. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Jeong, J.H.; Brown, A.L.; Lee, C.H.; Pandolfi, P.P.; Chung, J.H.; Kim, M.K. Promyelocytic leukemia activates Chk2 by mediating Chk2 autophosphorylation. J. Biol. Chem. 2006, 281, 26645–26654. [Google Scholar] [CrossRef] [PubMed]
- Alsheich-Bartok, O.; Haupt, S.; Alkalay-Snir, I.; Saito, S.; Appella, E.; Haupt, Y. PML enhances the regulation of p53 by CK1 in response to DNA damage. Oncogene 2008, 27, 3653–3661. [Google Scholar] [CrossRef]
- Bode, A.M.; Dong, Z. Post-translational modification of p53 in tumorigenesis. Nat. Rev. Cancer 2004, 4, 793–805. [Google Scholar] [CrossRef]
- Hofmann, T.G.; Moller, A.; Sirma, H.; Zentgraf, H.; Taya, Y.; Droge, W.; Will, H.; Schmitz, M.L. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat. Cell Biol. 2002, 4, 1–10. [Google Scholar] [CrossRef]
- D’Orazi, G.; Cecchinelli, B.; Bruno, T.; Manni, I.; Higashimoto, Y.; Saito, S.; Gostissa, M.; Coen, S.; Marchetti, A.; Del Sal, G.; et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat. Cell Biol. 2002, 4, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Moller, A.; Sirma, H.; Hofmann, T.G.; Rueffer, S.; Klimczak, E.; Droge, W.; Will, H.; Schmitz, M.L. PML is required for homeodomain-interacting protein kinase 2 (HIPK2)-mediated p53 phosphorylation and cell cycle arrest but is dispensable for the formation of HIPK domains. Cancer Res. 2003, 63, 4310–4314. [Google Scholar] [PubMed]
- Winter, M.; Sombroek, D.; Dauth, I.; Moehlenbrink, J.; Scheuermann, K.; Crone, J.; Hofmann, T.G. Control of HIPK2 stability by ubiquitin ligase Siah-1 and checkpoint kinases ATM and ATR. Nat. Cell Biol. 2008, 10, 812–824. [Google Scholar] [CrossRef] [PubMed]
- Rokudai, S.; Laptenko, O.; Arnal, S.M.; Taya, Y.; Kitabayashi, I.; Prives, C. MOZ increases p53 acetylation and premature senescence through its complex formation with PML. Proc. Natl. Acad. Sci. USA 2013, 110, 3895–3900. [Google Scholar] [CrossRef] [PubMed]
- Legube, G.; Linares, L.K.; Tyteca, S.; Caron, C.; Scheffner, M.; Chevillard-Briet, M.; Trouche, D. Role of the histone acetyl transferase Tip60 in the p53 pathway. J. Biol. Chem. 2004, 279, 44825–44833. [Google Scholar] [CrossRef]
