The Prognostic Value of a Single, Randomly Timed Circulating Tumor DNA Measurement in Patients with Metastatic Melanoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
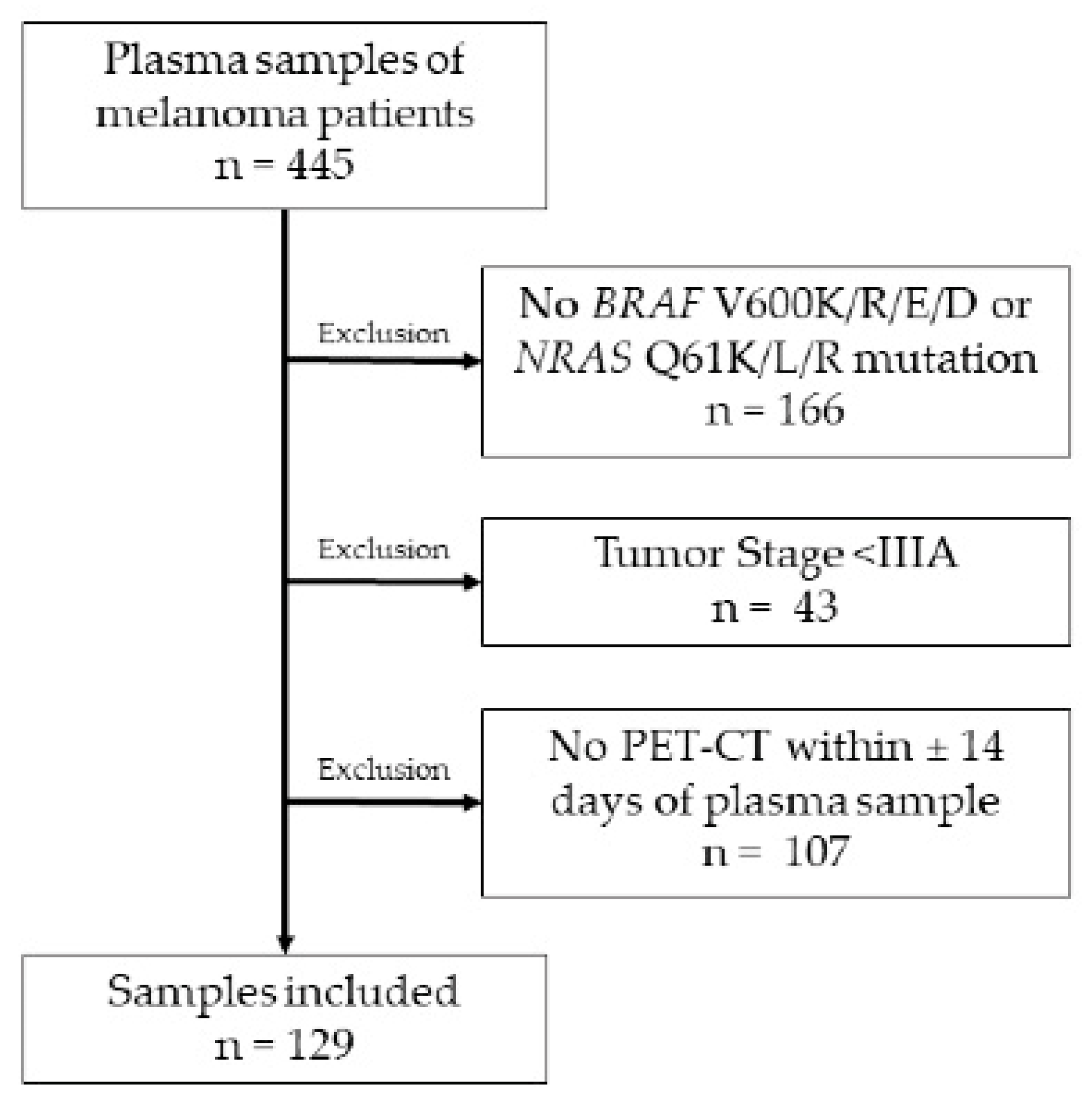
2.1. Patients
2.2. Routine Blood Markers
2.3. ctDNA Assessment
2.4. Disease Progression Assessment
2.5. Statistics
3. Results
3.1. Demographics
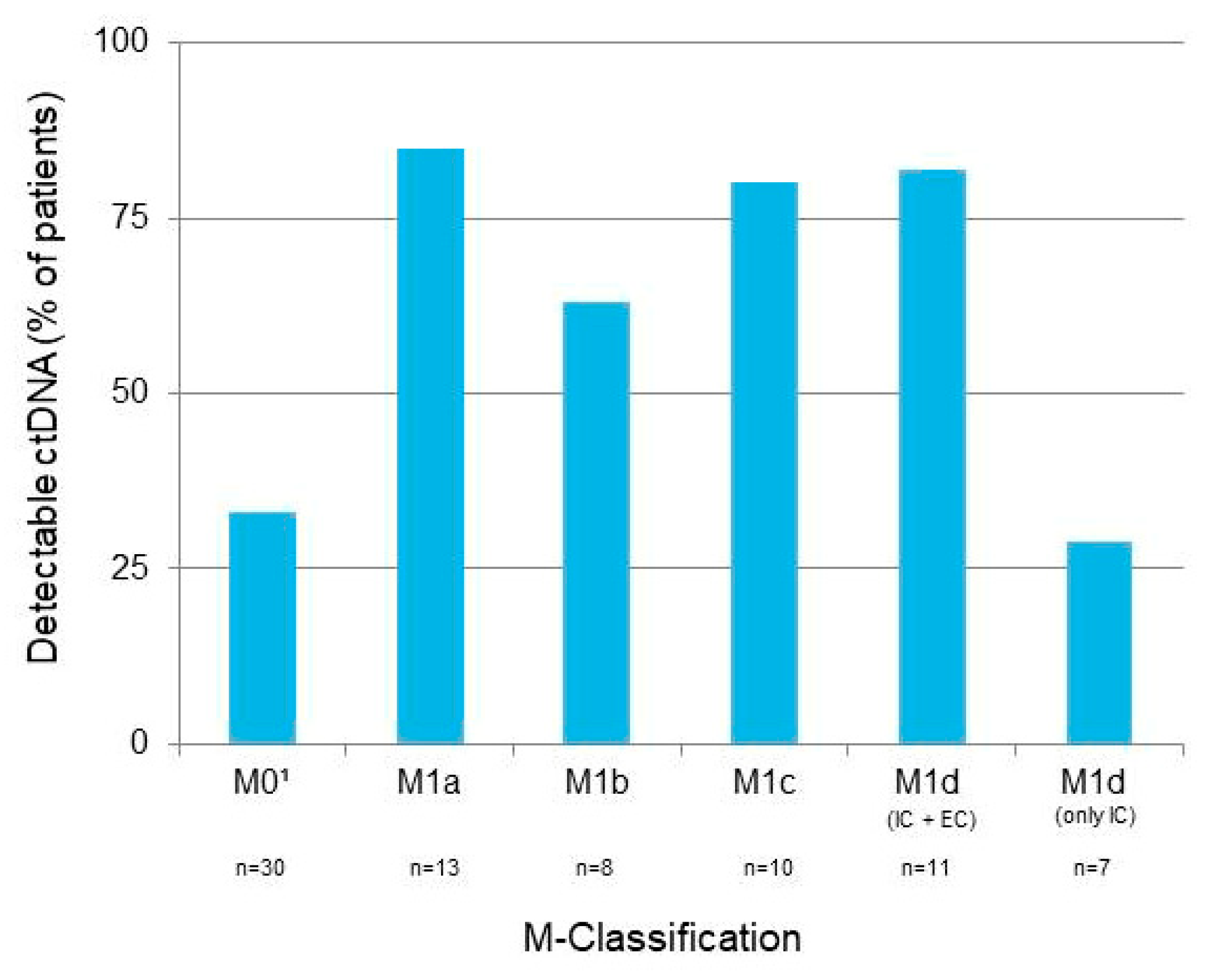
3.2. M-Classification and Metastasis Location at Time of Sample Collection
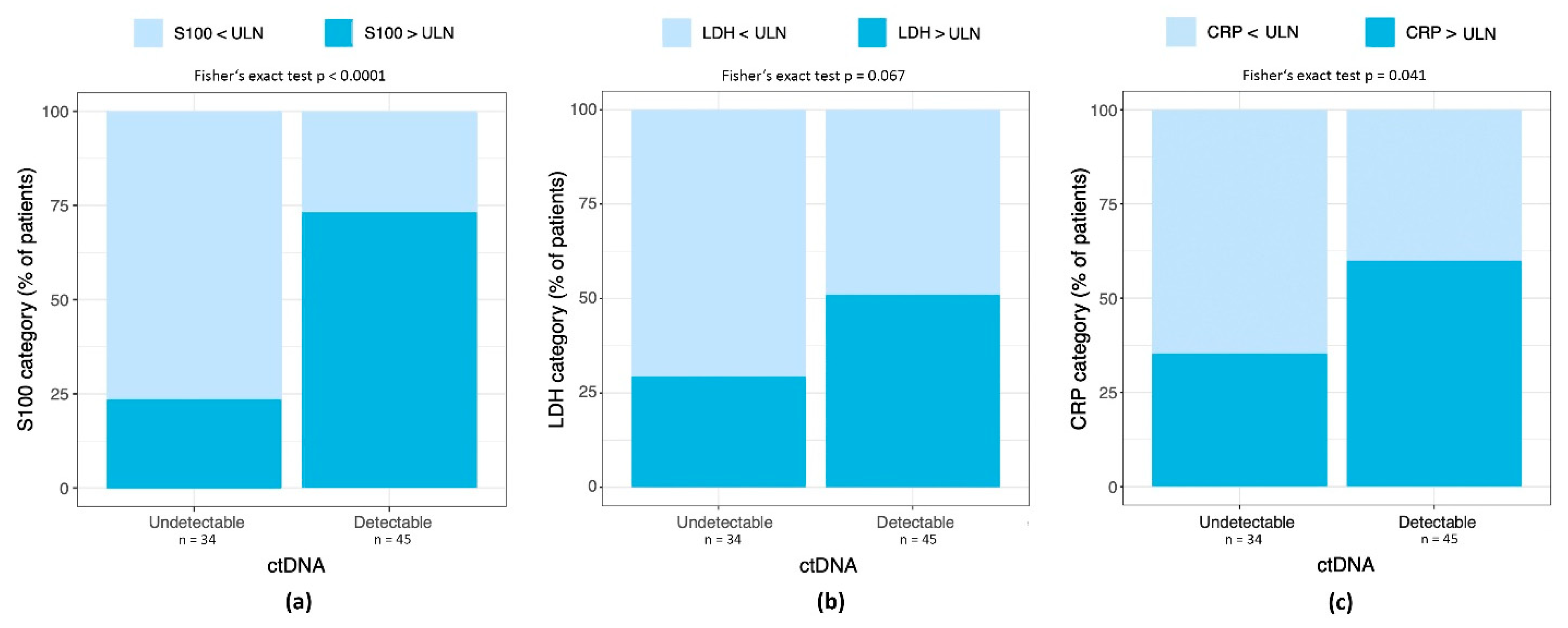
3.3. Detectable ctDNA Correlates with Elevated S100 and CRP, but Not LDH
3.4. ctDNA as a Predictor for Tumor Progression and Overall Survival
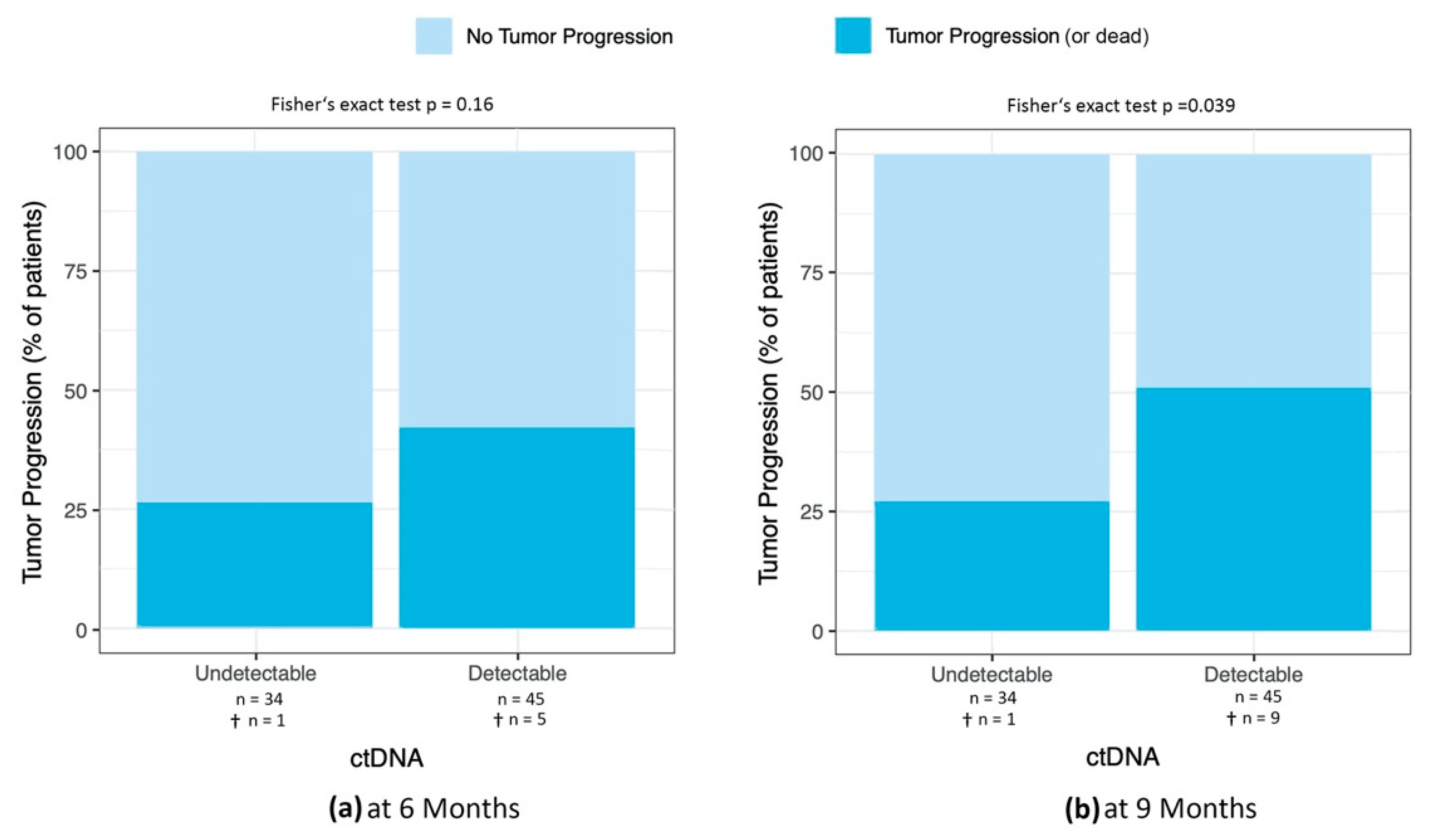
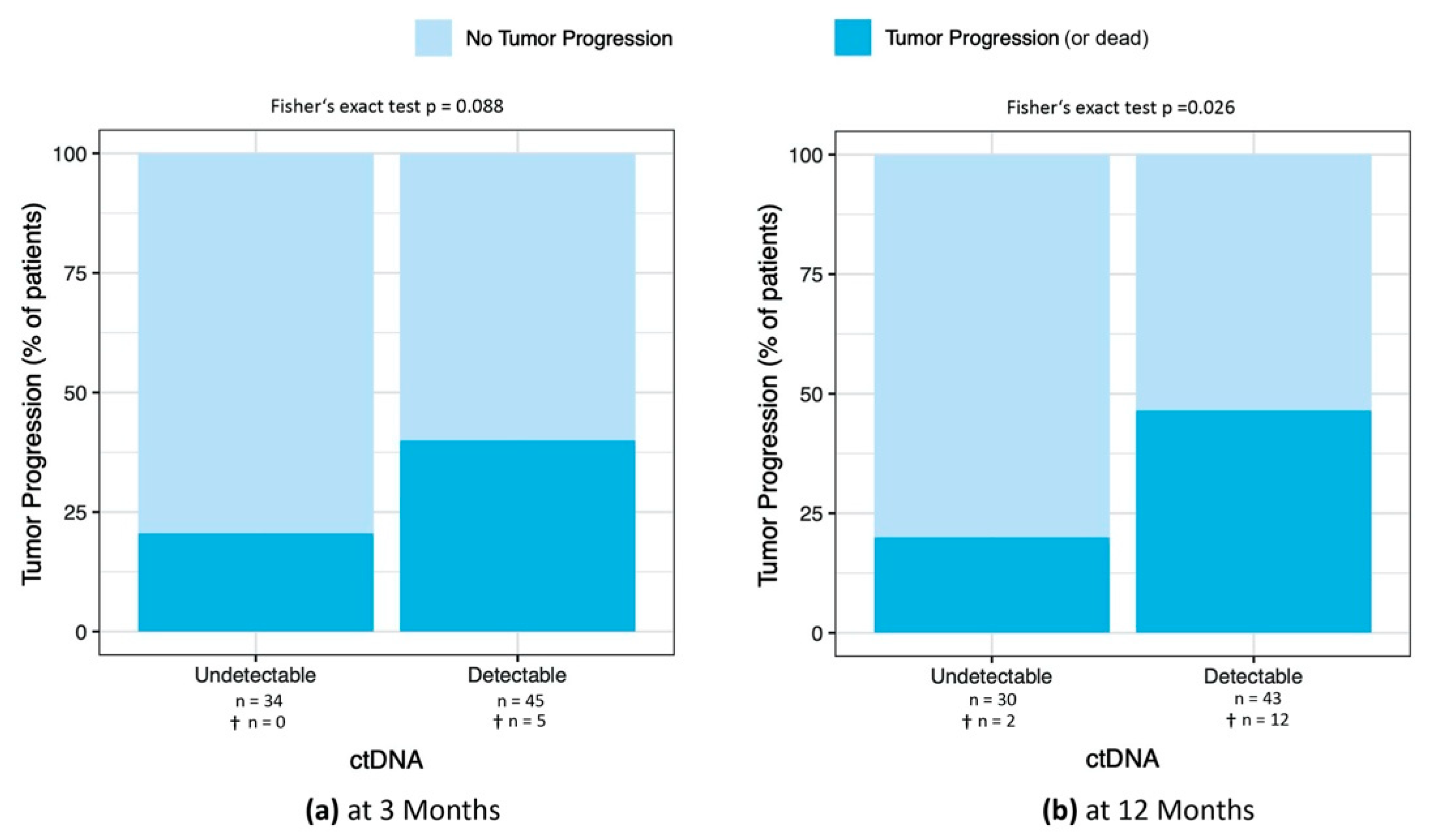
3.4.1. Percentage of Patients with Tumor Progression at 3, 6, 9, and 12 Months after ctDNA Sample
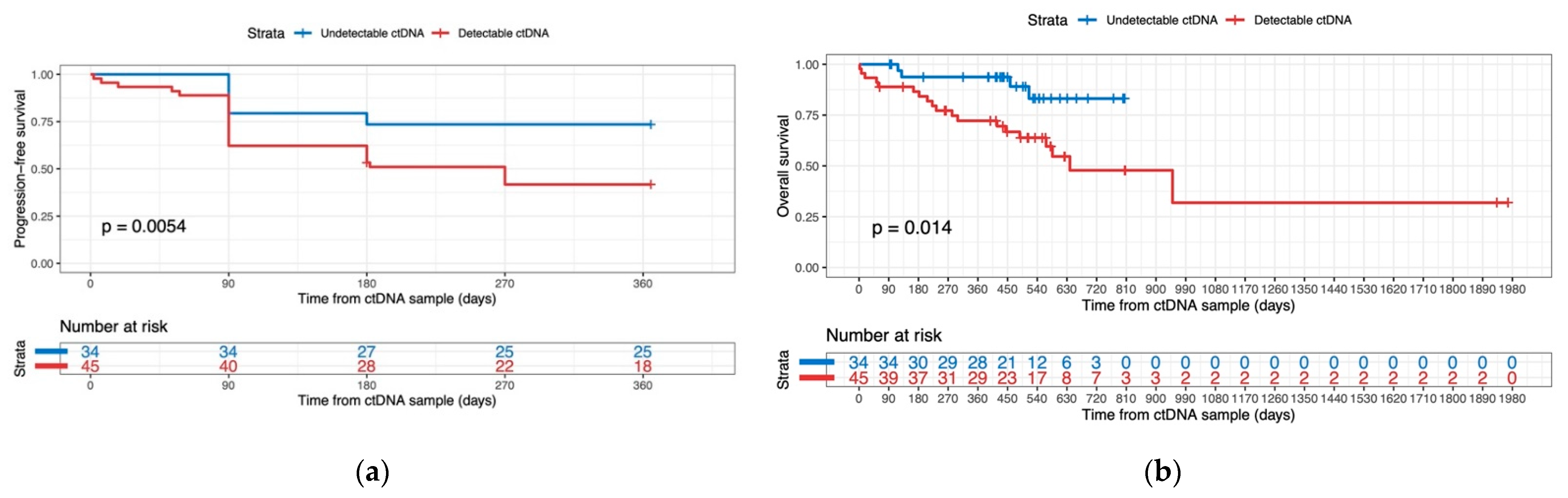
3.4.2. ctDNA Detectability as a Predictor for Progression-Free Survival and Overall Survival
3.5. Longitudinal Disease Monitoring with ctDNA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Percentage of Patients with Tumor Progression at 6 and 9 Months after ctDNA Sample
Appendix B
Longitudinal Disease Monitoring with ctDNA

References
- Gray, E.S.; Rizos, H.; Reid, A.L.; Boyd, S.C.; Pereira, M.R.; Lo, J.; Tembe, V.; Freeman, J.; Lee, J.H.; Scolyer, R.A.; et al. Circulating tumor DNA to monitor treatment response and detect acquired resistance in patients with metastatic melanoma. Oncotarget 2015, 6, 42008–42018. [Google Scholar] [CrossRef] [PubMed]
