Identification of EZH2 as Cancer Stem Cell Marker in Clear Cell Renal Cell Carcinoma and the Anti-Tumor Effect of Epigallocatechin-3-Gallate (EGCG)
Abstract
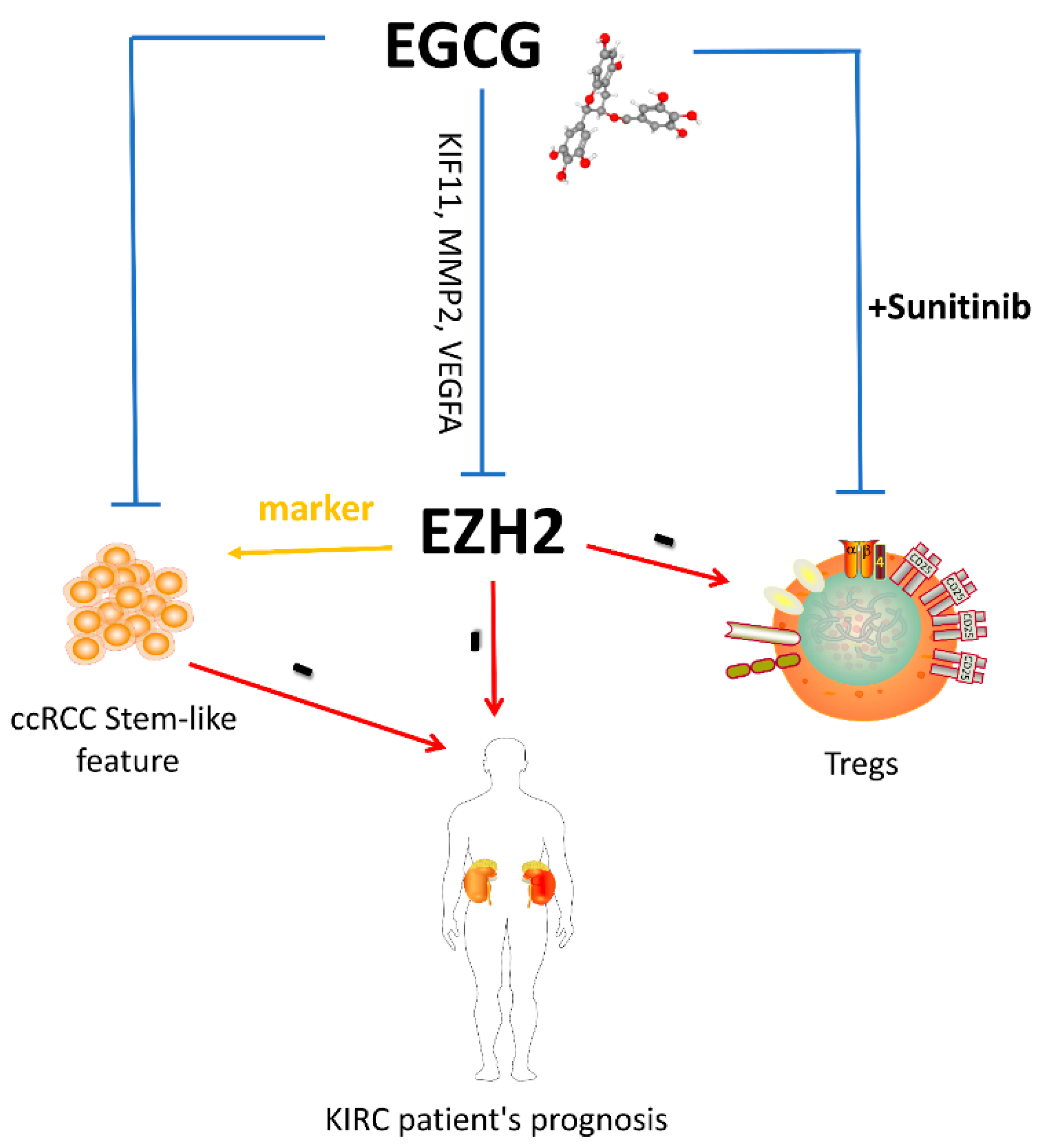
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Sphere Formation Assay
2.3. Quantitative Reverse Transcription PCR (RT-qPCR)
2.4. KIRC Data Download
2.5. Survival and Clinical Characteristics Analysis
2.6. Gene Expression in Tumor and Normal Tissue
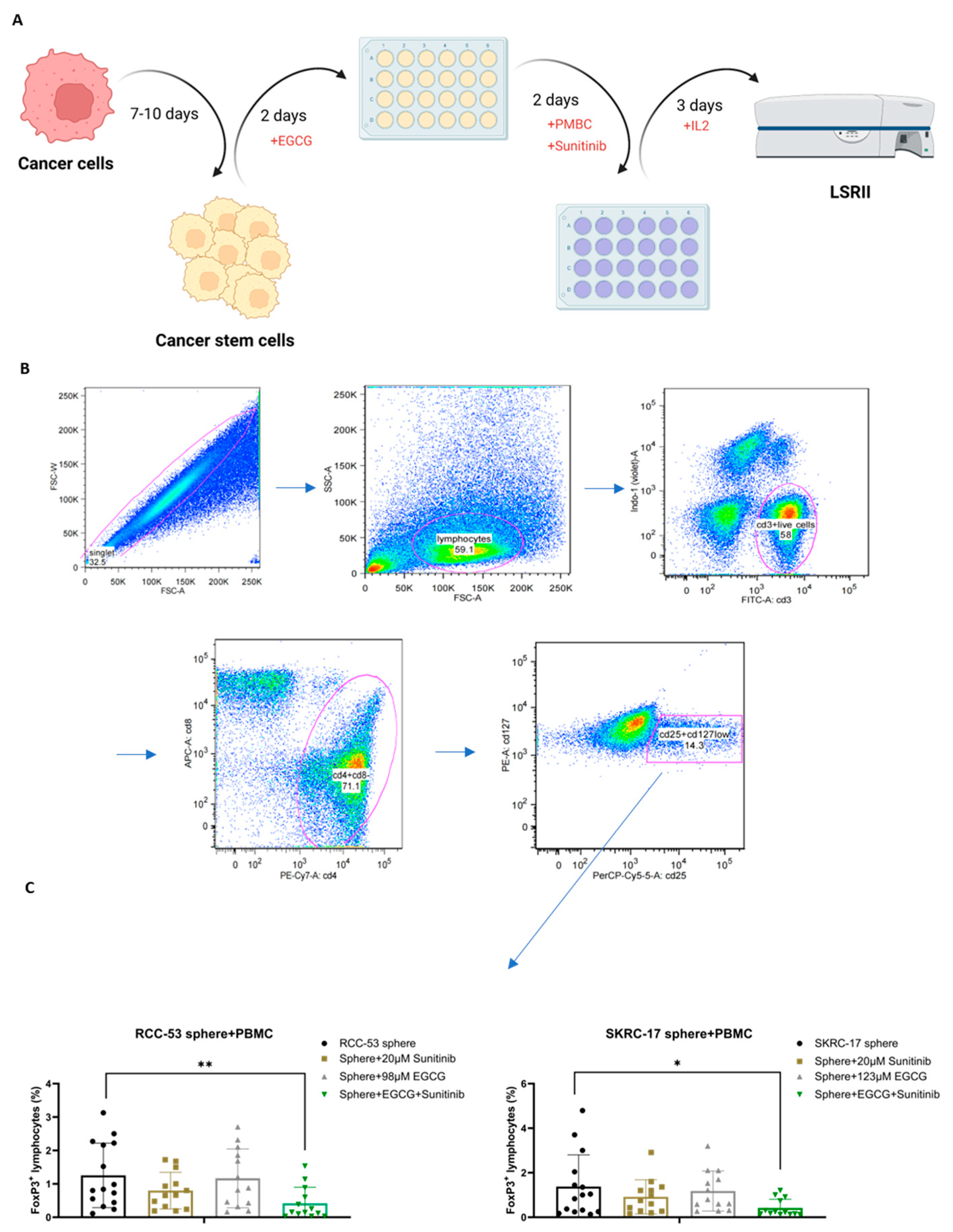
2.7. Mixed Lymphocyte Tumor Cell Culture (MLTC)
2.8. Flow Cytometry
2.9. COX Regression Analysis
2.10. TICs Profile
2.11. Gene Set Enrichment Analysis
2.12. Cell Viability Assay
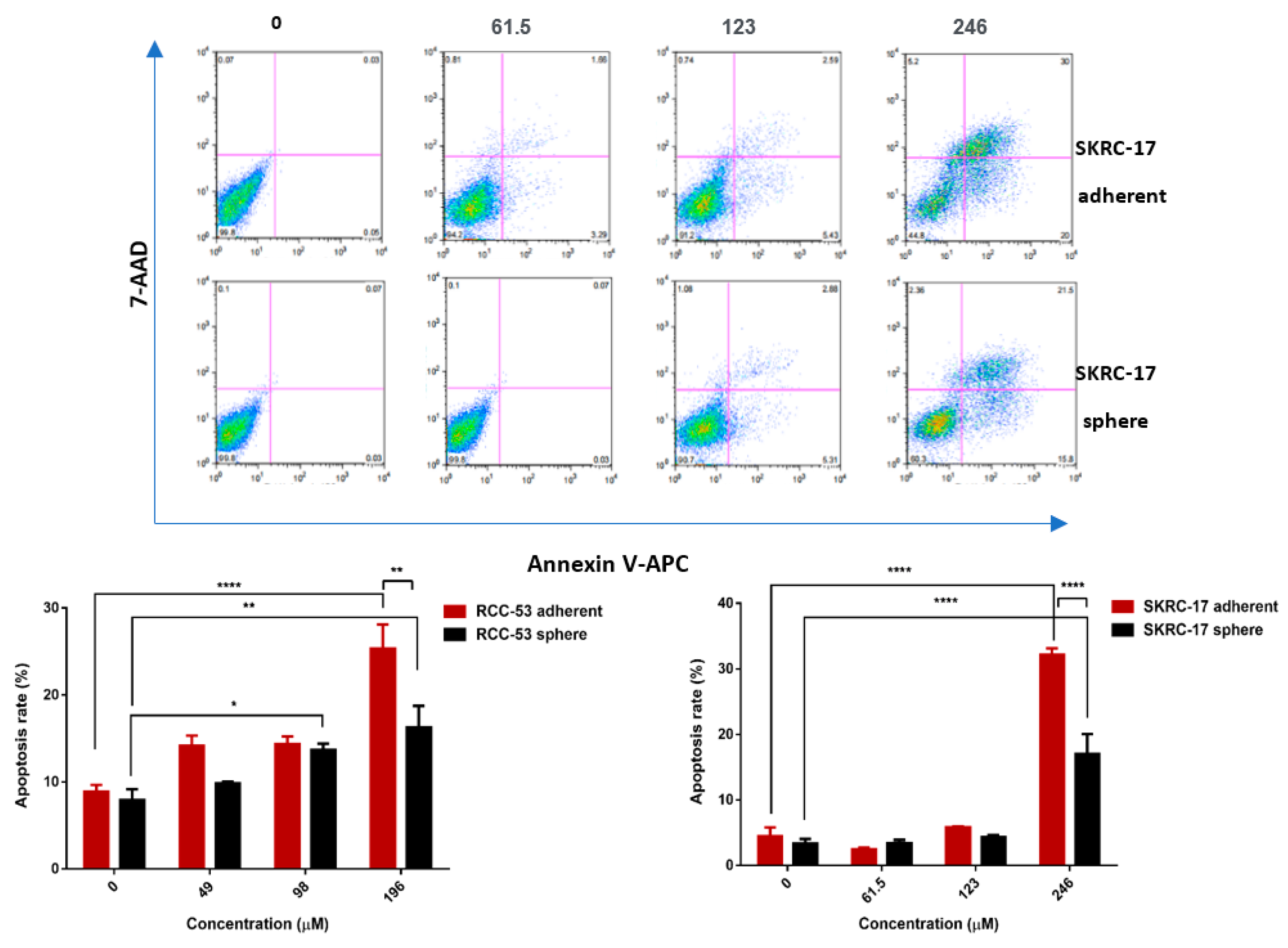
2.13. Apoptosis Assay
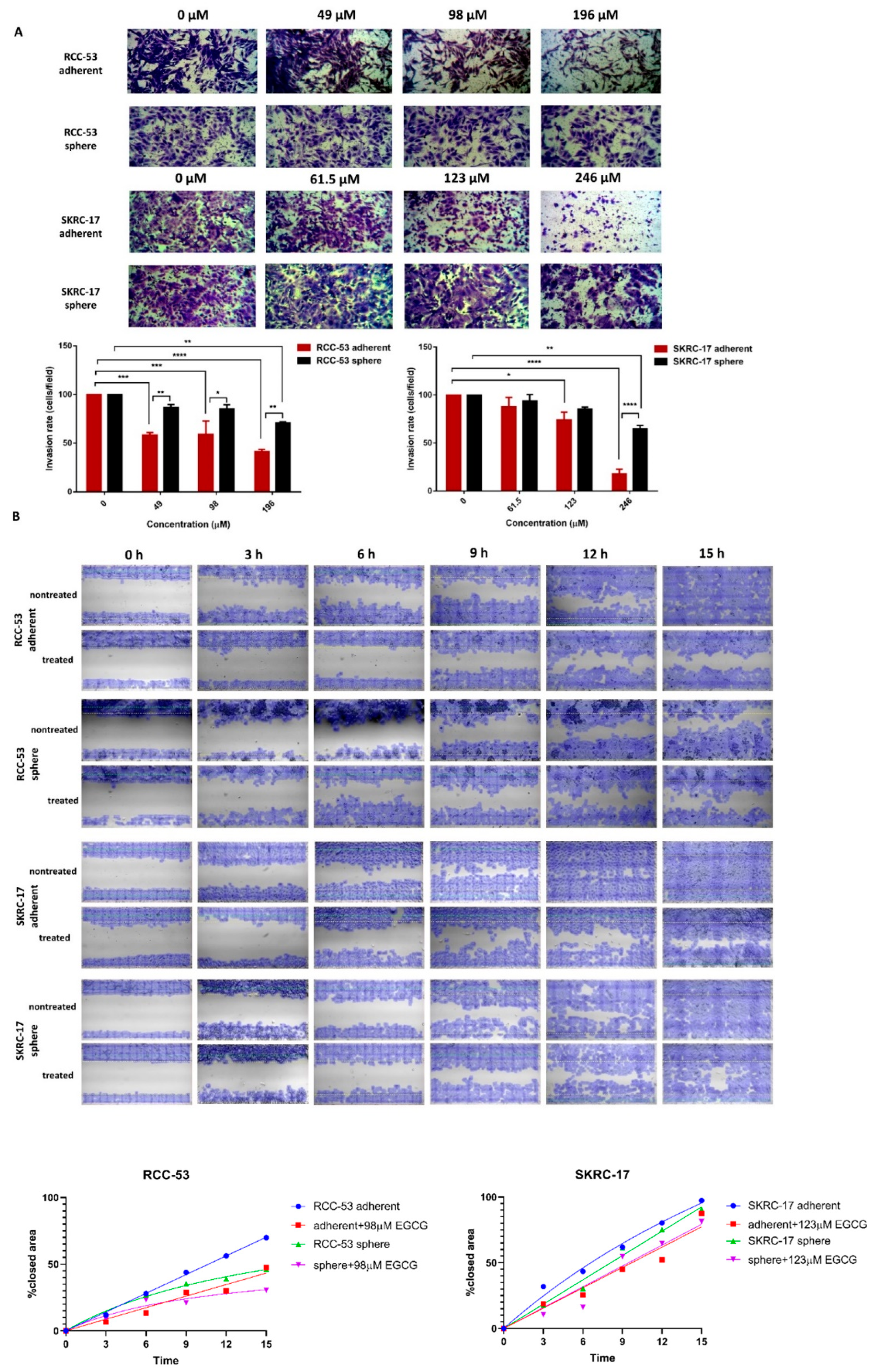
2.14. Migration Assay
2.15. Invasion Assay
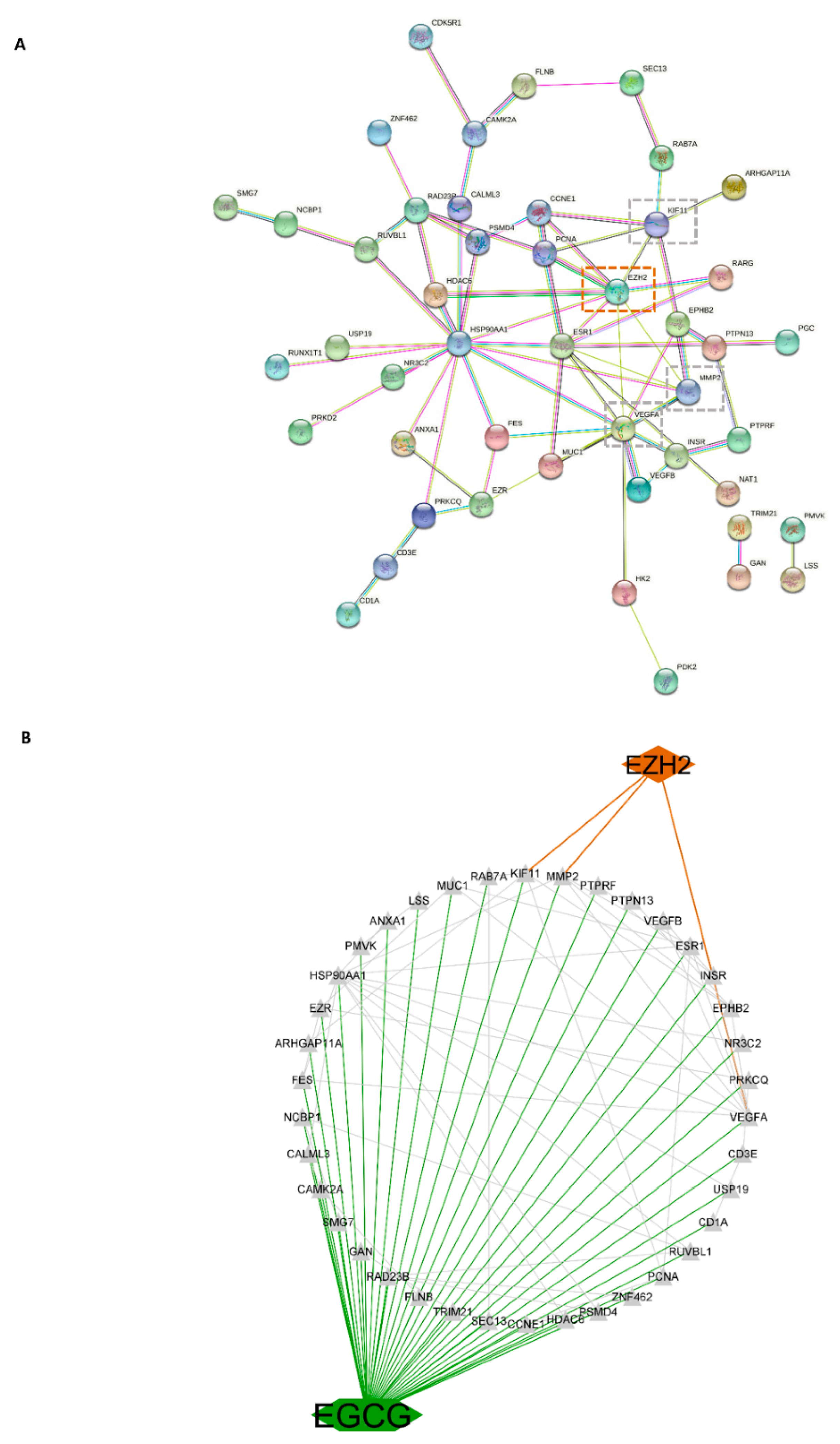
2.16. Prediction of EGCG Target Genes and Construction of Network
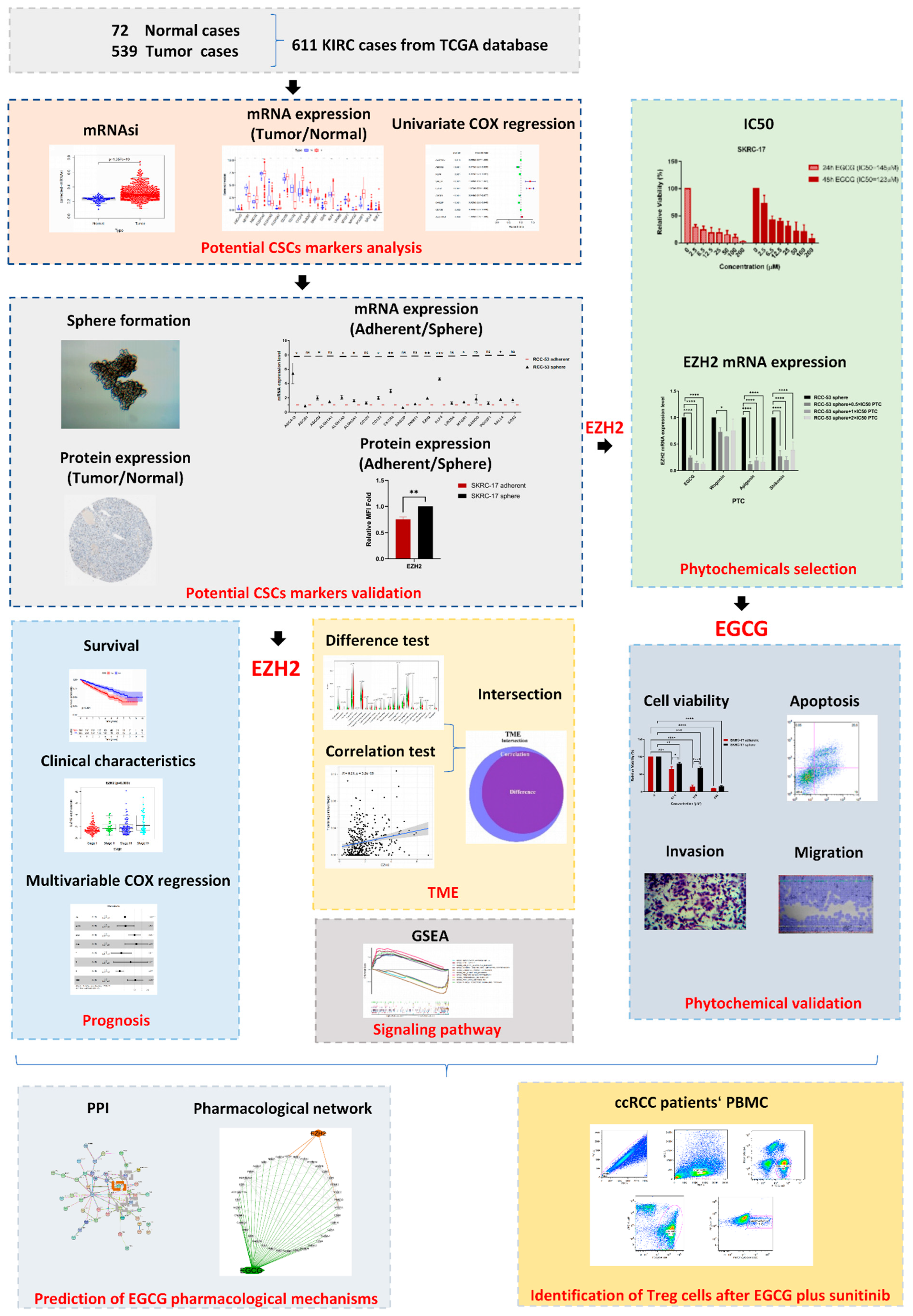
3. Results
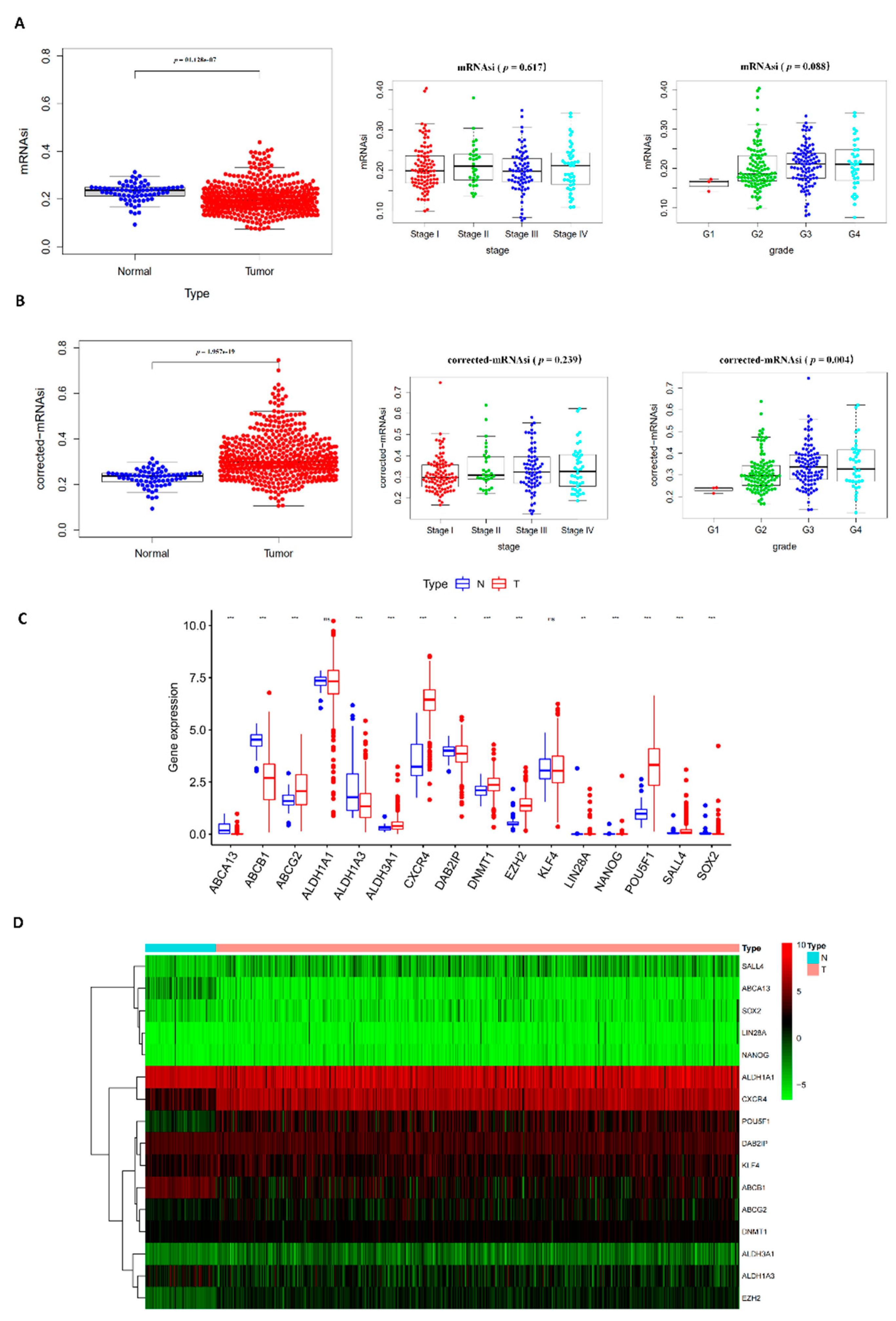
3.1. Corrected-mRNAsi for KIRC
3.2. Expression Analysis of 19 Potential CSC Marker Genes
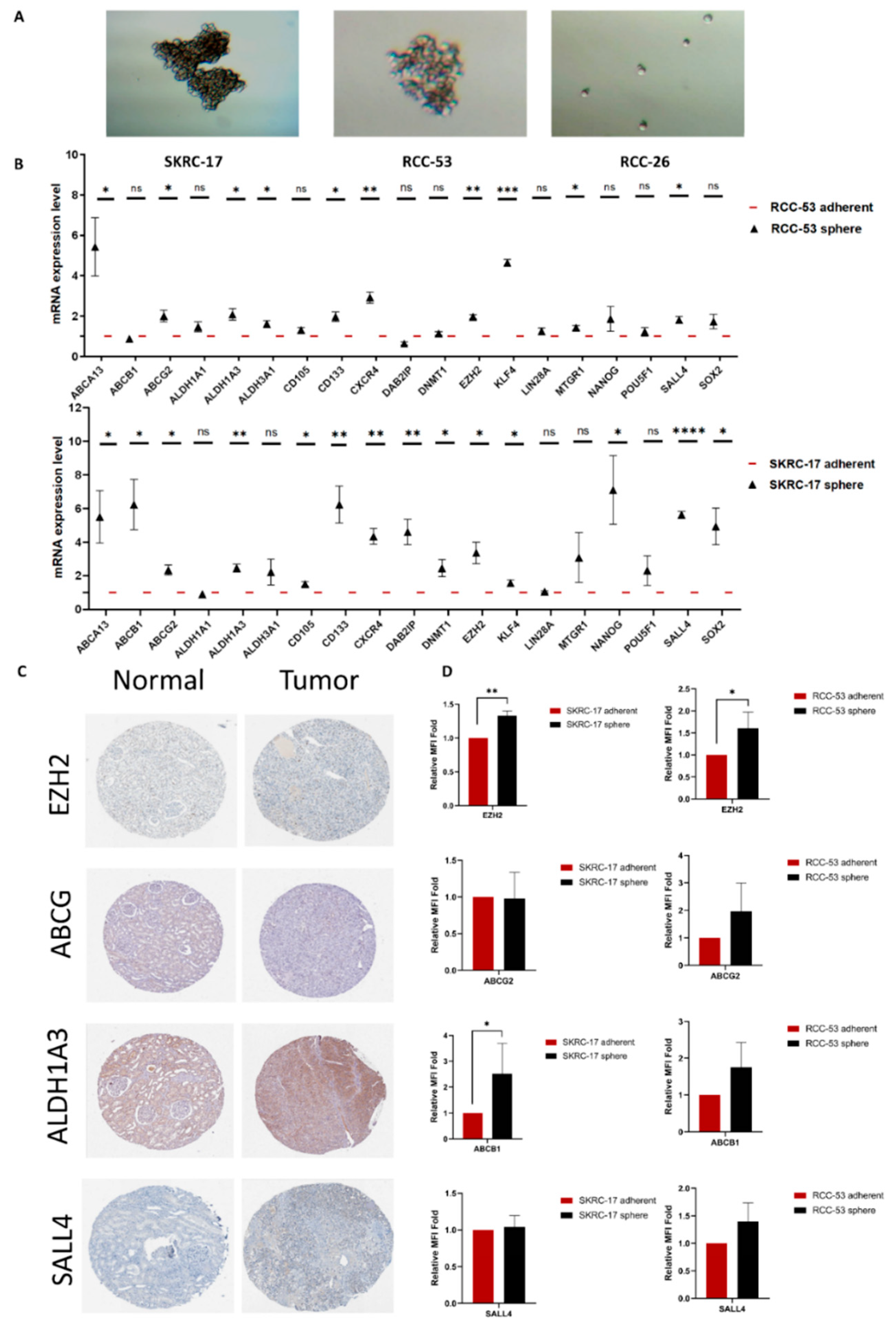
3.3. Validation of Potential CSC Markers on mRNA and Protein Level
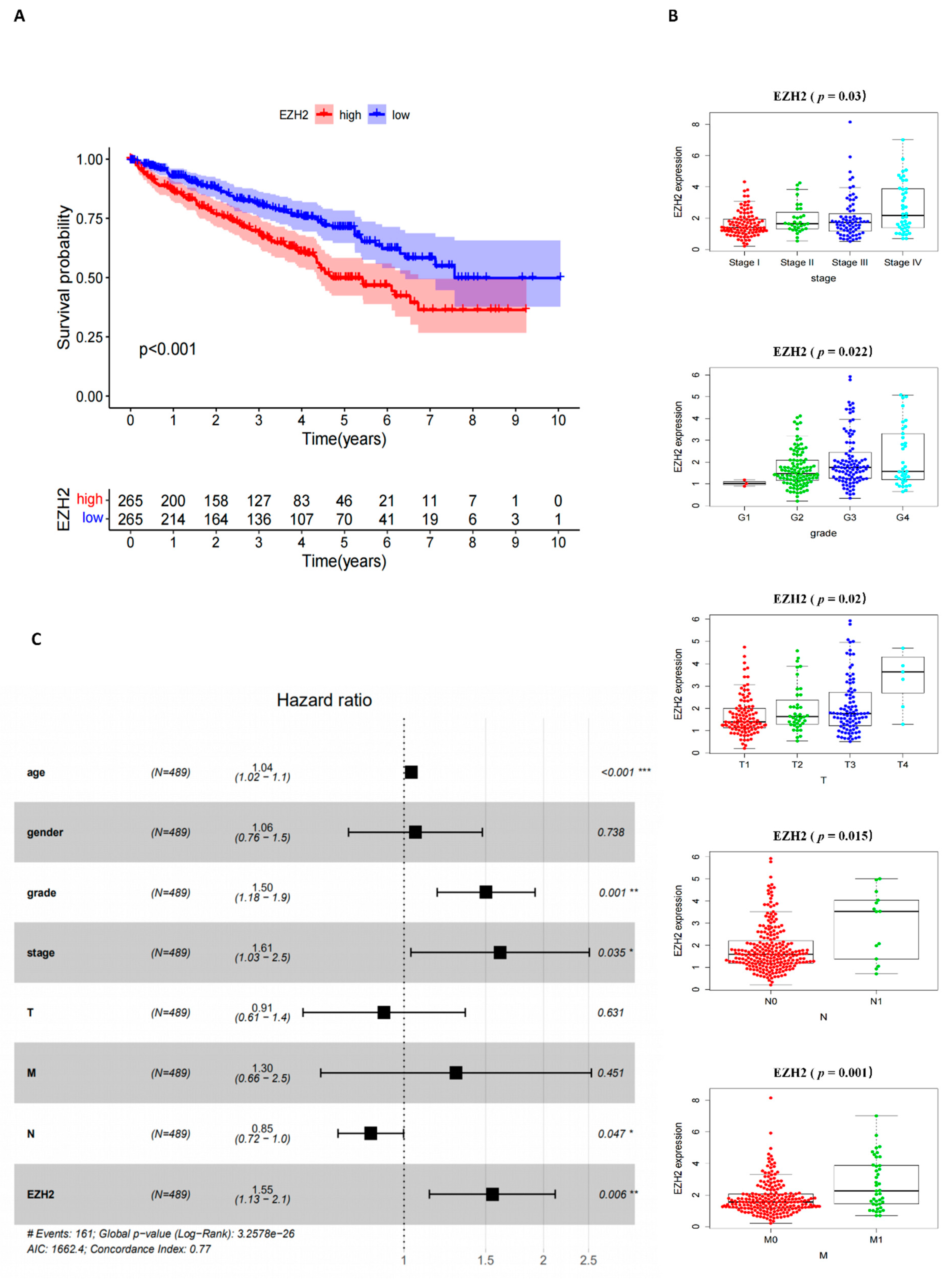
3.4. Correlation of EZH2 Expression with the Survival and Clinical Characteristics of KIRC Patients
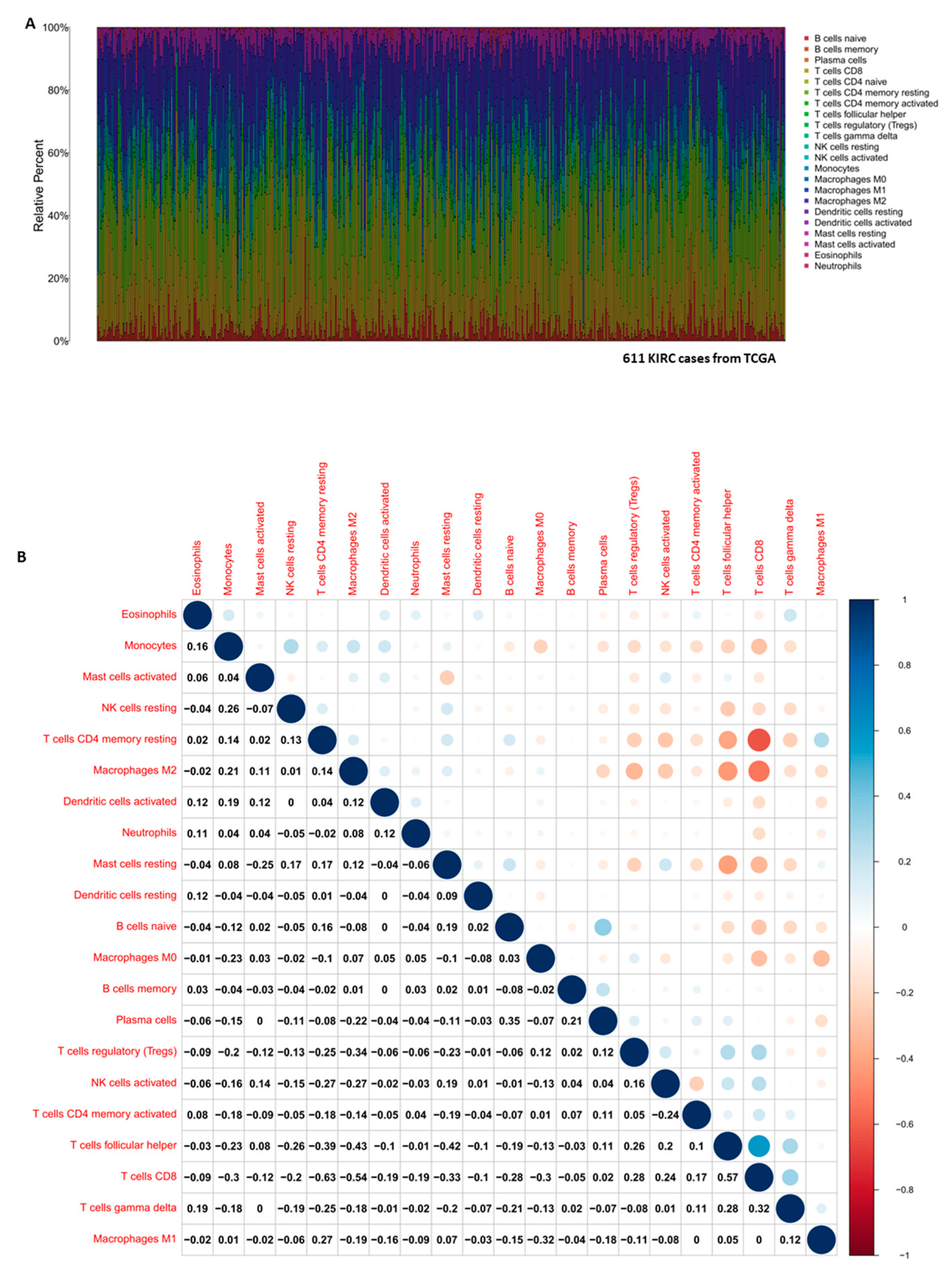
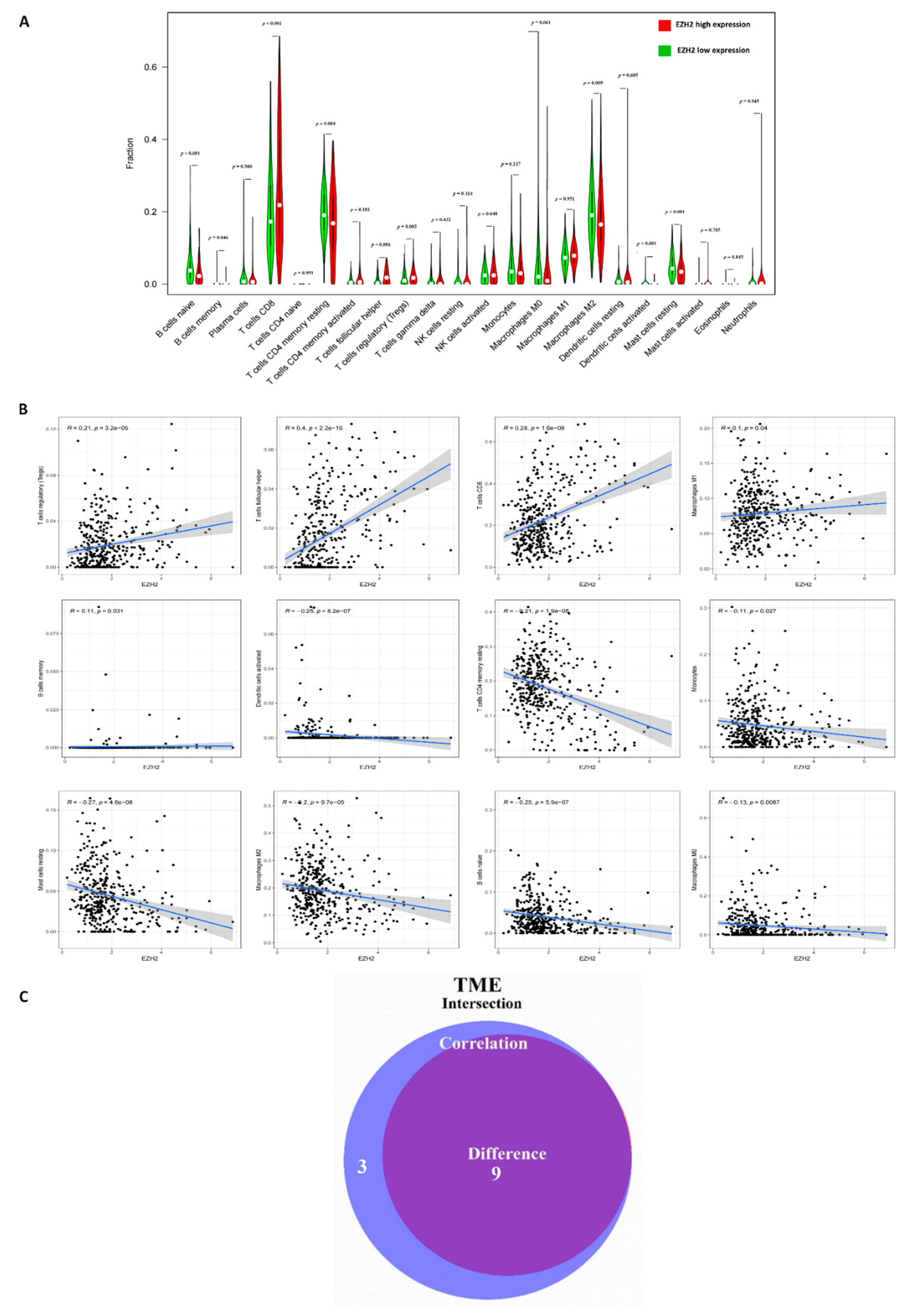
3.5. Correlation of EZH2 with the Composition of TICs
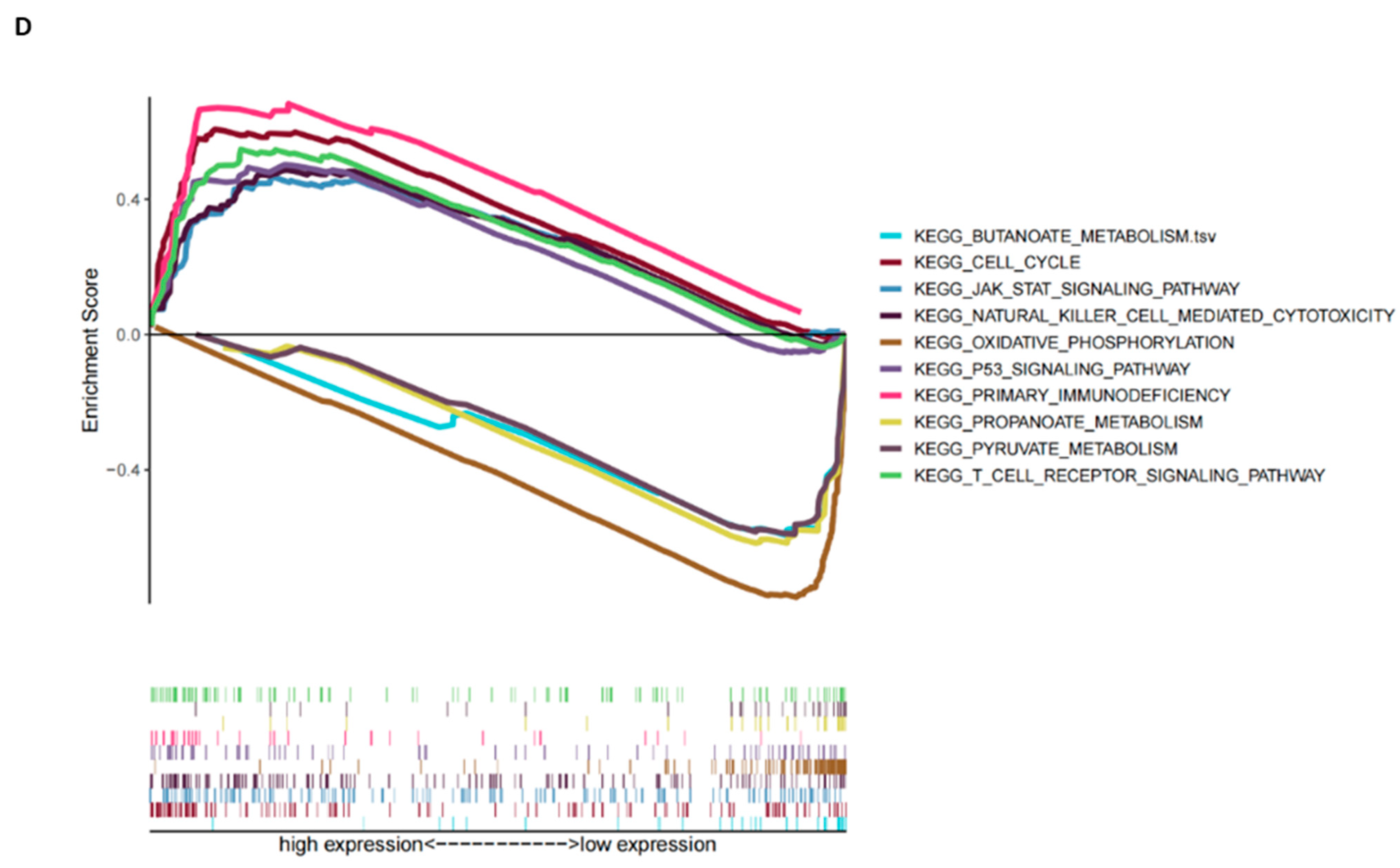
3.6. Functional Enrichment Analysis of EZH2
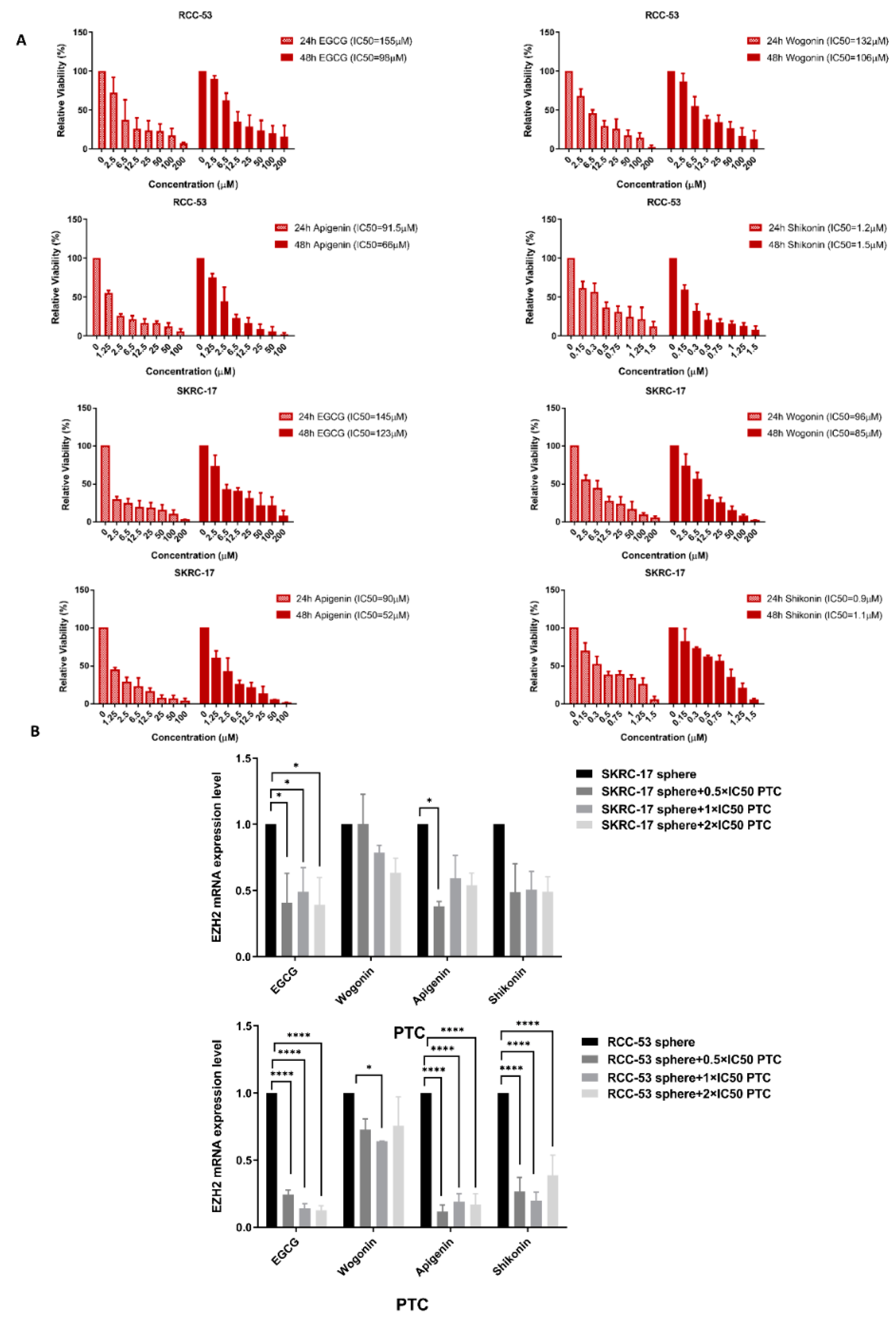
3.7. Influence of Four Different Phytochemicals on EZH2 mRNA Expression
3.8. EGCG Inhibits Cell Viability and Induces Apoptosis in RCC-53 and SKRC-17 Cell Lines
3.9. EGCG Inhibits Migration and Invasion of RCC-53 and SKRC-17 Cell Lines
3.10. Network of EGCG Target Genes and EZH2
3.11. The Effect of EGCG Combined with Sunitinib on Immune Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Capitanio, U.; Montorsi, F. Renal cancer. Lancet 2016, 387, 894–906. [Google Scholar] [CrossRef]
- Barata, P.C.; Rini, B.I. Treatment of renal cell carcinoma: Current status and future directions. CA Cancer J. Clin. 2017, 67, 507–524. [Google Scholar] [CrossRef] [PubMed]
- Motzer, R.J.; Jonasch, E.; Agarwal, N.; Beard, C.; Bhayani, S.; Bolger, G.B.; Chang, S.S.; Choueiri, T.K.; Costello, B.A.; Derweesh, I.H. Kidney cancer, version 3.2015. J. Natl. Compr. Cancer Netw. 2015, 13, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Koren, E.; Fuchs, Y. The bad seed: Cancer stem cells in tumor development and resistance. Drug Resist. Updates 2016, 28, 1–12. [Google Scholar] [CrossRef]
- Pan, X.-W.; Zhang, H.; Xu, D.; Chen, J.-X.; Chen, W.-J.; Gan, S.-S.; Qu, F.-J.; Chu, C.-M.; Cao, J.-W.; Fan, Y.-H.; et al. Identification of a novel cancer stem cell subpopulation that promotes progression of human fatal renal cell carcinoma by single-cell RNA-seq analysis. Int. J. Biol. Sci. 2020, 16, 3149. [Google Scholar] [CrossRef]
- Bussolati, B.; Bruno, S.; Grange, C.; Ferrando, U.; Camussi, G. Identification of a tumor-initiating stem cell population in human renal carcinomas. FASEB J. 2008, 22, 3696–3705. [Google Scholar] [CrossRef]