- Pearson, M.; Carbone, R.; Sebastiani, C.; Cioce, M.; Fagioli, M.; Saito, S.; Higashimoto, Y.; Appella, E.; Minucci, S.; Pandolfi, P.P.; et al. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 2000, 406, 207–210. [Google Scholar] [CrossRef]
- Langley, E.; Pearson, M.; Faretta, M.; Bauer, U.M.; Frye, R.A.; Minucci, S.; Pelicci, P.G.; Kouzarides, T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002, 21, 2383–2396. [Google Scholar] [CrossRef]
- Sombroek, D.; Hofmann, T.G. How cells switch HIPK2 on and off. Cell Death Differ. 2009, 16, 187–194. [Google Scholar] [CrossRef]
- Matt, S.; Hofmann, T.G. The DNA damage-induced cell death response: A roadmap to kill cancer cells. Cell Mol. Life Sci. 2016, 73, 2829–2850. [Google Scholar] [CrossRef] [PubMed]
- Conrad, E.; Polonio-Vallon, T.; Meister, M.; Matt, S.; Bitomsky, N.; Herbel, C.; Liebl, M.; Greiner, V.; Kriznik, B.; Schumacher, S.; et al. HIPK2 restricts SIRT1 activity upon severe DNA damage by a phosphorylation-controlled mechanism. Cell Death Differ. 2016, 23, 110–122. [Google Scholar] [CrossRef] [PubMed]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.J.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021, 30, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022. [Google Scholar] [CrossRef] [PubMed]
- Liebl, M.C.; Hofmann, T.G. Cell Fate Regulation upon DNA Damage: p53 Serine 46 Kinases Pave the Cell Death Road. Bioessays 2019, 41, e1900127. [Google Scholar] [CrossRef] [PubMed]
- Follis, A.V.; Llambi, F.; Merritt, P.; Chipuk, J.E.; Green, D.R.; Kriwacki, R.W. Pin1-Induced Proline Isomerization in Cytosolic p53 Mediates BAX Activation and Apoptosis. Mol. Cell 2015, 59, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, F.; Tocco, F.; Girardini, J.; Smith, P.; Gasco, M.; Lu, X.; Crook, T.; Del Sal, G. The prolyl isomerase Pin1 orchestrates p53 acetylation and dissociation from the apoptosis inhibitor iASPP. Nat. Struct. Mol. Biol. 2007, 14, 912–920. [Google Scholar] [CrossRef] [PubMed]
- Reineke, E.L.; Lam, M.; Liu, Q.; Liu, Y.; Stanya, K.J.; Chang, K.S.; Means, A.R.; Kao, H.Y. Degradation of the tumor suppressor PML by Pin1 contributes to the cancer phenotype of breast cancer MDA-MB-231 cells. Mol. Cell Biol. 2008, 28, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Bitomsky, N.; Conrad, E.; Moritz, C.; Polonio-Vallon, T.; Sombroek, D.; Schultheiss, K.; Glas, C.; Greiner, V.; Herbel, C.; Mantovani, F.; et al. Autophosphorylation and Pin1 binding coordinate DNA damage-induced HIPK2 activation and cell death. Proc. Natl. Acad. Sci. USA 2013, 110, E4203–E4212. [Google Scholar] [CrossRef]