- Schreuer, M.; Meersseman, G.; Van Den Herrewegen, S.; Jansen, Y.; Chevolet, I.; Bott, A.; Wilgenhof, S.; Seremet, T.; Jacobs, B.; Buyl, R.; et al. Quantitative assessment of BRAF V600 mutant circulating cell-free tumor DNA as a tool for therapeutic monitoring in metastatic melanoma patients treated with BRAF/MEK inhibitors. J. Transl. Med. 2016, 14, 95. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Pessoa, L.S.; Heringer, M.; Ferrer, V.P. ctDNA as a cancer biomarker: A broad overview. Crit. Rev. Oncol. Hematol. 2020, 155, 103109. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra224. [Google Scholar] [CrossRef]
- Gracie, L.; Pan, Y.; Atenafu, E.G.; Ward, D.G.; Teng, M.; Pallan, L.; Stevens, N.M.; Khoja, L. Circulating tumour DNA (ctDNA) in metastatic melanoma, a systematic review and meta-analysis. Eur. J. Cancer 2021, 158, 191–207. [Google Scholar] [CrossRef]
- Syeda, M.M.; Wiggins, J.M.; Corless, B.C.; Long, G.V.; Flaherty, K.T.; Schadendorf, D.; Nathan, P.D.; Robert, C.; Ribas, A.; Davies, M.A.; et al. Circulating tumour DNA in patients with advanced melanoma treated with dabrafenib or dabrafenib plus trametinib: A clinical validation study. Lancet Oncol. 2021, 22, 370–380. [Google Scholar] [CrossRef]
- Váraljai, R.; Wistuba-Hamprecht, K.; Seremet, T.; Diaz, J.M.S.; Nsengimana, J.; Sucker, A.; Griewank, K.; Placke, J.M.; Horn, P.A.; von Neuhoff, N.; et al. Application of Circulating Cell-Free Tumor DNA Profiles for Therapeutic Monitoring and Outcome Prediction in Genetically Heterogeneous Metastatic Melanoma. JCO Precis. Oncol. 2020, 3, PO.18.00229. [Google Scholar] [CrossRef]
