The Long and the Short of It: NEAT1 and Cancer Cell Metabolism
Abstract
Simple Summary
Abstract
1. Background
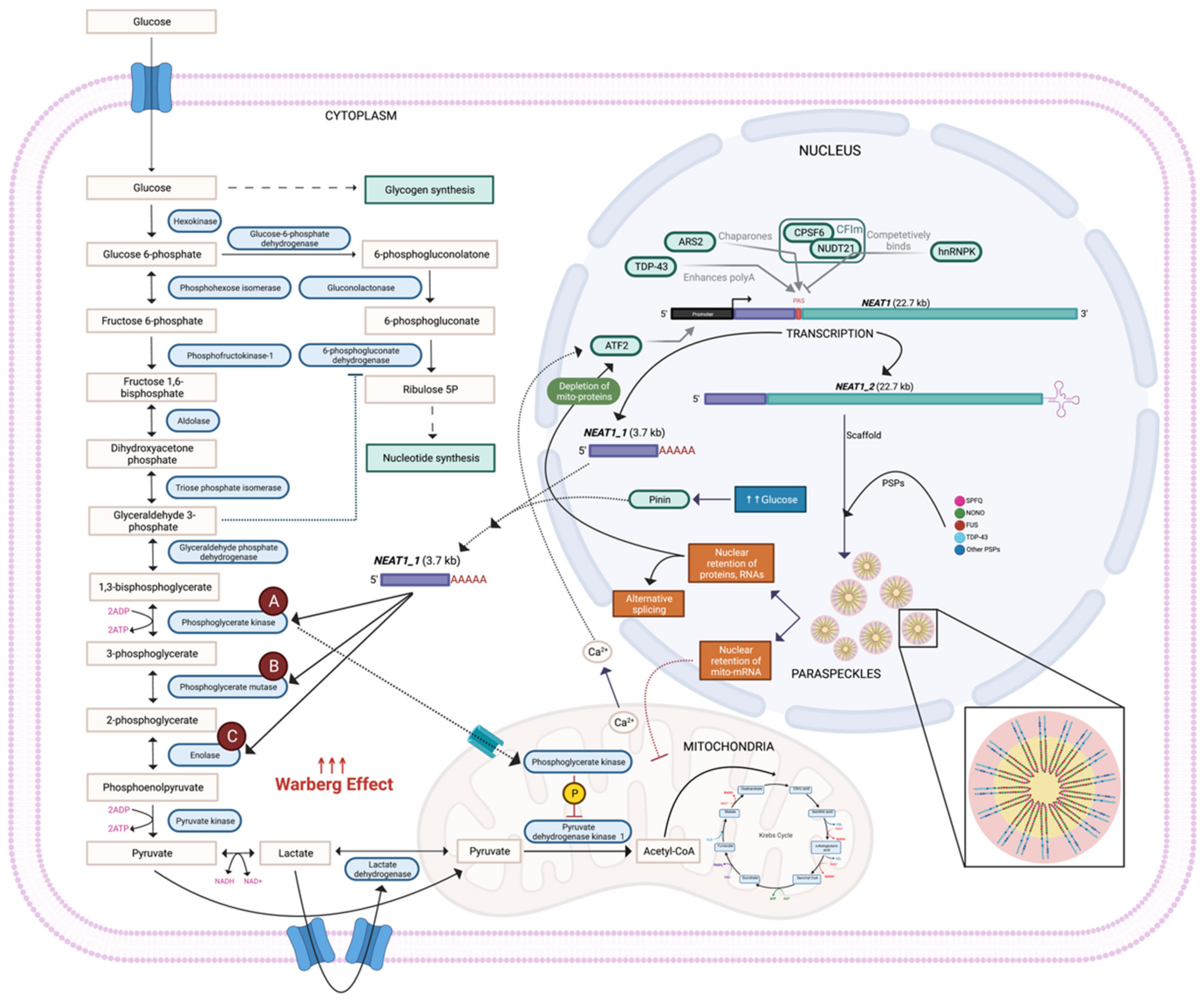
2. NEAT1_1 Enhances the Warburg Effect by Accelerating Glycolytic Metabolite Flux
3. NEAT1_2 and Paraspeckle Abundance Increase following Stress
4. What Is Driving the NEAT1 Isoform Switch?
5. NEAT1 and Paraspeckles Alter Metabolism via Mitochondria
6. Alternative Processing of lncRNAs in Cancer
7. Dysregulation of Both Short and Long NEAT1 in Cancer
8. Considerations for Isoform Detection of NEAT1
9. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ALL | Acute lymphoblastic leukaemia |
| AML | Acute myeloid leukaemia |
| APL | Acute promyelocytic leukaemia |
| ATC | Anaplastic thyroid carcinoma |
| ATR | Ataxia telangiectasia and Rad3-related |
| BC | Breast cancer |
| CBMs | Circulating blood monocytes |
| CC | Cervical cancer |
| CFIm | CPSF6-NUDT21 complex |
| Chk1 | Checkpoint kinase 1 |
| CLL | Chronic lymphocytic leukaemia |
| CML | Chronic myeloid leukaemia |
| CoA | Co-enzyme A |
| CPSF6 | Cleavage and polyadenylation-specific factor 6 |
| CRC | Colorectal cancer |
| DRP1 | Dynamin-related protein 1 |
| ECAR | Extracellular acidification rate |
| ENO1 | Alpha enolase |
| ETC | Electron transport chain |
| FCCp | Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone |
| FUS | Fused in sarcoma |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase |
| GC | Gastric cancer |
| HCC | Hepatocellular carcinoma |
| KD | Knockdown |
| KO | Knockout |
| LC | Laryngeal cancer |
| lncRNA | Long noncoding RNA |
| LSCC | Laryngeal squamous cell cancer |
| LUAD | Lung adenocarcinoma |
| mascRNA | MALAT1-associated cytoplasmic RNA |
| MFN1/2 | Mitofusion protein |
| MM | Multiple myeloma |
| mtDNA | Mitochondrial DNA |
| NADH | Nicotinamide adenine dinucleotide (NAD) + hydrogen |
| NEAT1 | Nuclear enriched abundant transcript 1 |
| NONO | Non-POU-domain-containing octamer-binding protein |
| NPC | Nasopharyngeal carcinoma |
| NSCLC | Non-small-cell lung cancer |
| NUDT21 | Nudix hydrolase 21 |
| OC | Ovarian cancer |
| OE | Over-expression |
| OSCC | Oesophageal squamous cell carcinoma |
| OXPHOS | Oxidative phosphorylation |
| p53 | Tumour suppressor protein 53 |
| PC | Prostate cancer |
| PDHK1 | Pyruvate dehydrogenase kinase isozyme 1 |
| PGAM1 | Phosphoglycerate mutase 1 |
| PGK1 | Phosphoglycerate kinase 1 |
| PTC | Papillary thyroid carcinoma |
| RAS | Rat sarcoma virus oncogene |
| RBP | RNA binding protein |
| ROS | Reactive oxygen species |
| SFPQ | Splicing factor proline and glutamine-rich |
| shRNA | Short hairpin RNA |
| TAMs | Tumour associated macrophages |
| TAR | Transactive response |
| TC | Thyroid carcinoma |
| TDP-43 | TAR DNA binding protein 43 kDa |
References
- Pavlova, N.N.; Zhu, J.; Thompson, C.B. The hallmarks of cancer metabolism: Still emerging. Cell Metab. 2022, 34, 355–377. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Warburg, O. On the Origin of Cancer Cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Koppenol, W.H.; Bounds, P.L.; Dang, C.V. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer 2011, 11, 325–337. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 Catalog of Human Long Noncoding RNAs: Analysis of Their Gene Structure, Evolution, and Expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, J.; Chen, C.; Zhang, R.; Wang, K. Pan-cancer analysis of long non-coding RNA NEAT1 in various cancers. Gene Funct. Dis. 2017, 5, 27–35. [Google Scholar] [CrossRef]
- Bu, F.; Wang, A.; Zhu, Y.; You, H.; Zhang, Y.; Meng, X.; Huang, C.; Li, J.; Zhu, Y. LncRNA NEAT1: Shedding light on mechanisms and opportunities in liver diseases. Liver Int. 2020, 40, 2612–2626. [Google Scholar] [CrossRef] [PubMed]
- McCluggage, F.; Fox, A.H. Paraspeckle nuclear condensates: Global sensors of cell stress? BioEssays 2021, 43, e2000245. [Google Scholar] [CrossRef]
- Wilusz, J.E.; JnBaptiste, C.K.; Lu, L.Y.; Kuhn, C.-D.; Joshua-Tor, L.; Sharp, P.A. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012, 26, 2392–2407. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hu, S.-B.; Wang, M.-R.; Yao, R.W.; Wu, D.; Yang, L.; Chen, L.-L. Genome-wide screening of NEAT1 regulators reveals cross-regulation between paraspeckles and mitochondria. Nat. Cell Biol. 2018, 20, 1145–1158. [Google Scholar] [CrossRef]
- Li, W.; Zhang, Z.; Liu, X.; Cheng, X.; Zhang, Y.; Han, X.; Zhang, Y.; Liu, S.; Yang, J.; Xu, B.; et al. The FOXN3-NEAT1-SIN3A repressor complex promotes progression of hormonally responsive breast cancer. J. Clin. Investig. 2017, 127, 3421–3440. [Google Scholar] [CrossRef] [PubMed]
- Park, M.K.; Zhang, L.; Min, K.-W.; Cho, J.-H.; Yeh, C.-C.; Moon, H.; Hormaechea-Agulla, D.; Mun, H.; Ko, S.; Lee, J.W.; et al. NEAT1 is essential for metabolic changes that promote breast cancer growth and metastasis. Cell Metab. 2021, 33, 2380–2397.e9. [Google Scholar] [CrossRef]
- Adriaens, C.; Rambow, F.; Bervoets, G.; Silla, T.; Mito, M.; Chiba, T.; Asahara, H.; Hirose, T.; Nakagawa, S.; Jensen, T.H.; et al. The long noncoding RNA NEAT1_1 is seemingly dispensable for normal tissue homeostasis and cancer cell growth. RNA 2019, 25, 1681–1695. [Google Scholar] [CrossRef]
- Alpatov, R.; Munguba, G.C.; Caton, P.; Joo, J.H.; Shi, Y.; Shi, Y.; Hunt, M.E.; Sugrue, S.P. Nuclear Speckle-Associated Protein Pnn/DRS Binds to the Transcriptional Corepressor CtBP and Relieves CtBP- Mediated Repression of the E-Cadherin Gene. Mol. Cell. Biol. 2004, 24, 10223–10235. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Liu, C.; Xu, D.; Xie, K.; Li, A. LncRNA NEAT1 facilitates glioma progression via stabilizing PGK1. J. Transl. Med. 2022, 20, 1–13. [Google Scholar] [CrossRef]
- Fu, Q.; Yu, Z. Phosphoglycerate kinase 1 (PGK1) in cancer: A promising target for diagnosis and therapy. Life Sci. 2020, 256, 117863. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Hunter, T. Metabolic Kinases Moonlighting as Protein Kinases. Trends Biochem. Sci. 2018, 43, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Jiang, Y.; Meisenhelder, J.; Yang, W.; Hawke, D.H.; Zheng, Y.; Xia, Y.; Aldape, K.; He, J.; Hunter, T.; et al. Mitochondria-Translocated PGK1 Functions as a Protein Kinase to Coordinate Glycolysis and the TCA Cycle in Tumorigenesis. Mol. Cell 2016, 61, 705–719. [Google Scholar] [CrossRef]
