Liquid Biopsies in Colorectal Liver Metastases: Towards the Era of Precision Oncologic Surgery
Abstract
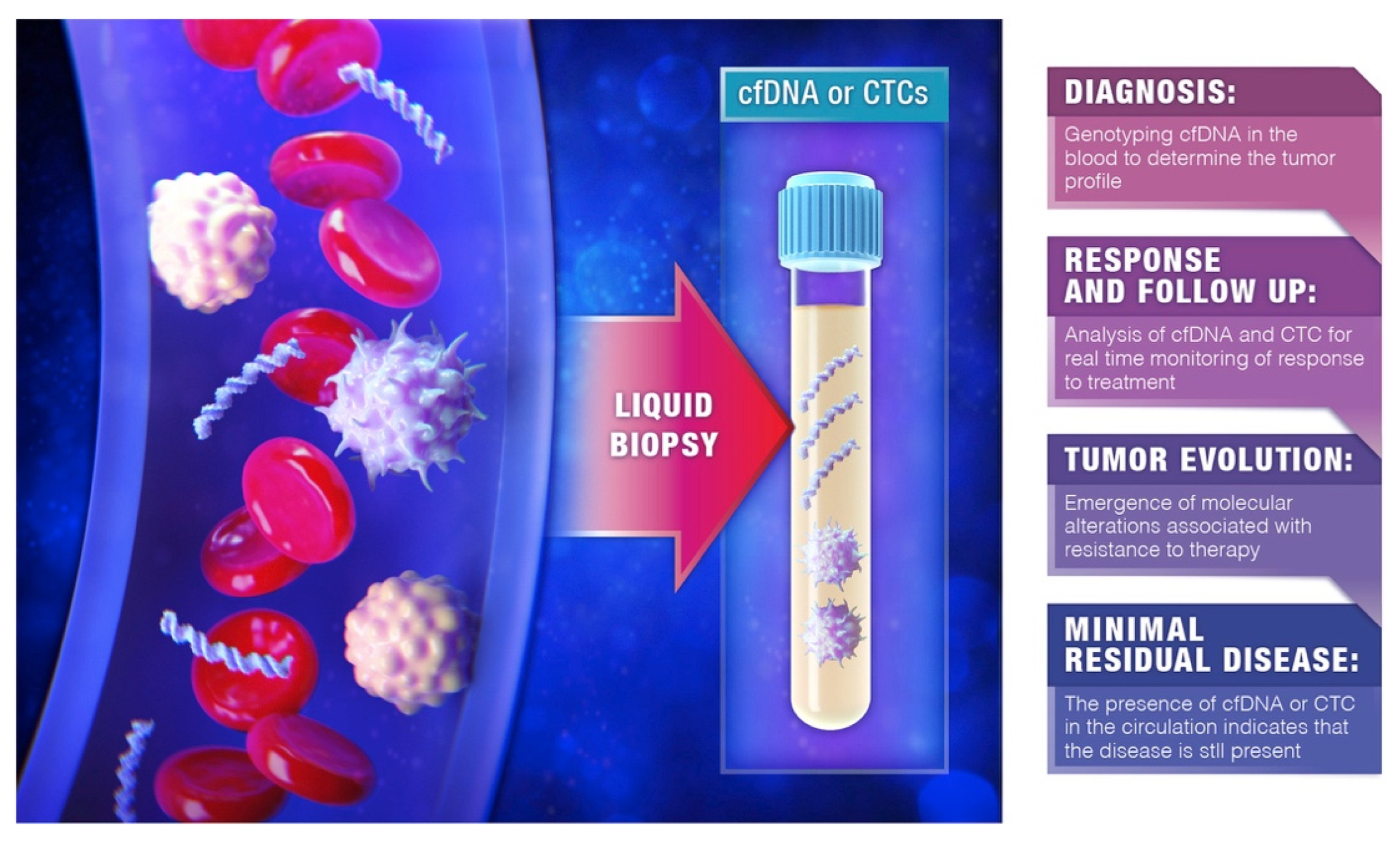
:Simple Summary
Abstract
1. Introduction
2. The Biology behind Liquid Biopsies: CTCs and ctDNA
2.1. CTCs
2.2. ctDNA
3. Liquid Biopsies in CRLM: Experience and Utility
3.1. Prediction of Recurrence following CRLM Resection
3.2. Prediction of Resistance to Treatment
3.3. Real-Time Monitoring of Response to Therapy
3.4. Concordance Rates between Liquid and Tissue Biopsies
4. Challenges and Conclusions
Funding
Conflicts of Interest
References
- Adam, R.; de Gramont, A.; Figueras, J.; Kokudo, N.; Kunstlinger, F.; Loyer, E.; Poston, G.; Rougier, P.; Rubbia-Brandt, L.; Sobrero, A.; et al. Managing synchronous liver metastases from colorectal cancer: A multidisciplinary international consensus. Cancer Treat. Rev. 2015, 41, 729–741. [Google Scholar] [CrossRef]
- Ali, S.M.; Pawlik, T.M.; Rodriguez-Bigas, M.A.; Monson, J.R.T.; Chang, G.J.; Larson, D.W. Timing of Surgical Resection for Curative Colorectal Cancer with Liver Metastasis. Ann. Surg. Oncol. 2018, 25, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Tsilimigras, D.I.; Brodt, P.; Clavien, P.A.; Muschel, R.J.; D’Angelica, M.I.; Endo, I.; Parks, R.W.; Doyle, M.; de Santibanes, E.; Pawlik, T.M. Liver metastases. Nat. Rev. Dis. Primers. 2021, 7, 27. [Google Scholar] [CrossRef] [PubMed]
- de Jong, M.C.; Pulitano, C.; Ribero, D.; Strub, J.; Mentha, G.; Schulick, R.D.; Choti, M.A.; Aldrighetti, L.; Capussotti, L.; Pawlik, T.M. Rates and patterns of recurrence following curative intent surgery for colorectal liver metastasis: An international multi-institutional analysis of 1669 patients. Ann. Surg. 2009, 250, 440–448. [Google Scholar] [CrossRef]
- Tsilimigras, D.I.; Ntanasis-Stathopoulos, I.; Bagante, F.; Moris, D.; Cloyd, J.; Spartalis, E.; Pawlik, T.M. Clinical significance and prognostic relevance of KRAS, BRAF, PI3K and TP53 genetic mutation analysis for resectable and unresectable colorectal liver metastases: A systematic review of the current evidence. Surg. Oncol. 2018, 27, 280–288. [Google Scholar] [CrossRef]
- Gao, W.; Chen, Y.; Yang, J.; Zhuo, C.; Huang, S.; Zhang, H.; Shi, Y. Clinical Perspectives on Liquid Biopsy in Metastatic Colorectal Cancer. Front. Genet. 2021, 12, 634642. [Google Scholar] [CrossRef]
- Raza, A.; Khan, A.Q.; Inchakalody, V.P.; Mestiri, S.; Yoosuf, Z.; Bedhiafi, T.; El-Ella, D.M.A.; Taib, N.; Hydrose, S.; Akbar, S.; et al. Dynamic liquid biopsy components as predictive and prognostic biomarkers in colorectal cancer. J. Exp. Clin. Cancer Res. 2022, 41, 99. [Google Scholar] [CrossRef]