- Knuever, J.; Weiss, J.; Persa, O.D.; Kreuzer, K.; Mauch, C.; Hallek, M.; Schlaak, M. The use of circulating cell-free tumor DNA in routine diagnostics of metastatic melanoma patients. Sci. Rep. 2020, 10, 4940. [Google Scholar] [CrossRef]
- McEvoy, A.C.; Pereira, M.R.; Reid, A.; Pearce, R.; Cowell, L.; Al-Ogaili, Z.; Khattak, M.A.; Millward, M.; Meniawy, T.M.; Gray, E.S.; et al. Monitoring melanoma recurrence with circulating tumor DNA: A proof of concept from three case studies. Oncotarget 2019, 10, 113–122. [Google Scholar] [CrossRef][Green Version]
- Garlan, F.; Blanchet, B.; Kramkimel, N.; Puszkiel, A.; Golmard, J.L.; Noe, G.; Dupin, N.; Laurent-Puig, P.; Vidal, M.; Taly, V.; et al. Circulating Tumor DNA Measurement by Picoliter Droplet-Based Digital PCR and Vemurafenib Plasma Concentrations in Patients with Advanced BRAF-Mutated Melanoma. Target Oncol. 2017, 12, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Braune, J.; Keller, L.; Schiller, F.; Graf, E.; Rafei-Shamsabadi, D.; Wehrle, J.; Follo, M.; Philipp, U.; Hussung, S.; Pfeifer, D.; et al. Circulating Tumor DNA Allows Early Treatment Monitoring in BRAF- and NRAS-Mutant Malignant Melanoma. JCO Precis. Oncol. 2020, 4, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Forthun, R.B.; Hovland, R.; Schuster, C.; Puntervoll, H.; Brodal, H.P.; Namløs, H.M.; Aasheim, L.B.; Meza-Zepeda, L.A.; Gjertsen, B.T.; Knappskog, S.; et al. ctDNA detected by ddPCR reveals changes in tumour load in metastatic malignant melanoma treated with bevacizumab. Sci. Rep. 2019, 9, 17471. [Google Scholar] [CrossRef] [PubMed]