- Sykes, S.M.; Mellert, H.S.; Holbert, M.A.; Li, K.; Marmorstein, R.; Lane, W.S.; McMahon, S.B. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell 2006, 24, 841–851. [Google Scholar] [CrossRef]
- Tang, Y.; Luo, J.; Zhang, W.; Gu, W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell 2006, 24, 827–839. [Google Scholar] [CrossRef] [PubMed]
- Brooks, C.L.; Gu, W. How does SIRT1 affect metabolism, senescence and cancer? Nat. Rev. Cancer 2009, 9, 123–128. [Google Scholar] [CrossRef]
- Ferbeyre, G.; de Stanchina, E.; Querido, E.; Baptiste, N.; Prives, C.; Lowe, S.W. PML is induced by oncogenic ras and promotes premature senescence. Genes Dev. 2000, 14, 2015–2027. [Google Scholar] [CrossRef]
- Ablain, J.; Rice, K.; Soilihi, H.; de Reynies, A.; Minucci, S.; de The, H. Activation of a promyelocytic leukemia-tumor protein 53 axis underlies acute promyelocytic leukemia cure. Nat. Med. 2014, 20, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Niwa-Kawakita, M.; Ferhi, O.; Soilihi, H.; Le Bras, M.; Lallemand-Breitenbach, V.; de The, H. PML is a ROS sensor activating p53 upon oxidative stress. J. Exp. Med. 2017, 214, 3197–3206. [Google Scholar] [CrossRef] [PubMed]
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.A.; et al. Mutational landscape and significance across 12 major cancer types. Nature 2013, 502, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Bouaoun, L.; Sonkin, D.; Ardin, M.; Hollstein, M.; Byrnes, G.; Zavadil, J.; Olivier, M. TP53 Variations in Human Cancers: New Lessons from the IARC TP53 Database and Genomics Data. Hum. Mutat. 2016, 37, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Li, L.; Zhang, D.; Wu, K.; Guo, P.; Zeng, J.; Wang, X.; He, D. Telomerase suppression initiates PML-dependent p53 activation to inhibit bladder cancer cell growth. Oncol. Rep. 2010, 24, 1551–1559. [Google Scholar] [CrossRef] [PubMed]
- Haupt, S.; di Agostino, S.; Mizrahi, I.; Alsheich-Bartok, O.; Voorhoeve, M.; Damalas, A.; Blandino, G.; Haupt, Y. Promyelocytic leukemia protein is required for gain of function by mutant p53. Cancer Res. 2009, 69, 4818–4826. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Choi, J.H.; Park, E.M.; Choi, Y.H. Interaction of promyelocytic leukemia/p53 affects signal transducer and activator of transcription-3 activity in response to oncostatin M. Korean J. Physiol. Pharm. 2020, 24, 203–212. [Google Scholar] [CrossRef]