- Zhou, H.; Zhu, L.; Song, J.; Wang, G.; Li, P.; Li, W.; Luo, P.; Sun, X.; Wu, J.; Liu, Y.; et al. Liquid biopsy at the frontier of detection, prognosis and progression monitoring in colorectal cancer. Mol. Cancer 2022, 21, 86. [Google Scholar] [CrossRef]
- Patelli, G.; Vaghi, C.; Tosi, F.; Mauri, G.; Amatu, A.; Massihnia, D.; Ghezzi, S.; Bonazzina, E.; Bencardino, K.; Cerea, G.; et al. Liquid Biopsy for Prognosis and Treatment in Metastatic Colorectal Cancer: Circulating Tumor Cells vs. Circulating Tumor DNA. Target. Oncol. 2021, 16, 309–324. [Google Scholar] [CrossRef]
- Peterson, J.E.; Zurakowski, D.; Italiano, J.E., Jr.; Michel, L.V.; Connors, S.; Oenick, M.; D’Amato, R.J.; Klement, G.L.; Folkman, J. VEGF, PF4 and PDGF are elevated in platelets of colorectal cancer patients. Angiogenesis 2012, 15, 265–273. [Google Scholar] [CrossRef]
- Plantureux, L.; Crescence, L.; Dignat-George, F.; Panicot-Dubois, L.; Dubois, C. Effects of platelets on cancer progression. Thromb. Res. 2018, 164 (Suppl. S1), S40–S47. [Google Scholar] [CrossRef] [PubMed]
- Pantel, K.; Alix-Panabieres, C. Liquid biopsy and minimal residual disease-latest advances and implications for cure. Nat. Rev. Clin. Oncol 2019, 16, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Cohen, J.D.; Lahouel, K.; Lo, S.N.; Wang, Y.; Kosmider, S.; Wong, R.; Shapiro, J.; Lee, M.; Harris, S.; et al. Circulating Tumor DNA Analysis Guiding Adjuvant Therapy in Stage II Colon Cancer. N. Engl. J. Med. 2022, 386, 2261–2272. [Google Scholar] [CrossRef] [PubMed]
- Larson, C.J.; Moreno, J.G.; Pienta, K.J.; Gross, S.; Repollet, M.; O’Hara, S.M.; Russell, T.; Terstappen, L.W. Apoptosis of circulating tumor cells in prostate cancer patients. Cytom. A 2004, 62, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Tripathy, D.; Frenkel, E.P.; Shete, S.; Naftalis, E.Z.; Huth, J.F.; Beitsch, P.D.; Leitch, M.; Hoover, S.; Euhus, D.; et al. Circulating tumor cells in patients with breast cancer dormancy. Clin. Cancer Res. 2004, 10, 8152–8162. [Google Scholar] [CrossRef]
- Deneve, E.; Riethdorf, S.; Ramos, J.; Nocca, D.; Coffy, A.; Daures, J.P.; Maudelonde, T.; Fabre, J.M.; Pantel, K.; Alix-Panabieres, C. Capture of viable circulating tumor cells in the liver of colorectal cancer patients. Clin. Chem. 2013, 59, 1384–1392. [Google Scholar] [CrossRef]
- Kang, Y.; Pantel, K. Tumor cell dissemination: Emerging biological insights from animal models and cancer patients. Cancer Cell 2013, 23, 573–581. [Google Scholar] [CrossRef]