- McEvoy, A.C.; Warburton, L.; Al-Ogaili, Z.; Celliers, L.; Calapre, L.; Pereira, M.R.; Khattak, M.A.; Meniawy, T.M.; Millward, M.; Ziman, M.; et al. Correlation between circulating tumour DNA and metabolic tumour burden in metastatic melanoma patients. BMC Cancer 2018, 18, 726. [Google Scholar] [CrossRef]
- Tolmeijer, S.H.; Koornstra, R.H.T.; de Groot, J.W.B.; Geerlings, M.J.; van Rens, D.H.; Boers-Sonderen, M.J.; Schalken, J.A.; Gerritsen, W.R.; Ligtenberg, M.J.L.; Mehra, N. Plasma BRAF Mutation Detection for the Diagnostic and Monitoring Trajectory of Patients with LDH-High Stage IV Melanoma. Cancers 2021, 13, 3913. [Google Scholar] [CrossRef]
- Seremet, T.; Jansen, Y.; Planken, S.; Njimi, H.; Delaunoy, M.; El Housni, H.; Awada, G.; Schwarze, J.K.; Keyaerts, M.; Everaert, H.; et al. Undetectable circulating tumor DNA (ctDNA) levels correlate with favorable outcome in metastatic melanoma patients treated with anti-PD1 therapy. J. Transl. Med. 2019, 17, 303. [Google Scholar] [CrossRef]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef]
- Gaynor, R.; Herschman, H.R.; Irie, R.; Jones, P.; Morton, D.; Cochran, A. S100 protein: A marker for human malignant melanomas? Lancet 1981, 1, 869–871. [Google Scholar] [CrossRef]
- Henze, G.; Dummer, R.; Joller-Jemelka, H.I.; Böni, R.; Burg, G. Serum S100—A marker for disease monitoring in metastatic melanoma. Dermatology 1997, 194, 208–212. [Google Scholar] [CrossRef]