- Hirata, A.; Hatano, Y.; Niwa, M.; Hara, A.; Tomita, H. Heterogeneity in colorectal cancer stem cells. Cancer Prev. Res. 2019, 12, 413–420. [Google Scholar] [CrossRef]
- Zhao, Y.; Ding, L.; Wang, D.; Ye, Z.; He, Y.; Ma, L.; Zhu, R.; Pan, Y.; Wu, Q.; Pang, K. EZH 2 cooperates with gain-of-function p53 mutants to promote cancer growth and metastasis. EMBO J. 2019, 38, e99599. [Google Scholar] [CrossRef]
- Wang, W.; Fang, F.; Ozes, A.; Nephew, K.P. Targeting Ovarian Cancer Stem Cells by Dual Inhibition of HOTAIR and DNA Methylation. Mol. Cancer Ther. 2021, 20, 1092–1101. [Google Scholar] [CrossRef]
- Wen, Y.; Hou, Y.; Yi, X.; Sun, S.; Guo, J.; He, X.; Li, T.; Cai, J.; Wang, Z. EZH2 activates CHK1 signaling to promote ovarian cancer chemoresistance by maintaining the properties of cancer stem cells. Theranostics 2021, 11, 1795. [Google Scholar] [CrossRef]
- Kempf, J.M.; Weser, S.; Bartoschek, M.D.; Metzeler, K.H.; Vick, B.; Herold, T.; Völse, K.; Mattes, R.; Scholz, M.; Wange, L.E. Loss-of-function mutations in the histone methyltransferase EZH2 promote chemotherapy resistance in AML. Sci. Rep. 2021, 11, 5838. [Google Scholar] [CrossRef] [PubMed]
- Suvà, M.-L.; Riggi, N.; Janiszewska, M.; Radovanovic, I.; Provero, P.; Stehle, J.-C.; Baumer, K.; Le Bitoux, M.-A.; Marino, D.; Cironi, L. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res. 2009, 69, 9211–9218. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Sun, Q.; Sun, Y.; Zhang, J.; Yuan, H.; Pang, S.; Qi, X.; Wang, H.; Zhang, M.; Zhang, H. MELK and EZH2 Cooperate to Regulate Medulloblastoma Cancer Stem-like Cell Proliferation and DifferentiationInteraction between MELK and EZH2 in Medulloblastoma. Mol. Cancer Res. 2017, 15, 1275–1286. [Google Scholar] [CrossRef] [PubMed]
- van Vlerken, L.E.; Kiefer, C.M.; Morehouse, C.; Li, Y.; Groves, C.; Wilson, S.D.; Yao, Y.; Hollingsworth, R.E.; Hurt, E.M. EZH2 is required for breast and pancreatic cancer stem cell maintenance and can be used as a functional cancer stem cell reporter. Stem Cells Transl. Med. 2013, 2, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Verma, A.; Singh, A.; Arya, R.K.; Maheshwari, S.; Chaturvedi, P.; Nengroo, M.A.; Saini, K.K.; Vishwakarma, A.L.; Singh, K. Salinomycin inhibits epigenetic modulator EZH2 to enhance death receptors in colon cancer stem cells. Epigenetics 2021, 16, 144–161. [Google Scholar] [CrossRef]
- Eichenauer, T.; Simmendinger, L.; Fraune, C.; Mandelkow, T.; Blessin, N.C.; Kluth, M.; Hube-Magg, C.; Möller, K.; Clauditz, T.; Weidemann, S. High level of EZH2 expression is linked to high density of CD8-positive T-lymphocytes and an aggressive phenotype in renal cell carcinoma. World J. Urol. 2021, 39, 481–490. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, B.; Zhao, X.; Liu, Z.; Wang, X.; Yao, X.; Dong, X.; Chi, J. Clinical significances and prognostic value of cancer stem-like cells markers and vasculogenic mimicry in renal cell carcinoma. J. Surg. Oncol. 2013, 108, 414–419. [Google Scholar] [CrossRef]