- Smith, H.A.; Kang, Y. The metastasis-promoting roles of tumor-associated immune cells. J. Mol. Med. 2013, 91, 411–429. [Google Scholar] [CrossRef]
- Gay, L.J.; Felding-Habermann, B. Contribution of platelets to tumour metastasis. Nat. Rev. Cancer 2011, 11, 123–134. [Google Scholar] [CrossRef]
- Bonecchi, R.; Galliera, E.; Borroni, E.M.; Corsi, M.M.; Locati, M.; Mantovani, A. Chemokines and chemokine receptors: An overview. Front. Biosci. 2009, 14, 540–551. [Google Scholar] [CrossRef] [Green Version]
- Galindo-Pumarino, C.; Collado, M.; Herrera, M.; Pena, C. Tumor Microenvironment in Metastatic Colorectal Cancer: The Arbitrator in Patients’ Outcome. Cancers 2021, 13, 1130. [Google Scholar] [CrossRef]
- Muller, V.; Stahmann, N.; Riethdorf, S.; Rau, T.; Zabel, T.; Goetz, A.; Janicke, F.; Pantel, K. Circulating tumor cells in breast cancer: Correlation to bone marrow micrometastases, heterogeneous response to systemic therapy and low proliferative activity. Clin. Cancer Res. 2005, 11, 3678–3685. [Google Scholar] [CrossRef] [PubMed]
- Sastre, J.; Orden, V.; Martinez, A.; Bando, I.; Balbin, M.; Bellosillo, B.; Palanca, S.; Peligros Gomez, M.I.; Mediero, B.; Llovet, P.; et al. Association Between Baseline Circulating Tumor Cells, Molecular Tumor Profiling, and Clinical Characteristics in a Large Cohort of Chemo-naive Metastatic Colorectal Cancer Patients Prospectively Collected. Clin. Colorectal Cancer 2020, 19, e110–e116. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabieres, C.; Pantel, K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov. 2016, 6, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabieres, C.; Pantel, K. Challenges in circulating tumour cell research. Nat. Rev. Cancer 2014, 14, 623–631. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabieres, C. Functional Studies on Viable Circulating Tumor Cells. Clin. Chem. 2016, 62, 328–334. [Google Scholar] [CrossRef]
- Alix-Panabieres, C.; Pantel, K. Liquid biopsy in cancer patients: Advances in capturing viable CTCs for functional studies using the EPISPOT assay. Expert Rev. Mol. Diagn. 2015, 15, 1411–1417. [Google Scholar] [CrossRef]
- Scher, H.I.; Lu, D.; Schreiber, N.A.; Louw, J.; Graf, R.P.; Vargas, H.A.; Johnson, A.; Jendrisak, A.; Bambury, R.; Danila, D.; et al. Association of AR-V7 on Circulating Tumor Cells as a Treatment-Specific Biomarker With Outcomes and Survival in Castration-Resistant Prostate Cancer. JAMA Oncol. 2016, 2, 1441–1449. [Google Scholar] [CrossRef]