- Liu, H.; Wang, X.; Shen, P.; Ni, Y.; Han, X. The basic functions of phosphoglycerate kinase 1 and its roles in cancer and other diseases. Eur. J. Pharmacol. 2022, 920, 174835. [Google Scholar] [CrossRef] [PubMed]
- Gou, R.; Hu, Y.; Liu, O.; Dong, H.; Gao, L.; Wang, S.; Zheng, M.; Li, X.; Lin, B. PGK1 Is a Key Target for Anti-Glycolytic Therapy of Ovarian Cancer: Based on the Comprehensive Analysis of Glycolysis-Related Genes. Front. Oncol. 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-L.; Jiang, Z.-R.; Hu, C.; Chen, C.; Hu, Z.-Q.; Wang, A.-L.; Wang, L.; Liu, J.; Liu, Q.-S. Pharmacologically inhibiting phosphoglycerate kinase 1 for glioma with NG52. Acta Pharmacol. Sin. 2020, 42, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Kondoh, H.; Lleonart, M.E.; Nakashima, Y.; Yokode, M.; Tanaka, M.; Bernard, D.; Gil, J.; Beach, D. A High Glycolytic Flux Supports the Proliferative Potential of Murine Embryonic Stem Cells. Antioxidants Redox Signal. 2007, 9, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Kondoh, H.; Lleonart, M.E.; Gil, J.; Wang, J.; Degan, P.; Peters, G.; Martinez, D.; Carnero, A.; Beach, D. Glycolytic Enzymes Can Modulate Cellular Life Span. Cancer Res. 2005, 65, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Hitosugi, T.; Zhou, L.; Elf, S.; Fan, J.; Kang, H.-B.; Seo, J.H.; Shan, C.; Dai, Q.; Zhang, L.; Xie, J.; et al. Phosphoglycerate Mutase 1 Coordinates Glycolysis and Biosynthesis to Promote Tumor Growth. Cancer Cell 2012, 22, 585–600. [Google Scholar] [CrossRef]
- Peng, X.; Gong, F.; Chen, Y.; Qiu, M.; Cheng, K.; Tang, J.; Ge, J.; Chen, N.; Zeng, H.; Liu, J. Proteomics identification of PGAM1 as a potential therapeutic target for urothelial bladder cancer. J. Proteom. 2015, 132, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Mikawa, T.; Shibata, E.; Shimada, M.; Ito, K.; Ito, T.; Kanda, H.; Takubo, K.; Lleonart, M.E.; Inagaki, N.; Yokode, M.; et al. Phosphoglycerate Mutase Cooperates with Chk1 Kinase to Regulate Glycolysis. iScience 2020, 23, 101306. [Google Scholar] [CrossRef] [PubMed]
- Cancemi, P.; Buttacavoli, M.; Roz, E.; Feo, S. Expression of Alpha-Enolase (ENO1), Myc Promoter-Binding Protein-1 (MBP-1) and Matrix Metalloproteinases (MMP-2 and MMP-9) Reflect the Nature and Aggressiveness of Breast Tumors. Int. J. Mol. Sci. 2019, 20, 3952. [Google Scholar] [CrossRef]
- Perconti, G.; Pratesi, F.; Angelotti, F.; Manca, L.; Puxeddu, I.; Rubino, P.; Maranto, C.; Giallongo, A.; Migliorini, P. Fingerprinting of anti-alpha enolase antibodies in systemic sclerosis. Clin. Exp. Rheumatol. 2020, 38 (Suppl. 125), 115–119. [Google Scholar]
- Li, M.; Li, J.; Wang, J.; Li, Y.; Yang, P. Serum level of anti-α-enolase antibody in untreated systemic lupus erythematosus patients correlates with 24-hour urine protein and D-dimer. Lupus 2018, 27, 139–142. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Um, J.; Lee, J.-H.; Kim, W.-H.; Kang, W.S.; Kim, S.H.; Ha, H.-H.; Kim, Y.-C.; Ahn, Y.-K.; Jung, D.-W.; et al. ENOblock, a unique small molecule inhibitor of the non-glycolytic functions of enolase, alleviates the symptoms of type 2 diabetes. Sci. Rep. 2017, 7, srep44186. [Google Scholar] [CrossRef]
- Butterfield, D.A.; Lange, M.L.B. Multifunctional roles of enolase in Alzheimer’s disease brain: Beyond altered glucose metabolism. J. Neurochem. 2009, 111, 915–933. [Google Scholar] [CrossRef]
- Altenberg, B.; Greulich, K. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics 2004, 84, 1014–1020. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.-Y.; Cao, J.; Gao, L.-J.; Zhang, F.-P.; Shen, J.; Zhou, L.; Shi, J.-Y.; Feng, Y.-L.; Yan, Z.; Wang, D.-P.; et al. Upregulation of α enolase (ENO1) crotonylation in colorectal cancer and its promoting effect on cancer cell metastasis. Biochem. Biophys. Res. Commun. 2021, 578, 77–83. [Google Scholar] [CrossRef]
- Hippner, M.; Majkowski, M.; Biecek, P.; Szkudlarek, T.; Simiczyjew, A.; Pieniazek, M.; Nowak, D.; Miazek, A.; Donizy, P. Alpha-Enolase (ENO1) Correlates with Invasiveness of Cutaneous Melanoma—An In Vitro and a Clinical Study. Diagnostics 2022, 12, 254. [Google Scholar] [CrossRef]
- Capello, M.; Ferri-Borgogno, S.; Riganti, C.; Chattaragada, M.S.; Principe, M.; Roux, C.; Zhou, W.; Petricoin, E.F.; Cappello, P.; Novelli, F. Targeting the Warburg effect in cancer cells through ENO1 knockdown rescues oxidative phosphorylation and induces growth arrest. Oncotarget 2015, 7, 5598–5612. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.S.; Wu, W.; Walsh, G.; Hong, W.K.; Mao, L. Enolase-alpha is frequently down-regulated in non-small cell lung cancer and predicts aggressive biological behavior. Clin. Cancer Res. 2003, 9, 3641–3644. [Google Scholar]
- Fu, Q.-F.; Liu, Y.; Fan, Y.; Hua, S.-N.; Qu, H.-Y.; Dong, S.-W.; Li, R.-L.; Zhao, M.-Y.; Zhen, Y.; Yu, X.-L.; et al. Alpha-enolase promotes cell glycolysis, growth, migration, and invasion in non-small cell lung cancer through FAK-mediated PI3K/AKT pathway. J. Hematol. Oncol. 2015, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Hirose, T.; Yamazaki, T.; Nakagawa, S. Molecular anatomy of the architectural NEAT1 noncoding RNA: The domains, interactors, and biogenesis pathway required to build phase-separated nuclear paraspeckles. Wiley Interdiscip. Rev. RNA 2019, 10, e1545. [Google Scholar] [CrossRef] [PubMed]
- Fox, A.H.; Lamond, A. Paraspeckles. Cold Spring Harb. Perspect. Biol. 2010, 2, a000687. [Google Scholar] [CrossRef] [PubMed]
- Fox, A.H.; Lam, Y.W.; Leung, A.K.; Lyon, C.E.; Andersen, J.; Mann, M.; Lamond, A.I. Paraspeckles: A Novel Nuclear Domain. Curr. Biol. 2002, 12, 13–25. [Google Scholar] [CrossRef]