- Ge, Y.; Weygant, N.; Qu, D.; May, R.; Berry, W.L.; Yao, J.; Chandrakesan, P.; Zheng, W.; Zhao, L.; Zhao, K.L. Alternative splice variants of DCLK1 mark cancer stem cells, promote self-renewal and drug-resistance, and can be targeted to inhibit tumorigenesis in kidney cancer. Int. J. Cancer 2018, 143, 1162–1175. [Google Scholar] [CrossRef]
- Gassenmaier, M.; Chen, D.; Buchner, A.; Henkel, L.; Schiemann, M.; Mack, B.; Schendel, D.J.; Zimmermann, W.; Pohla, H. CXC chemokine receptor 4 is essential for maintenance of renal cell carcinoma-initiating cells and predicts metastasis. Stem Cells 2013, 31, 1467–1476. [Google Scholar] [CrossRef]
- Malta, T.M.; Sokolov, A.; Gentles, A.J.; Burzykowski, T.; Poisson, L.; Weinstein, J.N.; Kamińska, B.; Huelsken, J.; Omberg, L.; Gevaert, O. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell 2018, 173, 338–354.e315. [Google Scholar] [CrossRef]
- Tian, Y.; Wang, J.; Qin, C.; Zhu, G.; Chen, X.; Chen, Z.; Qin, Y.; Wei, M.; Li, Z.; Zhang, X. Identifying 8-mRNAsi based signature for predicting survival in patients with head and neck squamous cell carcinoma via machine learning. Front. Genet. 2020, 11, 1296. [Google Scholar] [CrossRef] [PubMed]
- Ben-Porath, I.; Thomson, M.W.; Carey, V.J.; Ge, R.; Bell, G.W.; Regev, A.; Weinberg, R.A. An embryonic stem cell–like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008, 40, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.-H.; Li, K.-W.; Chen, X.; He, H.-X.; Peng, S.-M.; Peng, S.-R.; Wang, Q.; Li, Z.-A.; Tao, Y.-R.; Cai, W.-L. HHLA2 and PD-L1 co-expression predicts poor prognosis in patients with clear cell renal cell carcinoma. J. Immunother. Cancer 2020, 8, e000157. [Google Scholar] [CrossRef]
- Zhou, F.; Shen, D.; Xiong, Y.; Cheng, S.; Xu, H.; Wang, G.; Qian, K.; Ju, L.; Zhang, X. CTHRC1 is a prognostic biomarker and correlated with immune infiltrates in kidney renal papillary cell carcinoma and kidney renal clear cell carcinoma. Front. Oncol. 2020, 10, 3389. [Google Scholar] [CrossRef]
- Gomez, K.E.; Wu, F.; Keysar, S.B.; Morton, J.J.; Miller, B.; Chimed, T.-S.; Le, P.N.; Nieto, C.; Chowdhury, F.N.; Tyagi, A. Cancer Cell CD44 Mediates Macrophage/Monocyte-Driven Regulation of Head and Neck Cancer Stem Cells. Cancer Res. 2020, 80, 4185–4198. [Google Scholar] [CrossRef]
- Linette, G.P.; Carreno, B.M. Tumor-infiltrating lymphocytes in the checkpoint inhibitor era. Curr. Hematol. Malig. Rep. 2019, 14, 286–291. [Google Scholar] [CrossRef]
- Huang, Z.; Yu, H.; Zhang, J.; Jing, H.; Zhu, W.; Li, X.; Kong, L.; Xing, L.; Yu, J.; Meng, X. Correlation of cancer stem cell markers and immune cell markers in resected non-small cell lung cancer. J. Cancer 2017, 8, 3190. [Google Scholar] [CrossRef] [PubMed]
- Ghatalia, P.; Gordetsky, J.; Kuo, F.; Dulaimi, E.; Cai, K.Q.; Devarajan, K.; Bae, S.; Naik, G.; Chan, T.A.; Uzzo, R. Prognostic impact of immune gene expression signature and tumor infiltrating immune cells in localized clear cell renal cell carcinoma. J. Immunother. Cancer 2019, 7, 139. [Google Scholar] [CrossRef]
- Bromwich, E.; McArdle, P.; Canna, K.; McMillan, D.; McNicol, A.; Brown, M.; Aitchison, M. The relationship between T-lymphocyte infiltration, stage, tumour grade and survival in patients undergoing curative surgery for renal cell cancer. Br. J. Cancer 2003, 89, 1906–1908. [Google Scholar] [CrossRef]