- Armstrong, A.J.; Halabi, S.; Luo, J.; Nanus, D.M.; Giannakakou, P.; Szmulewitz, R.Z.; Danila, D.C.; Healy, P.; Anand, M.; Rothwell, C.J.; et al. Prospective Multicenter Validation of Androgen Receptor Splice Variant 7 and Hormone Therapy Resistance in High-Risk Castration-Resistant Prostate Cancer: The PROPHECY Study. J. Clin. Oncol. 2019, 37, 1120–1129. [Google Scholar] [CrossRef]
- Heidrich, I.; Ackar, L.; Mossahebi Mohammadi, P.; Pantel, K. Liquid biopsies: Potential and challenges. Int. J. Cancer 2021, 148, 528–545. [Google Scholar] [CrossRef]
- Lo, Y.M.; Zhang, J.; Leung, T.N.; Lau, T.K.; Chang, A.M.; Hjelm, N.M. Rapid clearance of fetal DNA from maternal plasma. Am. J. Hum. Genet. 1999, 64, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Elshimali, Y.I.; Khaddour, H.; Sarkissyan, M.; Wu, Y.; Vadgama, J.V. The clinical utilization of circulating cell free DNA (CCFDNA) in blood of cancer patients. Int. J. Mol. Sci. 2013, 14, 18925–18958. [Google Scholar] [CrossRef] [PubMed]
- Kirsch, C.; Weickmann, S.; Schmidt, B.; Fleischhacker, M. An improved method for the isolation of free-circulating plasma DNA and cell-free DNA from other body fluids. Ann. N. Y. Acad. Sci. 2008, 1137, 135–139. [Google Scholar] [CrossRef]
- Trejo-Becerril, C.; Perez-Cardenas, E.; Taja-Chayeb, L.; Anker, P.; Herrera-Goepfert, R.; Medina-Velazquez, L.A.; Hidalgo-Miranda, A.; Perez-Montiel, D.; Chavez-Blanco, A.; Cruz-Velazquez, J.; et al. Cancer progression mediated by horizontal gene transfer in an in vivo model. PLoS ONE 2012, 7, e52754. [Google Scholar] [CrossRef]
- Poulet, G.; Massias, J.; Taly, V. Liquid Biopsy: General Concepts. Acta Cytol. 2019, 63, 449–455. [Google Scholar] [CrossRef]
- Keller, L.; Belloum, Y.; Wikman, H.; Pantel, K. Clinical relevance of blood-based ctDNA analysis: Mutation detection and beyond. Br. J. Cancer 2021, 124, 345–358. [Google Scholar] [CrossRef]
- Murtaza, M.; Dawson, S.J.; Tsui, D.W.; Gale, D.; Forshew, T.; Piskorz, A.M.; Parkinson, C.; Chin, S.F.; Kingsbury, Z.; Wong, A.S.; et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013, 497, 108–112. [Google Scholar] [CrossRef]
- Heitzer, E.; Ulz, P.; Geigl, J.B. Circulating tumor DNA as a liquid biopsy for cancer. Clin. Chem. 2015, 61, 112–123. [Google Scholar] [CrossRef]
- Bardelli, A.; Pantel, K. Liquid Biopsies, What We Do Not Know (Yet). Cancer Cell 2017, 31, 172–179. [Google Scholar] [CrossRef]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef]