- Fox, A.H.; Nakagawa, S.; Hirose, T.; Bond, C.S. Paraspeckles: Where Long Noncoding RNA Meets Phase Separation. Trends Biochem. Sci. 2018, 43, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Souquere, S.; Beauclair, G.; Harper, F.; Fox, A.; Pierron, G. Highly Ordered Spatial Organization of the Structural Long Noncoding NEAT1 RNAs within Paraspeckle Nuclear Bodies. Mol. Biol. Cell 2010, 21, 4020–4027. [Google Scholar] [CrossRef] [PubMed]
- Grosch, M.; Ittermann, S.; Rusha, E.; Greisle, T.; Ori, C.; Truong, D.-J.J.; O’Neill, A.C.; Pertek, A.; Westmeyer, G.G.; Drukker, M. Nucleus size and DNA accessibility are linked to the regulation of paraspeckle formation in cellular differentiation. BMC Biol. 2020, 18, 42. [Google Scholar] [CrossRef]
- Naganuma, T.; Nakagawa, S.; Tanigawa, A.; Sasaki, Y.; Goshima, N.; Hirose, T. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012, 31, 4020–4034. [Google Scholar] [CrossRef]
- Kim, S.; Yamamoto, J.; Chen, Y.; Aida, M.; Wada, T.; Handa, H.; Yamaguchi, Y. Evidence that cleavage factor Im is a heterotetrameric protein complex controlling alternative polyadenylation. Genes Cells 2010, 15, 1003–1013. [Google Scholar] [CrossRef] [PubMed]
- Naganuma, T.; Hirose, T. Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol. 2013, 10, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Machitani, M.; Taniguchi, I.; Ohno, M. ARS2 Regulates Nuclear Paraspeckle Formation through 3′-End Processing and Stability of NEAT1 Long Noncoding RNA. Mol. Cell. Biol. 2020, 40, e00269-19. [Google Scholar] [CrossRef]
- Modic, M.; Grosch, M.; Rot, G.; Schirge, S.; Lepko, T.; Yamazaki, T.; Lee, F.C.; Rusha, E.; Shaposhnikov, D.; Palo, M.; et al. Cross-Regulation between TDP-43 and Paraspeckles Promotes Pluripotency-Differentiation Transition. Mol. Cell 2019, 74, 951–965.e13. [Google Scholar] [CrossRef]
- Tollervey, J.R.; Curk, T.; Rogelj, B.; Briese, M.; Cereda, M.; Kayikci, M.; König, J.; Hortobágyi, T.; Nishimura, A.L.; Župunski, V.; et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 2011, 14, 452–458. [Google Scholar] [CrossRef]
- Shelkovnikova, T.A.; Kukharsky, M.S.; An, H.; DiMasi, P.; Alexeeva, S.; Shabir, O.; Heath, P.R.; Buchman, V.L. Protective paraspeckle hyper-assembly downstream of TDP-43 loss of function in amyotrophic lateral sclerosis. Mol. Neurodegener. 2018, 13, 30. [Google Scholar] [CrossRef] [PubMed]
- Nishimoto, Y.; Nakagawa, S.; Okano, H. NEAT1 lncRNA and amyotrophic lateral sclerosis. Neurochem. Int. 2021, 150, 105175. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, E.N.; Wang, H.; Mitra, J.; Hegde, P.M.; Stowell, S.E.; Liachko, N.; Kraemer, B.C.; Garruto, R.M.; Rao, K.; Hegde, M.L. TDP-43/FUS in motor neuron disease: Complexity and challenges. Prog. Neurobiol. 2016, 145–146, 78–97. [Google Scholar] [CrossRef]
- Suzuki, H.; Shibagaki, Y.; Hattori, S.; Matsuoka, M. C9-ALS/FTD-linked proline–arginine dipeptide repeat protein associates with paraspeckle components and increases paraspeckle formation. Cell Death Dis. 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Ma, X.; Ying, Y.; Xie, H.; Liu, X.; Wang, X.; Li, J. The Regulatory Role of RNA Metabolism Regulator TDP-43 in Human Cancer. Front. Oncol. 2021, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Fan, Z.; McGee, W.; Chen, M.; Kong, R.; Wen, P.; Xiao, T.; Chen, X.; Liu, J.; Zhu, L.; et al. TDP-43 regulates cancer-associated microRNAs. Protein Cell 2018, 9, 848–866. [Google Scholar] [CrossRef]
- Spinelli, J.B.; Haigis, M.C. The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell Biol. 2018, 20, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Quiros, P.M.; Mottis, A.; Auwerx, J. Mitonuclear communication in homeostasis and stress. Nat. Rev. Mol. Cell Biol. 2016, 17, 213–226. [Google Scholar] [CrossRef]
- Rizzuto, R.; De Stefani, D.; Raffaello, A.; Mammucari, C. Mitochondria as sensors and regulators of calcium signalling. Nat. Rev. Mol. Cell Biol. 2012, 13, 566–578. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, J.; Zhong, Y.; Zhang, Y.; Yang, L.; Wu, P.; Hou, X.; Xiong, F.; Li, X.; Zhang, S.; Gong, Z.; et al. Long non-coding RNAs are involved in alternative splicing and promote cancer progression. Br. J. Cancer 2022, 126, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.; Kanhere, A. The Missing Link Between Cancer-Associated Variants and LncRNAs. Trends Genet. 2021, 37, 410–413. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Baas, M.; Diederichs, S. Noncoding RNA gene silencing through genomic integration of RNA destabilizing elements using zinc finger nucleases. Genome Res. 2011, 21, 1944–1954. [Google Scholar] [CrossRef] [PubMed]
- Meseure, D.; Vacher, S.; Lallemand, F.; Alsibai, K.D.; Hatem, R.; Chemlali, W.; Nicolas, A.; De Koning, L.; Pasmant, E.; Callens, C.; et al. Prognostic value of a newly identified MALAT1 alternatively spliced transcript in breast cancer. Br. J. Cancer 2016, 114, 1395–1404. [Google Scholar] [CrossRef]
- Wilusz, J.E.; Freier, S.M.; Spector, D.L. 3′ End Processing of a Long Nuclear-Retained Noncoding RNA Yields a tRNA-like Cytoplasmic RNA. Cell 2008, 135, 919–932. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Huang, J.; Wu, S.; Zheng, Q.; Liu, P.; Feng, H.; Su, X.; Fu, H.; Xi, Q.; Wang, G. The tRNA -like small noncoding RNA masc RNA promotes global protein translation. EMBO Rep. 2020, 21, e49684. [Google Scholar] [CrossRef]
- Xie, S.-J.; Diao, L.-T.; Cai, N.; Zhang, L.-T.; Xiang, S.; Jia, C.-C.; Qiu, D.-B.; Liu, C.; Sun, Y.-J.; Lei, H.; et al. mascRNA and its parent lncRNA MALAT1 promote proliferation and metastasis of hepatocellular carcinoma cells by activating ERK/MAPK signaling pathway. Cell Death Discov. 2021, 7, 1–14. [Google Scholar] [CrossRef]
- Hoffmann, M.J.; Dehn, J.; Droop, J.; Niegisch, G.; Niedworok, C.; Szarvas, T.; Schulz, W.A. Truncated Isoforms of lncRNA ANRIL Are Overexpressed in Bladder Cancer, But Do Not Contribute to Repression of INK4 Tumor Suppressors. Non-Coding RNA 2015, 1, 266–284. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Qiao, F.; Tu, J.; Xu, J.; Ding, F.; Liu, Y.; Akuo, B.A.; Hu, J.; Shao, S. High expression of long non-coding RNA NEAT1 indicates poor prognosis of human cancer. Oncotarget 2017, 8, 45918–45927. [Google Scholar] [CrossRef]