- Dandawate, P.R.; Subramaniam, D.; Jensen, R.A.; Anant, S. Targeting cancer stem cells and signaling pathways by phytochemicals: Novel approach for breast cancer therapy. Semin. Cancer Biol. 2016, 40–41, 192–208. [Google Scholar] [CrossRef]
- Naujokat, C.; McKee, D.L. The “Big Five” phytochemicals targeting cancer stem cells: Curcumin, EGCG, sulforaphane, resveratrol, and genistein. Curr. Med. Chem. 2020, 28, 4321–4342. [Google Scholar] [CrossRef] [PubMed]
- Namiki, K.; Wongsirisin, P.; Yokoyama, S.; Sato, M.; Rawangkan, A.; Sakai, R.; Iida, K.; Suganuma, M. (−)-Epigallocatechin gallate inhibits stemness and tumourigenicity stimulated by AXL receptor tyrosine kinase in human lung cancer cells. Sci. Rep. 2020, 10, 2444. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Song, J.; Li, E.; Geng, H.; Li, Y.; Yu, D.; Zhong, C. (-)-Epigallocatechin-3-gallate inhibits bladder cancer stem cells via suppression of sonic hedgehog pathway. Oncol. Rep. 2019, 42, 425–435. [Google Scholar] [CrossRef]
- Jiang, P.; Xu, C.; Chen, L.; Chen, A.; Wu, X.; Zhou, M.; ul Haq, I.; Mariyam, Z.; Feng, Q. EGCG inhibits CSC-like properties through targeting miR-485/CD44 axis in A549-cisplatin resistant cells. Mol. Carcinog. 2018, 57, 1835–1844. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.N.; Fu, J.; Nall, D.; Rodova, M.; Shankar, S.; Srivastava, R.K. Inhibition of sonic hedgehog pathway and pluripotency maintaining factors regulate human pancreatic cancer stem cell characteristics. Int. J. Cancer 2012, 131, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Xu, C.; Zhang, P.; Ren, J.; Mageed, F.; Wu, X.; Chen, L.; Zeb, F.; Feng, Q.; Li, S. Epigallocatechin-3-gallate inhibits self-renewal ability of lung cancer stem-like cells through inhibition of CLOCK. Int. J. Mol. Med. 2020, 46, 2216–2224. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-W.; Xu, J.; Zhu, G.-Y.; Huang, Z.-J.; Lu, Y.; Li, X.-Q.; Wang, N.; Zhang, F.-X. Apigenin suppresses the stem cell-like properties of triple-negative breast cancer cells by inhibiting YAP/TAZ activity. Cell Death Discov. 2018, 4, 105. [Google Scholar] [CrossRef]
- Chan, M.M.; Chen, R.; Fong, D. Targeting cancer stem cells with dietary phytochemical-repositioned drug combinations. Cancer Lett. 2018, 433, 53–64. [Google Scholar] [CrossRef]
- Wang, L.; Stadlbauer, B.; Lyu, C.; Buchner, A.; Pohla, H. Shikonin enhances the antitumor effect of cabazitaxel in prostate cancer stem cells and reverses cabazitaxel resistance by inhibiting ABCG2 and ALDH3A1. Am. J. Cancer Res. 2020, 10, 3784. [Google Scholar]
- Schendel, D.J.; Gansbacher, B.; Oberneder, R.; Kriegmair, M.; Hofstetter, A.; Riethmüller, G.; Segurado, O. Tumor-specific lysis of human renal cell carcinomas by tumor-infiltrating lymphocytes. I. HLA-A2-restricted recognition of autologous and allogeneic tumor lines. J. Immunol. 1993, 151, 4209–4220. [Google Scholar]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Zhan, Y.; Chen, X.; Wu, B.; Liu, B. Identification of biomarkers for controlling cancer stem cell characteristics in bladder cancer by network analysis of transcriptome data stemness indices. Front. Oncol. 2019, 9, 613. [Google Scholar] [CrossRef]