- Underwood, J.J.; Quadri, R.S.; Kalva, S.P.; Shah, H.; Sanjeevaiah, A.R.; Beg, M.S.; Sutphin, P.D. Liquid Biopsy for Cancer: Review and Implications for the Radiologist. Radiology 2020, 294, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabieres, C.; Pantel, K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov. 2021, 11, 858–873. [Google Scholar] [CrossRef] [PubMed]
- Osumi, H.; Shinozaki, E.; Yamaguchi, K.; Zembutsu, H. Clinical utility of circulating tumor DNA for colorectal cancer. Cancer Sci. 2019, 110, 1148–1155. [Google Scholar] [CrossRef] [PubMed]
- Ntanasis-Stathopoulos, I.; Gavriatopoulou, M.; Terpos, E.; Fotiou, D.; Kastritis, E.; Dimopoulos, M.A. Monitoring Plasma Cell Dyscrasias With Cell-free DNA Analysis. Clin. Lymphoma Myeloma Leuk. 2020, 20, e905–e909. [Google Scholar] [CrossRef]
- Garcia-Pardo, M.; Makarem, M.; Li, J.J.N.; Kelly, D.; Leighl, N.B. Integrating circulating-free DNA (cfDNA) analysis into clinical practice: Opportunities and challenges. Br. J. Cancer 2022, 127, 592–602. [Google Scholar] [CrossRef]
- Mason, M.C.; Tzeng, C.D.; Tran Cao, H.S.; Aloia, T.A.; Newhook, T.E.; Overman, M.J.; Kopetz, S.E.; Vauthey, J.N.; Chun, Y.S. Preliminary Analysis of Liquid Biopsy after Hepatectomy for Colorectal Liver Metastases. J. Am. Coll. Surg. 2021, 233, 82–89. [Google Scholar] [CrossRef]
- Nishioka, Y.; Chun, Y.S.; Overman, M.J.; Cao, H.S.T.; Tzeng, C.D.; Mason, M.C.; Kopetz, S.W.; Bauer, T.W.; Vauthey, J.N.; Newhook, T.E.; et al. Effect of Co-mutation of RAS and TP53 on Postoperative ctDNA Detection and Early Recurrence after Hepatectomy for Colorectal Liver Metastases. J. Am. Coll. Surg. 2022, 234, 474–483. [Google Scholar] [CrossRef]
- Reinert, T.; Petersen, L.M.S.; Henriksen, T.V.; Larsen, M.O.; Rasmussen, M.H.; Johansen, A.F.B.; Ogaard, N.; Knudsen, M.; Nordentoft, I.; Vang, S.; et al. Circulating tumor DNA for prognosis assessment and postoperative management after curative-intent resection of colorectal liver metastases. Int. J. Cancer 2022, 150, 1537–1548. [Google Scholar] [CrossRef]
- Scholer, L.V.; Reinert, T.; Orntoft, M.W.; Kassentoft, C.G.; Arnadottir, S.S.; Vang, S.; Nordentoft, I.; Knudsen, M.; Lamy, P.; Andreasen, D.; et al. Clinical Implications of Monitoring Circulating Tumor DNA in Patients with Colorectal Cancer. Clin. Cancer Res. 2017, 23, 5437–5445. [Google Scholar] [CrossRef]
- Ogaard, N.; Reinert, T.; Henriksen, T.V.; Frydendahl, A.; Aagaard, E.; Orntoft, M.W.; Larsen, M.O.; Knudsen, A.R.; Mortensen, F.V.; Andersen, C.L. Tumour-agnostic circulating tumour DNA analysis for improved recurrence surveillance after resection of colorectal liver metastases: A prospective cohort study. Eur. J. Cancer 2022, 163, 163–176. [Google Scholar] [CrossRef]