- Yang, C.; Li, Z.; Li, Y.; Xu, R.; Wang, Y.; Tian, Y.; Chen, W. Long non-coding RNA NEAT1 overexpression is associated with poor prognosis in cancer patients: A systematic review and meta-analysis. Oncotarget 2017, 8, 2672–2680. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, D.; Sboner, A.; Nair, S.S.; Giannopoulou, E.G.; Li, R.; Hennig, S.; Mosquera, J.M.; Pauwels, J.; Park, K.; Kossai, M.; et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat. Commun. 2014, 5, 5383. [Google Scholar] [CrossRef]
- Wu, Y.; Yang, L.; Zhao, J.; Li, C.; Nie, J.; Liu, F.; Zhuo, C.; Zheng, Y.; Li, B.; Wang, Z.; et al. Nuclear-enriched abundant transcript 1 as a diagnostic and prognostic biomarker in colorectal cancer. Mol. Cancer 2015, 14, 191. [Google Scholar] [CrossRef] [PubMed]
- Xianguo, C.; Zongyao, H.; Jun, Z.; Song, F.; Guangyue, L.; Ligang, Z.; Kaiping, Z.; Yangyang, Z.; ChaoZhao, L. Promoting progression and clinicopathological significance of NEAT1 over-expression in bladder cancer. Oncotarget 2016, 5. [Google Scholar] [CrossRef]
- Lu, Y.; Li, T.; Wei, G.; Liu, L.; Chen, Q.; Xu, L.; Zhang, K.; Zeng, D.; Liao, R. The long non-coding RNA NEAT1 regulates epithelial to mesenchymal transition and radioresistance in through miR-204/ZEB1 axis in nasopharyngeal carcinoma. Tumor Biol. 2016, 37, 11733–11741. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, Y.; Chen, W.; He, F.; Tan, Z.; Zheng, J.; Wang, W.; Zhao, Q.; Li, J. NEAT expression is associated with tumor recurrence and unfavorable prognosis in colorectal cancer. Oncotarget 2015, 6, 27641–27650. [Google Scholar] [CrossRef]
- Vancura, A.; Lanzós, A.; Bosch-Guiteras, N.; Esteban, M.T.; Gutierrez, A.H.; Haefliger, S.; Johnson, R. Cancer LncRNA Census 2 (CLC2): An enhanced resource reveals clinical features of cancer lncRNAs. NAR Cancer 2021, 3, zcab013. [Google Scholar] [CrossRef]
- Zeng, C.; Xu, Y.; Xu, L.; Yu, X.; Cheng, J.; Yang, L.; Chen, S.; Li, Y. Inhibition of long non-coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC Cancer 2014, 14, 693. [Google Scholar] [CrossRef] [PubMed]
- Adriaens, C.; Standaert, L.; Barra, J.; Latil, M.; Verfaillie, A.; Kalev, P.; Boeckx, B.; Wijnhoven, P.W.G.; Radaelli, E.; Vermi, W.; et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat. Med. 2016, 22, 861–868. [Google Scholar] [CrossRef] [PubMed]
- Ronchetti, D.; Favasuli, V.; Monti, P.; Cutrona, G.; Fabris, S.; Silvestris, I.; Agnelli, L.; Colombo, M.; Menichini, P.; Matis, S.; et al. NEAT1 Long Isoform Is Highly Expressed in Chronic Lymphocytic Leukemia Irrespectively of Cytogenetic Groups or Clinical Outcome. Non-Coding RNA 2020, 6, 11. [Google Scholar] [CrossRef]
- Zeng, C.; Liu, S.; Lu, S.; Yu, X.; Lai, J.; Wu, Y.; Chen, S.; Wang, L.; Yu, Z.; Luo, G.; et al. The c-Myc-regulated lncRNA NEAT1 and paraspeckles modulate imatinib-induced apoptosis in CML cells. Mol. Cancer 2018, 17, 130. [Google Scholar] [CrossRef]
- Duan, M.-Y.; Li, M.; Tian, H.; Tang, G.; Yang, Y.-C.; Peng, N.-C. Down-regulation of lncRNA NEAT1 regulated by miR-194-5p/DNMT3A facilitates acute myeloid leukemia. Blood Cells Mol. Dis. 2020, 82, 102417. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Liu, N.; Chen, X.; Liu, Y.; An, J. Long non-coding RNA NEAT1/miR-338-3p axis impedes the progression of acute myeloid leukemia via regulating CREBRF. Cancer Cell Int. 2020, 20, 112. [Google Scholar] [CrossRef]
- Taiana, E.; Ronchetti, D.; Favasuli, V.; Todoerti, K.; Manzoni, M.; Amodio, N.; Tassone, P.; Agnelli, L.; Neri, A. Long non-coding RNA NEAT1 shows high expression unrelated to molecular features and clinical outcome in multiple myeloma. Haematologica 2019, 104, e72–e76. [Google Scholar] [CrossRef]
- Blume, C.J.; Hotz-Wagenblatt, A.; Hüllein, J.; Sellner, L.; Jethwa, A.; Stolz, T.; Slabicki, M.; Lee, K.; Sharathchandra, A.; Benner, A.; et al. p53-dependent non-coding RNA networks in chronic lymphocytic leukemia. Leukemia 2015, 29, 2015–2023. [Google Scholar] [CrossRef]
- Zhao, L.; Zhou, N.; Zhao, P. Expression level of NEAT1 differentiates benign and malignant thyroid nodules by regulating NEAT1/miR-9/PTEN and NEAT1/miR-124/PDCD6 signalling. Int. J. Mol. Med. 2020, 46, 1661–1670. [Google Scholar] [CrossRef]
- Li, J.-H.; Zhang, S.-Q.; Qiu, X.-G.; Zhang, S.-J.; Zheng, S.-H.; Zhang, D.-H. Long non-coding RNA NEAT1 promotes malignant progression of thyroid carcinoma by regulating miRNA-214. Int. J. Oncol. 2016, 50, 708–716. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Wang, P.; Lou, J.; Zhao, J. Knockdown of lncRNA NEAT1 suppresses hypoxia-induced migration, invasion and glycolysis in anaplastic thyroid carcinoma cells through regulation of miR-206 and miR-599. Cancer Cell Int. 2020, 20, 132. [Google Scholar] [CrossRef]
- Zhang, H.; Cai, Y.; Zheng, L.; Zhang, Z.; Lin, X.; Jiang, N. Long noncoding RNA NEAT1 regulate papillary thyroid cancer progression by modulating miR-129-5p/ KLK7 expression. J. Cell. Physiol. 2018, 233, 6638–6648. [Google Scholar] [CrossRef] [PubMed]
- Naveed, A.; Cooper, J.A.; Li, R.; Hubbard, A.; Chen, J.; Liu, T.; Wilton, S.D.; Fletcher, S.; Fox, A.H. NEAT1 polyA-modulating antisense oligonucleotides reveal opposing functions for both long non-coding RNA isoforms in neuroblastoma. Cell. Mol. Life Sci. 2021, 78, 2213–2230. [Google Scholar] [CrossRef]
- Jiang, X.; Zhou, Y.; Sun, A.-J.; Xue, J.-L. NEAT1 contributes to breast cancer progression through modulating miR-448 and ZEB1. J. Cell. Physiol. 2018, 233, 8558–8566. [Google Scholar] [CrossRef] [PubMed]
- Knutsen, E.; Oslo Breast Cancer Research Consortium (OSBREAC); Lellahi, S.M.; Aure, M.R.; Nord, S.; Fismen, S.; Larsen, K.B.; Gabriel, M.T.; Hedberg, A.; Bjørklund, S.S.; et al. The expression of the long NEAT1_2 isoform is associated with human epidermal growth factor receptor 2-positive breast cancers. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Choudhry, H.; Albukhari, A.; Morotti, M.; Haider, S.; Moralli, D.; Smythies, J.; Schödel, J.; Green, C.; Camps, C.; Buffa, F.; et al. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2α dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene 2015, 34, 4482–4490. [Google Scholar] [CrossRef]