- Pei, J.; Wang, Y.; Li, Y. Identification of key genes controlling breast cancer stem cell characteristics via stemness indices analysis. J. Transl. Med. 2020, 18, 74. [Google Scholar] [CrossRef] [PubMed]
- Butte, A.; Aran, D.; Sirota, M.; Butte, A. Systematic pan-cancer analysis of tumour purity. Nat. Commun. 2015, 6, 8971. [Google Scholar]
- Corro, C.; Moch, H. Biomarker discovery for renal cancer stem cells. J. Pathol. Clin. Res. 2018, 4, 3–18. [Google Scholar] [CrossRef]
- Rasti, A.; Mehrazma, M.; Madjd, Z.; Abolhasani, M.; Saeednejad Zanjani, L.; Asgari, M. Co-expression of cancer stem cell markers OCT4 and NANOG predicts poor prognosis in renal cell carcinomas. Sci. Rep. 2018, 8, 11739. [Google Scholar] [CrossRef]
- Shi, D.; Che, J.; Yan, Y.; Peng, B.; Yao, X.; Guo, C. Expression and clinical value of CD105 in renal cell carcinoma based on data mining in The Cancer Genome Atlas. Exp. Ther. Med. 2019, 17, 4499–4505. [Google Scholar] [CrossRef]
- Abbaszadegan, M.R.; Bagheri, V.; Razavi, M.S.; Momtazi, A.A.; Sahebkar, A.; Gholamin, M. Isolation, identification, and characterization of cancer stem cells: A review. J. Cell. Physiol. 2017, 232, 2008–2018. [Google Scholar] [CrossRef]
- Plaks, V.; Kong, N.; Werb, Z. The cancer stem cell niche: How essential is the niche in regulating stemness of tumor cells? Cell Stem Cell 2015, 16, 225–238. [Google Scholar] [CrossRef]
- Koh, W.-J.; Abu-Rustum, N.R.; Bean, S.; Bradley, K.; Campos, S.M.; Cho, K.R.; Chon, H.S.; Chu, C.; Clark, R.; Cohn, D. Cervical cancer, version 3.2019, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Cancer Netw. 2019, 17, 64–84. [Google Scholar] [CrossRef] [PubMed]
- Anker, J.; Miller, J.; Taylor, N.; Kyprianou, N.; Tsao, C.-K. From bench to bedside: How the tumor microenvironment is impacting the future of immunotherapy for renal cell carcinoma. Cells 2021, 10, 3231. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.; Liu, X.; Liu, Y.; Zhang, Z.; Xie, L.; Tian, K.; Liu, J.; Yu, Y. Roles of the dynamic tumor immune microenvironment in the individualized treatment of advanced clear cell renal cell carcinoma. Front. Immunol. 2021, 12, 653358. [Google Scholar] [CrossRef] [PubMed]
- Ballesteros, P.Á.; Chamorro, J.; Román-Gil, M.S.; Pozas, J.; Gómez Dos Santos, V.; Granados, Á.R.; Grande, E.; Alonso-Gordoa, T.; Molina-Cerrillo, J. Molecular Mechanisms of Resistance to Immunotherapy and Antiangiogenic Treatments in Clear Cell Renal Cell Carcinoma. Cancers 2021, 13, 5981. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.-X.; Mo, J.; Zhao, G.; Shu, G.; Fu, H.-L.; Zhao, W. Targeting strategies for renal cell carcinoma: From renal cancer cells to renal cancer stem cells. Front. Pharmacol. 2016, 7, 423. [Google Scholar] [CrossRef]
- Fang, P.; Zhou, L.; Lim, L.Y.; Fu, H.; Yuan, Z.-X.; Lin, J. Targeting strategies for renal cancer stem cell therapy. Curr. Pharm. Des. 2020, 26, 1964–1978. [Google Scholar] [CrossRef]
- Singh, D. Current updates and future perspectives on the management of renal cell carcinoma. Life Sci. 2021, 264, 118632. [Google Scholar] [CrossRef]