- Marsavela, G.; McEvoy, A.C.; Pereira, M.R.; Reid, A.L.; Al-Ogaili, Z.; Warburton, L.; Khattak, M.A.; Abed, A.; Meniawy, T.M.; Millward, M.; et al. Detection of clinical progression through plasma ctDNA in metastatic melanoma patients: A comparison to radiological progression. Br. J. Cancer 2022, 126, 401–408. [Google Scholar] [CrossRef]
- Griewank, K.G.; Schilling, B. Next-Generation Sequencing to Guide Treatment of Advanced Melanoma. Am. J. Clin. Dermatol. 2017, 18, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Merker, J.D.; Oxnard, G.R.; Compton, C.; Diehn, M.; Hurley, P.; Lazar, A.J.; Lindeman, N.; Lockwood, C.M.; Rai, A.J.; Schilsky, R.L.; et al. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol. 2018, 36, 1631–1641. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Hoon, D.S.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Kustanovich, A.; Schwartz, R.; Peretz, T.; Grinshpun, A. Life and death of circulating cell-free DNA. Cancer Biol. Ther. 2019, 20, 1057–1067. [Google Scholar] [CrossRef] [PubMed]
- Peters, D.L.; Pretorius, P.J. Origin, translocation and destination of extracellular occurring DNA—A new paradigm in genetic behaviour. Clin. Chim. Acta 2011, 412, 806–811. [Google Scholar] [CrossRef]
- Muhanna, N.; Di Grappa, M.A.; Chan, H.H.L.; Khan, T.; Jin, C.S.; Zheng, Y.; Irish, J.C.; Bratman, S.V. Cell-Free DNA Kinetics in a Pre-Clinical Model of Head and Neck Cancer. Sci. Rep. 2017, 7, 16723. [Google Scholar] [CrossRef] [PubMed]