- Bhatt, U.; Kretzmann, A.L.; Guédin, A.; Ou, A.; Kobelke, S.; Bond, C.S.; Evans, C.W.; Hurley, L.H.; Mergny, J.-L.; Iyer, K.S.; et al. The role of G-Quadruplex DNA in Paraspeckle formation in cancer. Biochimie 2021, 190, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Harvey, A.R.; Hodgetts, S.I.; Fox, A.H. Functional dissection of NEAT1 using genome editing reveals substantial localization of the NEAT1_1 isoform outside paraspeckles. RNA 2017, 23, 872–881. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Zhao, L. lncRNA nuclear-enriched abundant transcript 1 promotes cell proliferation and invasion by targeting miR-186-5p/HIF-1α in osteosarcoma. J. Cell. Biochem. 2019, 120, 6502–6514. [Google Scholar] [CrossRef]
- Xu, H.; Sun, X.; Huang, Y.; Si, Q.; Li, M. Long non-coding RNA NEAT1 modifies cell proliferation, colony formation, apoptosis, migration and invasion via the miR-4500/BZW1 axis in ovarian cancer. Mol. Med. Rep. 2020, 22, 3347–3357. [Google Scholar] [CrossRef]
- An, J.; Lv, W.; Zhang, Y. LncRNA NEAT1 contributes to paclitaxel resistance of ovarian cancer cells by regulating ZEB1 expression via miR-194. OncoTargets Ther. 2017, 10, 5377–5390. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xu, W.; Sun, K.; Wang, F.; Wong, T.W.; Kong, A. Identification of novel biomarkers in prostate cancer diagnosis and prognosis. J. Biochem. Mol. Toxicol. 2022. epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; Wei, Y.; Zen, C.; Xiong, W.; Niu, Y.; Zhao, Y. Long non-coding RNA NEAT1 promotes bone metastasis of prostate cancer through N6-methyladenosine. Mol. Cancer 2020, 19, 171. [Google Scholar] [CrossRef] [PubMed]
- Shaker, O.G.; Abdelwahed, M.Y.; Ahmed, N.A.; Hassan, E.A.; Ahmed, T.I.; Abousarie, M.A.; Ayoub, S.E. Evaluation of serum long noncoding RNA NEAT and MiR-129-5p in hepatocellular carcinoma. IUBMB Life 2019, 71, 1571–1578. [Google Scholar] [CrossRef]
- Wang, Z.; Zou, Q.; Song, M.; Chen, J. NEAT1 promotes cell proliferation and invasion in hepatocellular carcinoma by negative regulating miR-613 expression. Biomed. Pharmacother. 2017, 94, 612–618. [Google Scholar] [CrossRef]
- Zheng, X.; Zhang, Y.; Liu, Y.; Fang, L.; Li, L.; Sun, J.; Pan, Z.; Xin, W.; Huang, P. HIF-2α activated lncRNA NEAT1 promotes hepatocellular carcinoma cell invasion and metastasis by affecting the epithelial-mesenchymal transition. J. Cell. Biochem. 2018, 119, 3247–3256. [Google Scholar] [CrossRef]
- Fu, J.-W.; Kong, Y.; Sun, X. Long noncoding RNA NEAT1 is an unfavorable prognostic factor and regulates migration and invasion in gastric cancer. J. Cancer Res. Clin. Oncol. 2016, 142, 1571–1579. [Google Scholar] [CrossRef] [PubMed]
- Jen, J.; Tang, Y.-A.; Lu, Y.-H.; Lin, C.-C.; Lai, W.-W.; Wang, Y.-C. Oct4 transcriptionally regulates the expression of long non-coding RNAs NEAT1 and MALAT1 to promote lung cancer progression. Mol. Cancer 2017, 16, 104. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, K.; Wang, R.; Chen, S.; Wu, J.; Li, X.; Ning, Q.; Yang, G.; Pang, Y. The interplay between ATF2 and NEAT1 contributes to lung adenocarcinoma progression. Cancer Cell Int. 2020, 20, 1–13. [Google Scholar] [CrossRef]
- Wu, F.; Mo, Q.; Wan, X.; Dan, J.; Hu, H. NEAT1/hsa-mir-98-5p/MAPK6 axis is involved in non–small-cell lung cancer development. J. Cell. Biochem. 2019, 120, 2836–2846. [Google Scholar] [CrossRef]
- Zhang, M.; Weng, W.; Zhang, Q.; Wu, Y.; Ni, S.; Tan, C.; Xu, M.; Sun, H.; Liu, C.; Wei, P.; et al. The lncRNA NEAT1 activates Wnt/β-catenin signaling and promotes colorectal cancer progression via interacting with DDX5. J. Hematol. Oncol. 2018, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Du, S.; Yin, K.; Ai, S.; Yu, M.; Liu, Y.; Shen, Y.; Liu, M.; Jiao, R.; Chen, X.; et al. Knockdown long noncoding RNA nuclear paraspeckle assembly transcript 1 suppresses colorectal cancer through modulating miR-193a-3p/KRAS. Cancer Med. 2019, 8, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Chen, J.-J.; Lv, Q.; Qin, J.; Huang, Y.-Z.; Yu, M.-H.; Zhong, M. Long non-coding RNA NEAT1 promotes colorectal cancer progression by competitively binding miR-34a with SIRT1 and enhancing the Wnt/β-catenin signaling pathway. Cancer Lett. 2019, 440, 11–22. [Google Scholar] [CrossRef]
- Liu, Z.; Wu, K.; Wu, J.; Tian, D.; Chen, Y.; Yang, Z.; Wu, A. NEAT1 is a potential prognostic biomarker for patients with nasopharyngeal carcinoma. J. Cell. Biochem. 2019, 120, 9831–9838. [Google Scholar] [CrossRef]
- Gao, C.; Zhang, Y.; Sun, H. Mechanism of miR-340–5p in laryngeal cancer cell proliferation and invasion through the lncRNA NEAT1/MMP11 axis. Pathol. Res. Pract. 2022, 236, 153912. [Google Scholar] [CrossRef]
- Wang, P.; Wu, T.; Zhou, H.; Jin, Q.; He, G.; Yu, H.; Xuan, L.; Wang, X.; Tian, L.; Sun, Y.; et al. Long noncoding RNA NEAT1 promotes laryngeal squamous cell cancer through regulating miR-107/CDK6 pathway. J. Exp. Clin. Cancer Res. 2016, 35, 22. [Google Scholar] [CrossRef]
- Li, Y.; Chen, D.; Gao, X.; Li, X.; Shi, G. LncRNA NEAT1 Regulates Cell Viability and Invasion in Esophageal Squamous Cell Carcinoma through the miR-129/CTBP2 Axis. Dis. Markers 2017, 2017, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mello, S.S.; Sinow, C.; Raj, N.; Mazur, P.K.; Bieging-Rolett, K.; Broz, D.K.; Imam, J.F.C.; Vogel, H.; Wood, L.D.; Sage, J.; et al. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev. 2017, 31, 1095–1108. [Google Scholar] [CrossRef]
- Olivier, M.; Hollstein, M.; Hainaut, P. TP53 Mutations in Human Cancers: Origins, Consequences, and Clinical Use. Cold Spring Harb. Perspect. Biol. 2010, 2, a001008. [Google Scholar] [CrossRef] [PubMed]