- Liu, L.; Xu, Z.; Zhong, L.; Wang, H.; Jiang, S.; Long, Q.; Xu, J.; Guo, J. Prognostic value of EZH2 expression and activity in renal cell carcinoma: A prospective study. PLoS ONE 2013, 8, e81484. [Google Scholar] [CrossRef][Green Version]
- Adelaiye-Ogala, R.; Budka, J.; Damayanti, N.P.; Arrington, J.; Ferris, M.; Hsu, C.-C.; Chintala, S.; Orillion, A.; Miles, K.M.; Shen, L. EZH2 modifies sunitinib resistance in renal cell carcinoma by kinome reprogramming. Cancer Res. 2017, 77, 6651–6666. [Google Scholar] [CrossRef]
- Wang, D.; Quiros, J.; Mahuron, K.; Pai, C.-C.; Ranzani, V.; Young, A.; Silveria, S.; Harwin, T.; Abnousian, A.; Pagani, M. Targeting EZH2 reprograms intratumoral regulatory T cells to enhance cancer immunity. Cell Rep. 2018, 23, 3262–3274. [Google Scholar] [CrossRef]
- Fioravanti, R.; Stazi, G.; Zwergel, C.; Valente, S.; Mai, A. Six years (2012–2018) of researches on catalytic ezh2 inhibitors: The boom of the 2-pyridone compounds. Chem. Rec. 2018, 18, 1818–1832. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhu, Y.; He, H.; Liu, J.; Xu, L.; Zhang, H.; Liu, H.; Liu, W.; Liu, Y.; Pan, D. Sorafenib suppresses growth and survival of hepatoma cells by accelerating degradation of enhancer of zeste homolog 2. Cancer Sci. 2013, 104, 750–759. [Google Scholar] [CrossRef] [PubMed]
- Bao, B.; Ali, S.; Banerjee, S.; Wang, Z.; Logna, F.; Azmi, A.S.; Kong, D.; Ahmad, A.; Li, Y.; Padhye, S. Curcumin Analogue CDF Inhibits Pancreatic Tumor Growth by Switching on Suppressor microRNAs and Attenuating EZH2 ExpressionTargeting miRNA-Mediated Inactivation of EZH2 by CDF. Cancer Res. 2012, 72, 335–345. [Google Scholar] [CrossRef]
- Hua, W.-F.; Fu, Y.-S.; Liao, Y.-J.; Xia, W.-J.; Chen, Y.-C.; Zeng, Y.-X.; Kung, H.-F.; Xie, D. Curcumin induces down-regulation of EZH2 expression through the MAPK pathway in MDA-MB-435 human breast cancer cells. Eur. J. Pharmacol. 2010, 637, 16–21. [Google Scholar] [CrossRef]
- Dimri, M.; Bommi, P.V.; Sahasrabuddhe, A.A.; Khandekar, J.D.; Dimri, G.P. Dietary omega-3 polyunsaturated fatty acids suppress expression of EZH2 in breast cancer cells. Carcinogenesis 2010, 31, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-G.; Guo, J.-F.; Liu, D.-L.; Liu, Q.; Wang, J.-J. MicroRNA-101 exerts tumor-suppressive functions in non-small cell lung cancer through directly targeting enhancer of zeste homolog 2. J. Thorac. Oncol. 2011, 6, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Ougolkov, A.V.; Bilim, V.N.; Billadeau, D.D. Regulation of pancreatic tumor cell proliferation and chemoresistance by the histone methyltransferase enhancer of zeste homologue 2. Clin. Cancer Res. 2008, 14, 6790–6796. [Google Scholar] [CrossRef]
- De Carvalho, D.; Binato, R.; Pereira, W.D.O.; Leroy, J.M.G.; Colassanti, M.; Proto-Siqueira, R.; Bueno-Da-Silva, A.; Zago, M.A.; Zanichelli, M.; Abdelhay, E. BCR–ABL-mediated upregulation of PRAME is responsible for knocking down TRAIL in CML patients. Oncogene 2011, 30, 223–233. [Google Scholar] [CrossRef]