- Muhanna, N.; Eu, D.; Chan, H.H.L.; Douglas, C.; Townson, J.L.; Di Grappa, M.A.; Mohamadi, R.M.; Kelley, S.O.; Bratman, S.V.; Irish, J.C. Cell-free DNA and circulating tumor cell kinetics in a pre-clinical head and neck Cancer model undergoing radiation therapy. BMC Cancer 2021, 21, 1075. [Google Scholar] [CrossRef]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef]
- Leung, F.; Kulasingam, V.; Diamandis, E.P.; Hoon, D.S.; Kinzler, K.; Pantel, K.; Alix-Panabieres, C. Circulating Tumor DNA as a Cancer Biomarker: Fact or Fiction? Clin. Chem. 2016, 62, 1054–1060. [Google Scholar] [CrossRef]
- Davies, M.A.; Liu, P.; McIntyre, S.; Kim, K.B.; Papadopoulos, N.; Hwu, W.J.; Hwu, P.; Bedikian, A. Prognostic factors for survival in melanoma patients with brain metastases. Cancer 2011, 117, 1687–1696. [Google Scholar] [CrossRef]
- Lee, J.H.; Menzies, A.M.; Carlino, M.S.; McEvoy, A.C.; Sandhu, S.; Weppler, A.M.; Diefenbach, R.J.; Dawson, S.J.; Kefford, R.F.; Millward, M.J.; et al. Longitudinal Monitoring of ctDNA in Patients with Melanoma and Brain Metastases Treated with Immune Checkpoint Inhibitors. Clin. Cancer Res. 2020, 26, 4064–4071. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; Mussolin, B.; Venesio, T.; Marsoni, S.; Seoane, J.; Dive, C.; Papadopoulos, N.; Kopetz, S.; Corcoran, R.B.; Siu, L.L.; et al. How liquid biopsies can change clinical practice in oncology. Ann. Oncol. 2019, 30, 1580–1590. [Google Scholar] [CrossRef] [PubMed]
| Characteristic | Total n = 79 | Detectable ctDNA n = 45 (57%) | Undetectable ctDNA n = 34 (43%) | p-Value 1 |
|---|---|---|---|---|
| Age (years) | 0.086 | |||