- Schulz-Heddergott, R.; Stark, N.; Edmunds, S.J.; Li, J.; Conradi, L.C.; Bohnenberger, H.; Ceteci, F.; Greten, F.R.; Dobbelstein, M.; Moll, U.M. Therapeutic Ablation of Gain-of-Function Mutant p53 in Colorectal Cancer Inhibits Stat3-Mediated Tumor Growth and Invasion. Cancer Cell 2018, 34, 298–314.e297. [Google Scholar] [CrossRef]
- Bischof, O.; Kirsh, O.; Pearson, M.; Itahana, K.; Pelicci, P.G.; Dejean, A. Deconstructing PML-induced premature senescence. EMBO J. 2002, 21, 3358–3369. [Google Scholar] [CrossRef]
- Gresko, E.; Ritterhoff, S.; Sevilla-Perez, J.; Roscic, A.; Frobius, K.; Kotevic, I.; Vichalkovski, A.; Hess, D.; Hemmings, B.A.; Schmitz, M.L. PML tumor suppressor is regulated by HIPK2-mediated phosphorylation in response to DNA damage. Oncogene 2009, 28, 698–708. [Google Scholar] [CrossRef]
- Ivanschitz, L.; Takahashi, Y.; Jollivet, F.; Ayrault, O.; Le Bras, M.; de The, H. PML IV/ARF interaction enhances p53 SUMO-1 conjugation, activation, and senescence. Proc. Natl. Acad. Sci. USA 2015, 112, 14278–14283. [Google Scholar] [CrossRef] [PubMed]
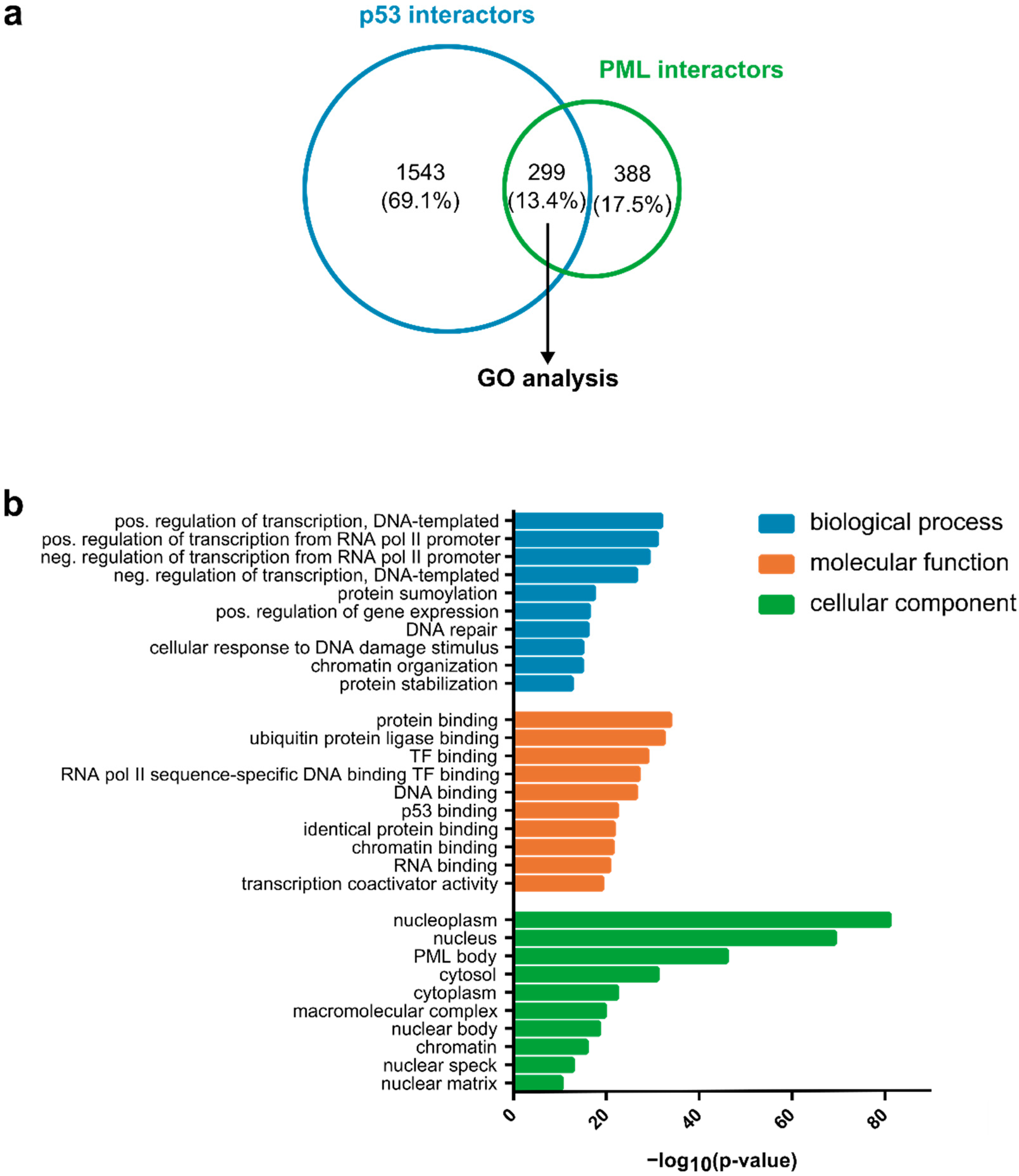

| ACACA | CUL1 | HNRNPM | NCOR1 | RFC4 | TDP2 |
| ACTG1 | DAXX | HNRNPR | NEDD1 | RNF125 | TES |
| ADD3 | DBN1 | HNRNPU | NFATC1 | RNF20 | TET2 |
| AHNAK | DCP1A | HOMER3 | NFRKB | RPL11 | TFCP2 |
| ANKRD2 | DCTN2 | HSF1 | NPM1 | RPL5 | TNRC6B |
| ANXA1 | DDX3X | HSP90AB1 | NR3C1 | RRM2 | TOP2B |
| ANXA2 | DDX50 | HSPA1A | NR4A1 | RTN4 | TOPBP1 |
| APEX1 | DGCR14 | HSPA5 | NUFIP2 | RUNX2 | TOPORS |
| ARID3A | DHX15 | HSPA6 | NUPR1 | RUNX3 | TP53 |
| ARIH2 | DIS3 | HSPA8 | PALLD | S100B | TP53BP1 |
| ARNT | DNAJB1 | HSPB1 | PARK7 | SAFB | TP63 |
| ASF1A | DNM2 | HTT | PARP1 | SART1 | TRIM24 |
| ATRX | ECT2 | IFI16 | PC | SATB1 | TRIM25 |
| ATXN3 | EEF1A1P5 | ILF3 | PCBP1 | SBNO1 | TRIM27 |
| AURKA | EGLN3 | JAK1 | PCCA | SENP1 | TRIM28 |
| AXIN1 | EHMT2 | JUN | PER2 | SEPT9 | TRIM33 |
| AZGP1 | EIF3C | KAT5 | PIAS1 | SFPQ | TRIM66 |
| BANP | EIF3F | KAT6A | PIAS2 | SIN3A | TRIM69 |
| BCL2 | EIF3G | KIF20A | PIAS3 | SIRT1 | TRIML2 |