- Avan, A.; Crea, F.; Paolicchi, E.; Funel, N.; Galvani, E.; Marquez, V.E.; Honeywell, R.J.; Danesi, R.; Peters, G.J.; Giovannetti, E. Molecular Mechanisms Involved in the Synergistic Interaction of the EZH2 Inhibitor 3-Deazaneplanocin A with Gemcitabine in Pancreatic Cancer CellsDZNeP/Gemcitabine Combination in Pancreatic Cancer. Mol. Cancer Ther. 2012, 11, 1735–1746. [Google Scholar] [CrossRef]
- Fiskus, W.; Rao, R.; Balusu, R.; Ganguly, S.; Tao, J.; Sotomayor, E.; Mudunuru, U.; Smith, J.E.; Hembruff, S.L.; Atadja, P. Superior Efficacy of a Combined Epigenetic Therapy against Human Mantle Cell Lymphoma CellsCombined Epigenetic Therapy against Human MCL Cells. Clin. Cancer Res. 2012, 18, 6227–6238. [Google Scholar] [CrossRef]
- Hayden, A.; Johnson, P.W.; Packham, G.; Crabb, S.J. S-adenosylhomocysteine hydrolase inhibition by 3-deazaneplanocin A analogues induces anti-cancer effects in breast cancer cell lines and synergy with both histone deacetylase and HER2 inhibition. Breast Cancer Res. Treat. 2011, 127, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Chan, E.; Wu, Z.; Yang, X.; Marquez, V.E.; Yu, Q. Combinatorial pharmacologic approaches target EZH2-mediated gene repression in breast cancer cells. Mol. Cancer Ther. 2009, 8, 3191–3202. [Google Scholar] [CrossRef]
- Lv, Y.; Yuan, C.; Xiao, X.; Wang, X.; Ji, X.; Yu, H.; Wu, Z.; Zhang, J. The expression and significance of the enhancer of zeste homolog 2 in lung adenocarcinoma. Oncol. Rep. 2012, 28, 147–154. [Google Scholar]
- Hu, S.; Yu, L.; Li, Z.; Shen, Y.; Wang, J.; Cai, J.; Xiao, L.; Wang, Z. Overexpression of EZH2 contributes to acquired cisplatin resistance in ovarian cancer cells in vitro and in vivo. Cancer Biol. Ther. 2010, 10, 788–795. [Google Scholar] [CrossRef] [PubMed]
- Straining, R.; Eighmy, W. Tazemetostat: EZH2 Inhibitor. J. Adv. Pract. Oncol. 2022, 13, 158. [Google Scholar] [CrossRef]
- Wen, L.; Tao, S.-H.; Guo, F.; Li, L.-Z.; Yang, H.-L.; Liang, Y.; Zhang, L.-D.; Ma, L.; Fu, P. Selective EZH2 inhibitor zld1039 alleviates inflammation in cisplatin-induced acute kidney injury partially by enhancing RKIP and suppressing NF-κB p65 pathway. Acta Pharmacol. Sin. 2021, 43, 2067–2080. [Google Scholar] [CrossRef] [PubMed]
- Dorff, T.B.; Longmate, J.A.; Pal, S.K.; Stadler, W.M.; Fishman, M.N.; Vaishampayan, U.N.; Rao, A.; Pinksi, J.K.; Hu, J.S.; Quinn, D.I. Bevacizumab alone or in combination with TRC105 for patients with refractory metastatic renal cell cancer. Cancer 2017, 123, 4566–4573. [Google Scholar] [CrossRef] [PubMed]
- Hainsworth, J.D.; Reeves, J.A.; Mace, J.R.; Crane, E.J.; Hamid, O.; Stille, J.R.; Flynt, A.; Roberson, S.; Polzer, J.; Arrowsmith, E.R. A randomized, open-label phase 2 study of the CXCR4 inhibitor LY2510924 in combination with sunitinib versus sunitinib alone in patients with metastatic renal cell carcinoma (RCC). Target Oncol. 2016, 11, 643–653. [Google Scholar] [CrossRef]
- Choueiri, T.; Kocsis, J.; Pachynski, R.; Poprach, A.; Deshazo, M.; Zakharia, Y.; Lara, P.; Pal, S.; Geczi, L.; Ho, T. Results of the phase II TRAXAR study: A randomized phase II trial of axitinib and TRC105 (TRAX) versus axitinib (AX) alone in patients with advanced or metastatic renal cell carcinoma (mRCC). Ann. Oncol. 2019, 30, v362–v363. [Google Scholar] [CrossRef]