| Mean (±SD) | 63.3 (±14.3) | 65.8 (±13.4) | 60.1 (±14.9) | |
| Median (Min., Max.) | 62.0 (24, 88) | 67.0 (31, 88) | 59.5 (24, 87) | |
| Gender | 0.831 | |||
| Female | 28 (35%) | 15 (33%) | 13 (38%) | |
| Male | 51 (65%) | 30 (67%) | 21 (62%) | |
| Tumor Stage | 0.308 | |||
| IIIB | 6 (8%) | 2 (4%) | 4 (12%) | |
| IIIC | 9 (11%) | 4 (9%) | 5 (15%) | |
| IV | 64 (81%) | 39 (87%) | 25 (73%) | |
| Breslow Thickness | 0.818 | |||
| Mean (±SD) | 3.08 (±2.97) | 2.99 (±2.08) | 3.17 (±3.75) | |
| Median (Min., Max.) | 2.10 (0.6, 20.0) | 2.66 (0.6, 9.0) | 1.92 (0.6, 20.0) | |
| Missing | 18 (23%) | 13 (29%) | 5 (15%) | |
| Ulceration | 0.668 | |||
| No | 42 (53%) | 21 (47%) | 21 (62%) | |
| Yes | 22 (28%) | 13 (29%) | 9 (26%) | |
| Missing | 15 (19%) | 11 (24%) | 4 (12%) | |
| Driver Mutation | 0.843 | |||
| BRAF p.V600E | 43 (55%) | 25 (56%) | 18 (53%) | |
| BRAF p.V600K | 9 (11%) | 5 (11%) | 4 (11%) | |
| NRAS p.Q61K | 16 (20%) | 10 (22%) | 6 (18%) | |
| NRAS p.Q61R | 11 (14%) | 5 (11%) | 6 (18%) | |
| Therapy at Sampling | 0.184 | |||
| Targeted therapy 2 | 14 (18%) | 9 (20%) | 5 (15%) | |
| Immunotherapy 3 | 43 (54%) | 20 (44%) | 23 (67%) | |
| Chemotherapy | 3 (4%) | 3 (7%) | 0 | |
| Local therapy 4 | 15 (19%) | 11 (25%) | 4 (12%) | |
| None | 4 (5%) | 2 (4%) | 2 (6%) |
| Coef | HR (95% CI) | SE (coef) | z | p-Value | |
|---|---|---|---|---|---|
| Progression-free survival | |||||
| Female Gender | −0.87 | 0.42 (0.20–0.88) | 0.38 | −2.30 | 0.021 |
| S100 > ULN | 1.55 | 4.74 (2.18–10.27) | 0.39 | 3.94 | <0.0001 |