- Botcheva, K.; McCorkle, S.; McCombie, W.R.; Dunn, J.J.; Anderson, C.W. Distinct p53 genomic binding patterns in normal and cancer-derived human cells. Cell Cycle 2011, 10, 4237–4249. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nik-Zainal, S.; Davies, H.; Staaf, J.; Ramakrishna, M.; Glodzik, D.; Zou, X.; Martincorena, I.; Alexandrov, L.B.; Martin, S.; Wedge, D.C.; et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016, 534, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Rheinbay, E.; Parasuraman, P.; Grimsby, J.; Tiao, G.; Engreitz, J.M.; Kim, J.; Lawrence, M.S.; Taylor-Weiner, A.; Rodriguez-Cuevas, S.; Rosenberg, M.; et al. Recurrent and functional regulatory mutations in breast cancer. Nature 2017, 547, 55–60. [Google Scholar] [CrossRef]
- Wedge, D.C.; CAMCAP Study Group; Gundem, G.; Mitchell, T.; Woodcock, D.J.; Martincorena, I.; Ghori, M.; Zamora, J.; Butler, A.; Whitaker, H.; et al. Sequencing of prostate cancers identifies new cancer genes, routes of progression and drug targets. Nat. Genet. 2018, 50, 682–692. [Google Scholar] [CrossRef] [PubMed]
- Rheinbay, E.; Nielsen, M.M.; Abascal, F.; Wala, J.A.; Shapira, O.; Tiao, G.; Hornshøj, H.; Hess, J.M.; Juul, R.I.; Lin, Z.; et al. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature 2020, 578, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Kolenda, T.; Ryś, M.; Guglas, K.; Teresiak, A.; Bliźniak, R.; Mackiewicz, J.; Lamperska, K. Quantification of long non-coding RNAs using qRT-PCR: Comparison of different cDNA synthesis methods and RNA stability. Arch. Med Sci. 2021, 17, 1006–1015. [Google Scholar] [CrossRef] [PubMed]
- Chujo, T.; Yamazaki, T.; Kawaguchi, T.; Kurosaka, S.; Takumi, T.; Nakagawa, S.; Hirose, T. Unusual semi-extractability as a hallmark of nuclear body-associated architectural noncoding RNAs. EMBO J. 2017, 36, 1447–1462. [Google Scholar] [CrossRef]
- Gleeson, J.; Leger, A.; Prawer, Y.D.J.; A Lane, T.; Harrison, P.J.; Haerty, W.; Clark, M.B. Accurate expression quantification from nanopore direct RNA sequencing with NanoCount. Nucleic Acids Res. 2022, 50, e19. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.-Z.; Wang, Y.; Li, S.-Q.; Yao, R.-W.; Luan, P.-F.; Wu, H.; Carmichael, G.G.; Chen, L.-L. Dynamic Imaging of RNA in Living Cells by CRISPR-Cas13 Systems. Mol. Cell 2019, 76, 981–997.e7. [Google Scholar] [CrossRef] [PubMed]

| Cancer Type | n/Cell Line/Sample Type | NEAT1 Isoforms Investigated | Major Findings | RT-PCR Control Gene | Year | Ref |
|---|---|---|---|---|---|---|
| Multiple myeloma (MM) | n = 46 MM, n = 14 plasma cell leukaemia (PCL) n = 628 from publicly available datasets: #GSE5900 (44 MM, 12 MGUS, 22 healthy donors) #GSE2658 and #GSE24080 | Total NEAT1 and NEAT1_2 NEAT1_1 and NEAT1_2 with RNAseq | RNAseq allowed estimated isoform abundance was based on unambiguously mapped reads. NEAT1 in tumour samples when compared to healthy controls 90% of total NEAT1 was NEAT1_1. A negative correlation was found between—4NEAT1 and UPR. Neither total NEAT1 nor NEAT1_2 correlated with overall survival or time-to-next-treatment. NEAT1 was not found to be differentially expressed in diverse cell types, i.e., primary vs. secondary cell leukaemia. | Undisclosed | 2019 | [81] |
| n = 82 blood samples | Total NEAT1 and NEAT1_2 | NEAT1_1, inferred by the difference in Ct value between total NEAT1 and NEAT1_2. | GAPDH | 2020 | [77] | |
| B-cell acute lymphoblastic leukaemia (ALL) | n = 16 blood samples | NEAT1_1, inferred by the difference in Ct value between total NEAT1 and NEAT1_2. NEAT1_1 and NEAT1_2. | ||||
| Acute myeloid leukaemia (AML) | n = 20 blood samples | NEAT1_1, inferred by the difference in Ct value between total NEAT1 and NEAT1_2. NEAT1_1 and NEAT1_2. | ||||
| Chronic lymphocytic leukaemia (CLL) | n = 310 blood samples | NEAT1_1, inferred by the difference in Ct value between total NEAT1 and NEAT1_2. Stable total NEAT1 but NEAT1_2 (40% of total NEAT1). NEAT1_2 in patients with IGHV gene mutations. NEAT1_2 in patients with Trisomy 12. | ||||
| n = 72 peripheral blood samples | Total NEAT1 | p53 binds to the NEAT1 promotor in CLL and lymphoma. p21 and NEAT1 expression levels significantly correlated after irradiation. Nutlin-3 induced NEAT1 expression 2.3-fold in WT p53 primary CLL cells, compared to 1.2-fold in p53 mutant cells. | Lamin B1 | 2015 | [82] | |
| Chronic myeloid leukaemia (CML) | n = 26 peripheral blood samples | Total NEAT1 and NEAT1_2 | Total NEAT1 and NEAT1_2. Silencing BCR-ABL expression total NEAT1 and NEAT1_2 in CML cell line K562, suggesting NEAT1 may regulate BCR-ABL mediated pathways. c-myc represses NEAT1 transcription by binding to promotor. | ACTB | 2018 | [78] |
| Acute promyelocytic leukaemia (APL) | n = 31 APL and n = 12 normal blood samples NB4, NB4-R2, and U937-PR9 cell lines | Total NEAT1 and NEAT1_2 | Total NEAT1 and NEAT1_2 in APL patient samples when compared to normal granulocytes. NEAT1 expression is repressed by PML-RAR fusion gene. NEAT1 expression is involved in the differentiation of APL cells. | ACTB | 2014 | [75] |
| Thyroid carcinoma (TC) | n = 98 Peripheral blood and thyroid tissue samples (malignant n = 52, benign n = 46) | NEAT1_2 | NEAT1_2 in benign vs. malignant thyroid nodules. | GAPDH | 2020 | [83] |
| Circulating blood monocytes (CBMs) and tumour-associated macrophages (TAMs) n = undisclosed TPC-1 cell line Bone marrow-derived macrophages and macrophages | Undefined | NEAT1 expression in TAMs, compared to CBMs. NEAT1 is a direct target of miR-214 in TC cell lines. Knockdown of NEAT1 impairs malignant progression of thyroid papillary carcinoma and tumour growth in vivo. | GAPDH | 2017 | [84] | |
| Anaplastic thyroid carcinoma (ATC) | n = 25 matched samples SW1736 and KAT-18 cell lines | Total NEAT1 | NEAT1 in ATC tissues and cells exposed to hypoxic conditions. | GAPDH | 2020 | [85] |
| Papillary thyroid carcinoma (PTC) | n = 20 matched samples NPA87, TPC-1, KAT-5, and HT-ori3 (control) cell lines | Total NEAT1 | NEAT1 expression in patient PTC samples when compared to adjacent normal tissues. NEAT1 expression in PTC cell lines compared to control cells. | GAPDH | 2018 | [86] |