- Bolhuis, K.; van ‘t Erve, I.; Mijnals, C.; Delis-Van Diemen, P.M.; Huiskens, J.; Komurcu, A.; Lopez-Yurda, M.; van den Broek, D.; Swijnenburg, R.J.; Meijer, G.A.; et al. Postoperative circulating tumour DNA is associated with pathologic response and recurrence-free survival after resection of colorectal cancer liver metastases. EBioMedicine 2021, 70, 103498. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Nakamura, Y.; Taniguchi, H.; Odegaard, J.I.; Nomura, S.; Kojima, M.; Sugimoto, M.; Konishi, M.; Gotohda, N.; Takahashi, S.; et al. Impact of Preoperative Circulating Tumor DNA Status on Survival Outcomes After Hepatectomy for Resectable Colorectal Liver Metastases. Ann. Surg. Oncol. 2021, 28, 4744–4755. [Google Scholar] [CrossRef] [PubMed]
- Polivka, J.; Windrichova, J.; Pesta, M.; Houfkova, K.; Rezackova, H.; Macanova, T.; Vycital, O.; Kucera, R.; Slouka, D.; Topolcan, O. The Level of Preoperative Plasma KRAS Mutations and CEA Predict Survival of Patients Undergoing Surgery for Colorectal Cancer Liver Metastases. Cancers 2020, 12, 2434. [Google Scholar] [CrossRef] [PubMed]
- Groot Koerkamp, B.; Rahbari, N.N.; Buchler, M.W.; Koch, M.; Weitz, J. Circulating tumor cells and prognosis of patients with resectable colorectal liver metastases or widespread metastatic colorectal cancer: A meta-analysis. Ann. Surg. Oncol. 2013, 20, 2156–2165. [Google Scholar] [CrossRef]
- Connor, A.A.; McNamara, K.; Al-Sukhni, E.; Diskin, J.; Chan, D.; Ash, C.; Lowes, L.E.; Allan, A.L.; Zogopoulos, G.; Moulton, C.A.; et al. Central, But Not Peripheral, Circulating Tumor Cells are Prognostic in Patients Undergoing Resection of Colorectal Cancer Liver Metastases. Ann. Surg. Oncol. 2016, 23, 2168–2175. [Google Scholar] [CrossRef]
- van ‘t Erve, I.; Greuter, M.J.E.; Bolhuis, K.; Vessies, D.C.L.; Leal, A.; Vink, G.R.; van den Broek, D.; Velculescu, V.E.; Punt, C.J.A.; Meijer, G.A.; et al. Diagnostic Strategies toward Clinical Implementation of Liquid Biopsy RAS/BRAF Circulating Tumor DNA Analyses in Patients with Metastatic Colorectal Cancer. J. Mol. Diagn. 2020, 22, 1430–1437. [Google Scholar] [CrossRef]
- Yao, J.; Zang, W.; Ge, Y.; Weygant, N.; Yu, P.; Li, L.; Rao, G.; Jiang, Z.; Yan, R.; He, L.; et al. RAS/BRAF Circulating Tumor DNA Mutations as a Predictor of Response to First-Line Chemotherapy in Metastatic Colorectal Cancer Patients. Can. J. Gastroenterol. Hepatol. 2018, 2018, 4248971. [Google Scholar] [CrossRef]