| Transcript | Primer | Sequence (5′-3′) | Product Size (bp) |
|---|---|---|---|
| GAPDH | GAPDH-F | CATGGGTGTGAACCATGA | 104 |
| GAPDH-R | TGTCATGGATGACCTTGG | ||
| ACTB | ACTB-F | CTGCCCTGAGGCACTC | 197 |
| ACTB-R | GTGCCAGGGCAGTGAT | ||
| ABCA13 | ABCA13-f | AGGAGTGTGAGGCTCTTTGC | 207 |
| ABCA13-r | TCAGGTGCTGTCCCTTGAAC | ||
| ABCB1 | ABCB1-f | GGAGGCCAACATACATGCCT | 205 |
| ABCB1-r | CAGGGCTTCTTGGACAACCT | ||
| ABCG2 | ABCG2-f | CATCAACTTTCCGGGGGTGA | 266 |
| ABCG2-r | CACTGGTTGGTCGTCAGGAA | ||
| ALDH1A1 | ALDH1A1-f | TGTTAGCTGATGCCGACTTG | 154 |
| ALDH1A1-r | TTCTTAGCCCGCTCAACACT | ||
| ALDH1A3 | ALDH1A3-f | GAGGAGATTTTCGGGCCAGT | 186 |
| ALDH1A3-r | GAGGGCGTTGTAGCAGTTGA | ||
| ALDH3A1 | ALDH1A3-f | GCAGACCTGCACAAGAATGA | 186 |
| ALDH1A3-r | TGTAGAGCTCGTCCTGCTGA | ||
| CD105 | ENG-f | TCACCACAGCGGAAAAAGGT | 141 |
| ENG-r | GGACACTCTGACCTGCACAA | ||
| CD133 | PROM1-f | TTGCGGTAAAACTGGCTAAG | 155 |
| PROM1-r | TGGGCTTGTCATAACAGGAT | ||
| CXCR4 | CXCR4-f | TGGGTGGTTGTGTTCCAGTTT | 80 |
| CXCR4-r | ATGCAATAGCAGGACAGGATG | ||
| DAB2IP | DAB2IP-f | TGTCGCCCTCACTCTTCAAC | 225 |
| DAB2IP-r | CGGCTGTATTGGAGAGGGTC | ||
| DNMT1 | DNMT1-f | GGCAGACCATCAGGCATTCT | 220 |
| DNMT1-r | ACCATGTCCTTGCAGGCTTT | ||
| EZH2 | hEZH2-F | AGGACGGCTCCTCTAACCAT | 179 |
| EZH2-R | CTTGGTGTTGCACTGTGCTT | ||
| KLF4 | KLF4-f | TCCCATCTTTCTCCACGTTC | 239 |
| KLF4-r | GGTCTCTCTCCGAGGTAGGG | ||
| LIN28A | LIN28A-f | TTCGGCTTCCTGTCCATGAC | 124 |
| LIN28A-r | CCACTGCCTCACCCTCCTT | ||
| MTGR1 | MTGR1-f | CCTCCTACCCTGAATGGTGC | 214 |
| MTGR1-r | GTGCAAGAACAAGAGTCCGC | ||
| NANOG | NANOG-f | TGTGTTCTCTTCCACCCAGC | 205 |
| NANOG-r | CTTCTGCGTCACACCATTGC | ||
| POU5F1 | POU5F1-f | CCCTGGGGGTTCTATTTGGG | 231 |
| POU5F1-r | TCTCCAGGTTGCCTCTCACT | ||
| SALL4 | SALL4-f | GCTCTGTTAGGTACGGACGG | 96 |
| SALL4-r | CTGGTTCCACACAACAGGGT | ||
| SOX2 | SOX-2-f | CATCACCCACAGCAAATGAC | 258 |
| SOX-2-r | GCAAACTTCCTGCAAAGCTC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lyu, C.; Wang, L.; Stadlbauer, B.; Noessner, E.; Buchner, A.; Pohla, H. Identification of EZH2 as Cancer Stem Cell Marker in Clear Cell Renal Cell Carcinoma and the Anti-Tumor Effect of Epigallocatechin-3-Gallate (EGCG). Cancers 2022, 14, 4200. https://doi.org/10.3390/cancers14174200
Lyu C, Wang L, Stadlbauer B, Noessner E, Buchner A, Pohla H. Identification of EZH2 as Cancer Stem Cell Marker in Clear Cell Renal Cell Carcinoma and the Anti-Tumor Effect of Epigallocatechin-3-Gallate (EGCG). Cancers. 2022; 14(17):4200. https://doi.org/10.3390/cancers14174200
Chicago/Turabian StyleLyu, Chen, Lili Wang, Birgit Stadlbauer, Elfriede Noessner, Alexander Buchner, and Heike Pohla. 2022. "Identification of EZH2 as Cancer Stem Cell Marker in Clear Cell Renal Cell Carcinoma and the Anti-Tumor Effect of Epigallocatechin-3-Gallate (EGCG)" Cancers 14, no. 17: 4200. https://doi.org/10.3390/cancers14174200
APA StyleLyu, C., Wang, L., Stadlbauer, B., Noessner, E., Buchner, A., & Pohla, H. (2022). Identification of EZH2 as Cancer Stem Cell Marker in Clear Cell Renal Cell Carcinoma and the Anti-Tumor Effect of Epigallocatechin-3-Gallate (EGCG). Cancers, 14(17), 4200. https://doi.org/10.3390/cancers14174200