| BCL6 | EIF4B | KIF5B | PIAS4 | SKI | TUBA1C |
| BCOR | EP400 | KMT2A | PIN1 | SKP1 | TUBB |
| BHLHE40 | EPB41L2 | KPNA4 | PIP | SLAIN2 | UBA52 |
| BLM | EPB41L3 | LDHB | PKM | SLC1A5 | UBC |
| BRCA1 | ERCC3 | LIG3 | PLAGL1 | SLC3A2 | UBE2I |
| BRCC3 | ERCC6 | LIMA1 | PLCG1 | SMAD2 | UBE3A |
| BRD1 | EXOSC9 | LMNA | PLEKHA4 | SMAD3 | UHRF1 |
| BRD4 | EZR | LMNB1 | PLOD3 | SMARCA4 | UIMC1 |
| BRD8 | FAM50A | LYZ | PML | SMC5 | UPF1 |
| BUB3 | FBXW7 | MAGEA2 | PMS1 | SMTN | USP10 |
| CALD1 | FLNA | MAGED2 | POLK | SNW1 | USP11 |
| CASP8 | FOS | MAP1LC3B | PPARG | SP1 | USP7 |
| CCNT1 | FOXK1 | MAP4 | PPARGC1A | SP100 | VIM |
| CCT6A | FUBP1 | MAPK1 | PPP1R13L | SP3 | WDR5 |
| CCT8 | FXR1 | MAPK3 | PPWD1 | SPAG9 | WRN |
| CDK1 | GATAD2A | MAPK7 | PRDX1 | SPTA1 | XAB2 |
| CDK2 | GATAD2B | MAVS | PRPF3 | SQSTM1 | XRCC1 |
| CDK6 | GTF2I | MDC1 | PSMC3 | STAT3 | YAP1 |
| CDK7 | GTF3C4 | MDM2 | PSMD2 | STX5 | YEATS2 |
| CDKN2A | H2AFX | MED1 | PSME3 | SUMO1 | YTHDF2 |
| CHD4 | HADHB | MED23 | PYHIN1 | SUMO2 | YTHDF3 |
| CHD8 | HBS1L | MIF | RAD51 | SUMO3 | YWHAZ |
| CHEK2 | HCFC1 | MTOR | RAD54L2 | SUZ12 | ZBTB16 |
| CMTR1 | HDAC1 | MYC | RANBP2 | SYNCRIP | ZBTB33 |
| CORO7 | HDAC2 | MYH9 | RANGAP1 | SYNE2 | ZBTB5 |
| CREBBP | HDLBP | MYO6 | RB1 | TAB1 | ZC3HAV1 |
| CSDE1 | HELLS | NAA40 | RBCK1 | TAF6 | ZMYM2 |
| CSNK1D | HIPK2 | NAB1 | RBX1 | TAF9 | ZNF148 |
| CSNK2A1 | HIST1H4A | NAP1L1 | RDX | TARS | ZNF451 |
| CSNK2B | HNF4A | NBN | RECQL | TCERG1 | ZYX |
| CSTA | HNRNPK | NCOA2 | RELA | TDG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liebl, M.C.; Hofmann, T.G. Regulating the p53 Tumor Suppressor Network at PML Biomolecular Condensates. Cancers 2022, 14, 4549. https://doi.org/10.3390/cancers14194549
Liebl MC, Hofmann TG. Regulating the p53 Tumor Suppressor Network at PML Biomolecular Condensates. Cancers. 2022; 14(19):4549. https://doi.org/10.3390/cancers14194549
Chicago/Turabian StyleLiebl, Magdalena C., and Thomas G. Hofmann. 2022. "Regulating the p53 Tumor Suppressor Network at PML Biomolecular Condensates" Cancers 14, no. 19: 4549. https://doi.org/10.3390/cancers14194549
APA StyleLiebl, M. C., & Hofmann, T. G. (2022). Regulating the p53 Tumor Suppressor Network at PML Biomolecular Condensates. Cancers, 14(19), 4549. https://doi.org/10.3390/cancers14194549