- Wang, D.S.; Yang, H.; Liu, X.Y.; Chen, Z.G.; Wang, Y.; Fong, W.P.; Hu, M.T.; Zheng, Y.C.; Zheng, Y.; Li, B.K.; et al. Dynamic monitoring of circulating tumor DNA to predict prognosis and efficacy of adjuvant chemotherapy after resection of colorectal liver metastases. Theranostics 2021, 11, 7018–7028. [Google Scholar] [CrossRef]
- Tie, J.; Wang, Y.; Cohen, J.; Li, L.; Hong, W.; Christie, M.; Wong, H.L.; Kosmider, S.; Wong, R.; Thomson, B.; et al. Circulating tumor DNA dynamics and recurrence risk in patients undergoing curative intent resection of colorectal cancer liver metastases: A prospective cohort study. PLoS Med. 2021, 18, e1003620. [Google Scholar] [CrossRef]
- Bidard, F.C.; Kiavue, N.; Ychou, M.; Cabel, L.; Stern, M.H.; Madic, J.; Saliou, A.; Rampanou, A.; Decraene, C.; Bouche, O.; et al. Circulating Tumor Cells and Circulating Tumor DNA Detection in Potentially Resectable Metastatic Colorectal Cancer: A Prospective Ancillary Study to the Unicancer Prodige-14 Trial. Cells 2019, 8, 516. [Google Scholar] [CrossRef] [Green Version]
- Bhangu, J.S.; Beer, A.; Mittlbock, M.; Tamandl, D.; Pulverer, W.; Schonthaler, S.; Taghizadeh, H.; Stremitzer, S.; Kaczirek, K.; Gruenberger, T.; et al. Circulating Free Methylated Tumor DNA Markers for Sensitive Assessment of Tumor Burden and Early Response Monitoring in Patients Receiving Systemic Chemotherapy for Colorectal Cancer Liver Metastasis. Ann. Surg. 2018, 268, 894–902. [Google Scholar] [CrossRef]
- Bachet, J.B.; Bouche, O.; Taieb, J.; Dubreuil, O.; Garcia, M.L.; Meurisse, A.; Normand, C.; Gornet, J.M.; Artru, P.; Louafi, S.; et al. RAS mutation analysis in circulating tumor DNA from patients with metastatic colorectal cancer: The AGEO RASANC prospective multicenter study. Ann. Oncol. 2018, 29, 1211–1219. [Google Scholar] [CrossRef] [PubMed]
- Mas, L.; Bachet, J.B.; Taly, V.; Bouche, O.; Taieb, J.; Cohen, R.; Meurisse, A.; Normand, C.; Gornet, J.M.; Artru, P.; et al. BRAF Mutation Status in Circulating Tumor DNA from Patients with Metastatic Colorectal Cancer: Extended Mutation Analysis from the AGEO RASANC Study. Cancers 2019, 11, 998. [Google Scholar] [CrossRef] [PubMed]
- Kagawa, Y.; Elez, E.; Garcia-Foncillas, J.; Bando, H.; Taniguchi, H.; Vivancos, A.; Akagi, K.; Garcia, A.; Denda, T.; Ros, J.; et al. Combined Analysis of Concordance between Liquid and Tumor Tissue Biopsies for RAS Mutations in Colorectal Cancer with a Single Metastasis Site: The METABEAM Study. Clin. Cancer Res. 2021, 27, 2515–2522. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Ma, X.; Chen, K.; Liu, F.; Cai, S.; Han-Zhang, H.; Hou, T.; Xiang, J.; Peng, J. Perioperative Circulating Tumor DNA in Colorectal Liver Metastases: Concordance with Metastatic Tissue and Predictive Value for Tumor Burden and Prognosis. Cancer Manag. Res. 2020, 12, 1621–1630. [Google Scholar] [CrossRef]