| CRP > ULN | 1.18 | 3.27 (1.49–7.18) | 0.40 | 2.95 | 0.003 |
| Presence of Intracranial Metastases | 0.97 | 2.65 (1.32–5.32) | 0.36 | 2.74 | 0.006 |
| Overall Survival | |||||
| Detectable ctDNA | 1.17 | 3.06 (1.03–9.06) | 0.55 | 2.02 | 0.044 |
| CRP > ULN | 1.65 | 4.36 (1.71–12.07) | 0.62 | 2.19 | 0.013 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boerlin, A.; Bellini, E.; Turko, P.; Cheng, P.F.; Levesque, M.P.; Dummer, R.; Ramelyte, E. The Prognostic Value of a Single, Randomly Timed Circulating Tumor DNA Measurement in Patients with Metastatic Melanoma. Cancers 2022, 14, 4158. https://doi.org/10.3390/cancers14174158
Boerlin A, Bellini E, Turko P, Cheng PF, Levesque MP, Dummer R, Ramelyte E. The Prognostic Value of a Single, Randomly Timed Circulating Tumor DNA Measurement in Patients with Metastatic Melanoma. Cancers. 2022; 14(17):4158. https://doi.org/10.3390/cancers14174158
Chicago/Turabian StyleBoerlin, Aurelio, Elisa Bellini, Patrick Turko, Phil F. Cheng, Mitchell P. Levesque, Reinhard Dummer, and Egle Ramelyte. 2022. "The Prognostic Value of a Single, Randomly Timed Circulating Tumor DNA Measurement in Patients with Metastatic Melanoma" Cancers 14, no. 17: 4158. https://doi.org/10.3390/cancers14174158
APA StyleBoerlin, A., Bellini, E., Turko, P., Cheng, P. F., Levesque, M. P., Dummer, R., & Ramelyte, E. (2022). The Prognostic Value of a Single, Randomly Timed Circulating Tumor DNA Measurement in Patients with Metastatic Melanoma. Cancers, 14(17), 4158. https://doi.org/10.3390/cancers14174158