| Neuroblastoma | Publicly available datasets (total n = 1062): Versteeg (n = 88), Kocak (n = 476), and SEQC (n = 498) | Total NEAT1 and NEAT1_2 (RT-PCR and RNA-FISH) | NEAT1_1 abundance inferred by subtracting NEAT1_2 levels from total NEAT1 levels. NEAT1_1:NEAT1_2 in aggressive neuroblastoma. NEAT1_2 and paraspeckles in nonaggressive neuroblastoma. | RPLP0 | 2021 | [87] |
| Breast cancer (BC) | MCF-7, MDA-MB-453, MDA-MB-231, SKBR3, and MCF-10A (control) cell lines | Total NEAT1 | NEAT1 in all cancer cell lines when compared to control cell line. NEAT1 was negatively correlated with miR-448. | GAPDH | 2018 | [88] |
| n = 1065 post-data filtering of TCGA (n = 526), Oslo2 (n = 378), and METABRIC (n = 1904) cohorts. BT474, BT549, HCC1569, Hs578T, MDA-MB-231, MDA-MB-468, MCF7, SK-BR-3, and T-47D cell lines n = 74 BC biopsies and n = 27 non-malignant biopsies | NEAT1_2 NEAT1_1 with RNAseq NEAT1_2 with RNA-FISH | NEAT1_1 expression level determined from polyA-selected RNAseq data from TCGA cohort. NEAT1_1 expression is highest in ER-positive luminal A and B breast cancer. NEAT1_2 and paraspeckle abundance correlate with high-grade disease (RNA-FISH). NEAT1_2 in HER2-enriched and luminal B BC in all three cohorts. NEAT1_2 is not expressed at RNA-FISH-detectable levels in normal breast tissue. | Geometric mean of GAPDH, B2M, and RPLP0 | 2020 | [89] | |
| MCF-7, MDA-MB-231, and MDA-MB-468 cell lines Gene expression data n = 2000 | NEAT1_1 and NEAT1_2 | NEAT1_1 transcription was analysed by using a polyA primer for cDNA generation before RT-qPCR, using primers targeting total NEAT1. NEAT1_2 transcription was analysed by using random primers for cDNA generation before primers specifically targeting the NEAT1_2 region of the transcript. NEAT1 associated with poor patient prognosis. | RPL11 | 2015 | [90] | |
| MDA-MB-231 and MCF-10A (control) cell lines | NEAT1_2 with RNA-FISH | Paraspeckle formation in MCF-7 cell lines when compared to MCF-10A cells. NEAT1_2 expression after G-quadruplex (G4)-specific stabilization with small molecules. NEAT1_2 expression could be regulated by a G4s. | GAPDH and ACTB | 2021 | [91] | |
| Osteosarcoma (OS) | U2OS cell line | Total NEAT1 and NEAT1_2 (RT-qPCR and RNA-FISH). NEAT1_1 in NEAT1_2 KO cells | NEAT1 isoform-specific KO cell lines were achieved using CRISPR-Cas9 technologies. NEAT1_1 levels were unaltered or increased in some NEAT1_2−/− lines. NEAT1_1 localises to nuclear speckles, independent of paraspeckles. | RPLP0 | 2017 | [92] |
| n = 47 biopsies and adjacent matched tissues HOS, SaOS2, MG63, U2OS, and hFOB1.19 (control) cell lines | Total NEAT1 | NEAT1 expression HIF-1 in MG63 cells, and this NEAT1-mediated HIF-1 expression was reversed by miR-186-5p in HOS cells. NEAT1 in OS tissues and cell lines. NEAT1 associated with advanced clinicopathologic features and poor overall survival. NEAT1 promotes proliferation, invasion, and EMT in cell lines. NEAT1 promoted growth in vivo. miR-186-5p is a downstream target of NEAT1 in osteosarcoma. | GAPDH | 2019 | [93] | |
| U2OS cell line | Total NEAT1 and NEAT1_2 | Total NEAT1 levels were slightly higher in CBP80-KD and ARS2-KD cells when compared to control KD cells. NEAT1_2 alone 5-fold in ARS2-KD cells, but not in CBP80- or PHAX-KD cells. ARS2 suppresses the formation of paraspeckles. | GAPDH | 2020 | [47] | |
| Ovarian cancer (OC) | n = 30 paired tissue samples; CAOV3, ES-2, and IOSE80 (control) cell lines | Total NEAT1 | NEAT1 in patient samples and OC cell lines. NEAT1 knockdown with siRNA increased apoptosis and decreased proliferation, colony formation, migration, invasion, and glycolysis. | GAPDH | 2020 | [94] |
| ovarian carcinoma patient specimens (n = 18 responsive, n = 14 resistant) SKOV3 HeyA-8, PTX-resistant, SKOV3/PTX, and HeyA-8/PTX cell lines. n = 10 BALB/c athymic mice | Total NEAT1 | NEAT1 in treatment-resistant patients when compared to treatment-responsive patients. NEAT1 knockdown enhanced PTX sensitivity in PTX-resistant OC cells. NEAT1 negatively regulates miR-194 expression. NEAT1 sponges miR-194, leading to upregulation of ZEB1 expression. NEAT1 knockdown improved sensitivity to PTX in OC in vivo. | GAPDH | 2017 | [95] | |
| Prostate cancer (PC) | LNCaP, DU145, and RWPE-1 (control) cell lines | Total NEAT1 | NEAT1 in PCa cells. NEAT1 negatively regulates hsa-miR-218-5p and has-miR-483-3p when compared to normal prostate epithelial cells. | GAPDH | 2022 | [96] |
| Explant cultures from primary, patient-derived bone metastatic prostate panel Primary prostate and bone metastatic tissues Patient-derived xenograft TCGA datasets | Total NEAT1 Total NEAT1_1 (RNA-FISH) | NEAT1 in prostate cancer when compared to normal tissues (from TCGA datasets). NEAT1_1 predicts poor patient prognosis. NEAT1_1 enhances prostate-patient-derived xenograft growth through the post-transcriptional RNA modification N6-methyladenosine (m6A). m6A level of NEAT1_1 correlated to prostate cancer progression and bone metastasis, and negatively correlated to patient survival. | GAPDH | 2020 | [97] | |
| Hepatocellular carcinoma (HCC) | blood samples (n = 36 HCC, n = 36 controls) | Total NEAT1 | NEAT1 in HCC patient samples when compared to healthy controls. miR-129-5p negatively correlated to NEAT1 levels. | GAPDH | 2019 | [98] |
| n = 62 matched biopsies MHCC97H, MHCC97L, SMCC7721 and LO2 (control) cell lines | Total NEAT1 | NEAT1 in HCC tissues compared to adjacent tissues. NEAT1 correlated with tumour size and vascular invasion. NEAT1 knockdown inhibits proliferation, colony formation, and cell invasion in HCC. miR-613 is a target of NEAT1 in HCC. | GAPDH | 2017 | [99] | |
| n = 28 biopsies and adjacent tissues HepG2, MHCC97L, MHCC97H, and LO2 (control) cell lines | Total NEAT1 | NEAT1 expression compared to matched tumour samples. Patients with NEAT1 expression had HIF-2 expression, whilst patients with—NEAT1 expression (though still significantly higher than matched samples) had ¯ HIF-2 expression. | GAPDH | 2018 | [100] | |