- Furuki, H.; Yamada, T.; Takahashi, G.; Iwai, T.; Koizumi, M.; Shinji, S.; Yokoyama, Y.; Takeda, K.; Taniai, N.; Uchida, E. Evaluation of liquid biopsies for detection of emerging mutated genes in metastatic colorectal cancer. Eur. J. Surg. Oncol. 2018, 44, 975–982. [Google Scholar] [CrossRef]
- Mostert, B.; Jiang, Y.; Sieuwerts, A.M.; Wang, H.; Bolt-de Vries, J.; Biermann, K.; Kraan, J.; Lalmahomed, Z.; van Galen, A.; de Weerd, V.; et al. KRAS and BRAF mutation status in circulating colorectal tumor cells and their correlation with primary and metastatic tumor tissue. Int. J. Cancer 2013, 133, 130–141. [Google Scholar] [CrossRef]
- Onstenk, W.; Sieuwerts, A.M.; Mostert, B.; Lalmahomed, Z.; Bolt-de Vries, J.B.; van Galen, A.; Smid, M.; Kraan, J.; Van, M.; de Weerd, V.; et al. Molecular characteristics of circulating tumor cells resemble the liver metastasis more closely than the primary tumor in metastatic colorectal cancer. Oncotarget 2016, 7, 59058–59069. [Google Scholar] [CrossRef]
- Kidess-Sigal, E.; Liu, H.E.; Triboulet, M.M.; Che, J.; Ramani, V.C.; Visser, B.C.; Poultsides, G.A.; Longacre, T.A.; Marziali, A.; Vysotskaia, V.; et al. Enumeration and targeted analysis of KRAS, BRAF and PIK3CA mutations in CTCs captured by a label-free platform: Comparison to ctDNA and tissue in metastatic colorectal cancer. Oncotarget 2016, 7, 85349–85364. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsilimigras, D.I.; Ntanasis-Stathopoulos, I.; Pawlik, T.M. Liquid Biopsies in Colorectal Liver Metastases: Towards the Era of Precision Oncologic Surgery. Cancers 2022, 14, 4237. https://doi.org/10.3390/cancers14174237
Tsilimigras DI, Ntanasis-Stathopoulos I, Pawlik TM. Liquid Biopsies in Colorectal Liver Metastases: Towards the Era of Precision Oncologic Surgery. Cancers. 2022; 14(17):4237. https://doi.org/10.3390/cancers14174237
Chicago/Turabian StyleTsilimigras, Diamantis I., Ioannis Ntanasis-Stathopoulos, and Timothy M. Pawlik. 2022. "Liquid Biopsies in Colorectal Liver Metastases: Towards the Era of Precision Oncologic Surgery" Cancers 14, no. 17: 4237. https://doi.org/10.3390/cancers14174237
APA StyleTsilimigras, D. I., Ntanasis-Stathopoulos, I., & Pawlik, T. M. (2022). Liquid Biopsies in Colorectal Liver Metastases: Towards the Era of Precision Oncologic Surgery. Cancers, 14(17), 4237. https://doi.org/10.3390/cancers14174237