| Gastric cancer (GC) | n = 140 samples and n = 20 adjacent tissues NCI-N87, SGC-7901, MKN-45, AGS, and GES-1 (control) cell lines | Total NEAT1 | NEAT1 expression in GC cell lines compared to control cell line. NEAT1 regulates expression of EMT-associated genes in GC cells; in vimentin and N-cadherin, in Zo-1 and E-cadherin; suggests KD of NEAT1 may inhibit EMT. | GAPDH | 2016 | [101] |
| Lung adenocarcinoma (LUAD) | n = 124 biopsies and adjacent tissues A549, CL1-0, and BEAS-2B (control) cell lines | Total NEAT1 | Overexpression rate of NEAT1 in lung cancer samples was 90.3%. Significant positive correlations found between NEAT1 and Oct4 mRNA expression levels. Oct4 directly binds to NEAT1 promoter. Lung cancer cell lines A549 and CL1-0 transiently overexpressing Oct4 induced NEAT1 promoter activity. | GAPDH | 2017 | [102] |
| A549, H460, H1650, H1975, H1299, and NHBE (control) cell lines TCGA database: n = 687 | Total NEAT1 | NEAT1 expression in all cell lines and patient samples when compared to normal tissue and control cell lines. Positive correlation between ATF2 and NEAT1 expression in LUAD tissues. | ACTB | 2020 | [103] | |
| Non-small-cell lung cancer (NSCLC) | A549, H1299, H460, H1975, and BES-2B (control) cell lines | Total NEAT1 | NEAT1 expression in all carcinoma cell lines when compared to control cell lines. NEAT1 promotes growth, migration, and invasion of A549 and H460 cells. NEAT1 directly targets hsa-miR-98-5p, and its expression was significantly downregulated in NSCLC cell lines when compared to normal lung epithelial cell line. MAPK6 is a direct target of hsa-miR-98-5p in NSCLC cells. | GAPDH | 2019 | [104] |
| CRC | n = 30 blood samples, n = 30 controls; validation in n = 100 patients, n = 100 controls. n = 29 matched tissue samples, n = 19, whole blood and tissue samples. HCT116 and LOVO cell lines | Total NEAT1 and NEAT1_2 | Details as to how NEAT1_1 expression was measured were not disclosed. NEAT1 in whole blood of CRC patients when compared to normal controls. Total NEAT1 and NEAT1_2 expression found to be highly accurate in distinguishing CRC patients from normal controls. KD of NEAT1_1 inhibits proliferation and invasion. KD of NEAT1_2 promoted growth. NEAT1 expression was elevated in neutrophils in CRC patients. NEAT1_2 correlated with better overall survival. | ACTB | 2015 | [71] |
| n = 71 tissue samples and n = 61 normal tissue samples from publicly available dataset, RKO, CACO2, SW1116, LOVO, SW480, SW620, HT29, and HCT116 cell lines BALB/c nude mice | Total NEAT1 | NEAT1 associated with poor prognosis in CRC patients. NEAT1 mediates cell proliferation in vitro and tumorigenicity in vivo. KD of NEAT1 significantly inhibited flattening and spreading abilities of HCT116 and SW1116 cells, whilst overexpressing NEAT1 strongly promoted these abilities in HT29 cells. E-cadherin and—N-cadherin expressed at both mRNA and protein levels in NEAT1 KD cells. NEAT1 OE recovered proliferation potential of CRC cell lines which were impaired by simultaneous downregulation of DDX5. DDX5 correlated with NEAT1 expression in 71 CRC samples. | GAPDH | 2018 | [105] | |
| n = 12 paired patient samples SW480, HT29, and Caco2 cell lines Nude mice (n = 5–7 per group) | NEAT1_2 | NEAT1 in CRC tissues is negatively correlated with miR-193a-3p. NEAT1 KD miR-193a-3p expression and attenuates CRC cells. KRAS acts as a target of miR-193a-3p. | GAPDH | 2019 | [106] | |
| GEO databases GSE20916 and GSE9348 n = 100 and adjacent tissue samples SW620, SW480, HCT116, HT29, CaCo-2, LOVO, and Colo205 cell lines | Total NEAT1 | NEAT1 in tumour tissue, when compared to normal tissue, in both the independent datasets and in the matched tissue samples. NEAT1 expression correlated with carcinoembryonic antigen (CEA) levels, tumour size, and distant metastasis. NEAT1 predicts overall survival in CRC patients. NEAT1 regulates cell proliferation and invasion through miR-34a. | GAPDH | 2019 | [107] | |
| Nasopharyngeal carcinoma (NPC) | n = 96 NPC and n = 32 nasopharyngeal epilethium tissues | Total NEAT1 | NEAT1 expression in patient samples when compared to normal tissues. NEAT1 expression was negatively correlated with overall survival of NPC patients. | ACTB | 2019 | [108] |
| Laryngeal cancer (LC) | n = 50 paired patient samples TU686, TU177, AMC-HN-8, and 16HBE (control) cell lines | Total NEAT1 | miR-340-5p OENEAT1 stability via direct binding and consequently NEAT1 expression in LC cells. NEAT1 OE reversed repression of miR-340-5p OE on LC cell proliferation and invasion. | GAPDH | 2022 | [109] |
| Laryngeal squamous cell cancer (LSCC) | n = 52 paired tissue samples Hep-2 cell line | Total NEAT1 | NEAT1 expression in LSCC tumour tissues compared to nonneoplastic tissues. NEAT1 expression correlated with T grade, neck nodal metastasis, and clinical stages of LSCC. NEAT1 knockdown inhibited the growth of LSCC xenografts in mice. NEAT1 knockdown induced apoptosis in LSCC cells in vivo. | ACTB | 2016 | [110] |
| Oesophageal squamous cell carcinoma (OSCC) | EC109, EC9706, and HET-1A (control) cell lines | Total NEAT1 | NEAT1 expression in EC109 and EC9706 cell lines. NEAT1 functions as an endogenous sponge for miR-129. | GAPDH | 2017 | [111] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, N.E.; Spencer-Merris, P.; Fox, A.H.; Petersen, J.; Michael, M.Z. The Long and the Short of It: NEAT1 and Cancer Cell Metabolism. Cancers 2022, 14, 4388. https://doi.org/10.3390/cancers14184388
Smith NE, Spencer-Merris P, Fox AH, Petersen J, Michael MZ. The Long and the Short of It: NEAT1 and Cancer Cell Metabolism. Cancers. 2022; 14(18):4388. https://doi.org/10.3390/cancers14184388
Chicago/Turabian StyleSmith, Nadine E., Phaedra Spencer-Merris, Archa Hannah Fox, Janni Petersen, and Michael Z. Michael. 2022. "The Long and the Short of It: NEAT1 and Cancer Cell Metabolism" Cancers 14, no. 18: 4388. https://doi.org/10.3390/cancers14184388
APA StyleSmith, N. E., Spencer-Merris, P., Fox, A. H., Petersen, J., & Michael, M. Z. (2022). The Long and the Short of It: NEAT1 and Cancer Cell Metabolism. Cancers, 14(18), 4388. https://doi.org/10.3390/cancers14184388









