Clinically Applicable Pathological Diagnosis System for Cell Clumps in Endometrial Cancer Screening via Deep Convolutional Neural Networks
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement and Patients
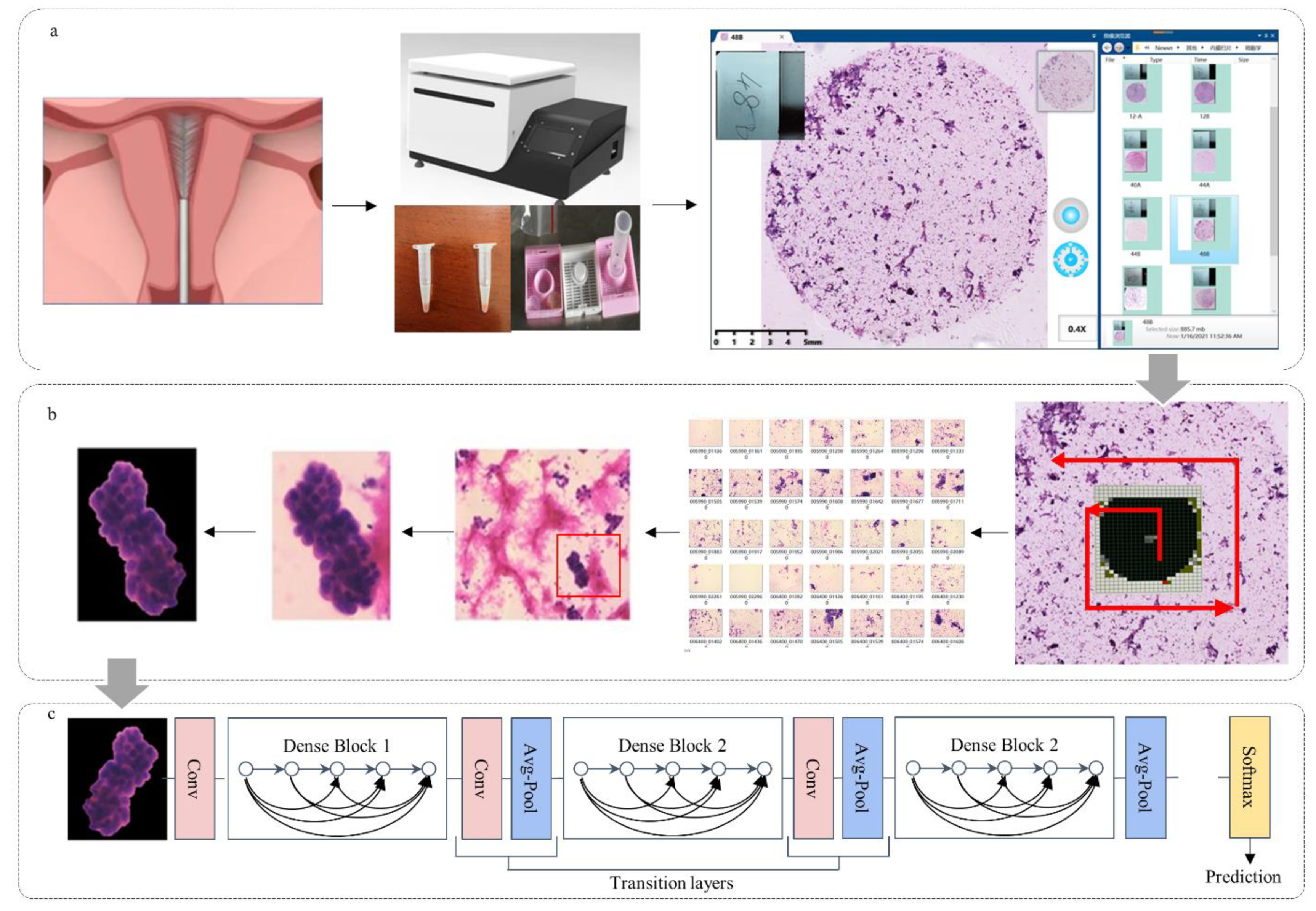
2.2. Preparation of Pathological Slides
2.3. Cytopathological Image Acquisition
2.4. ECCs Image Annotation
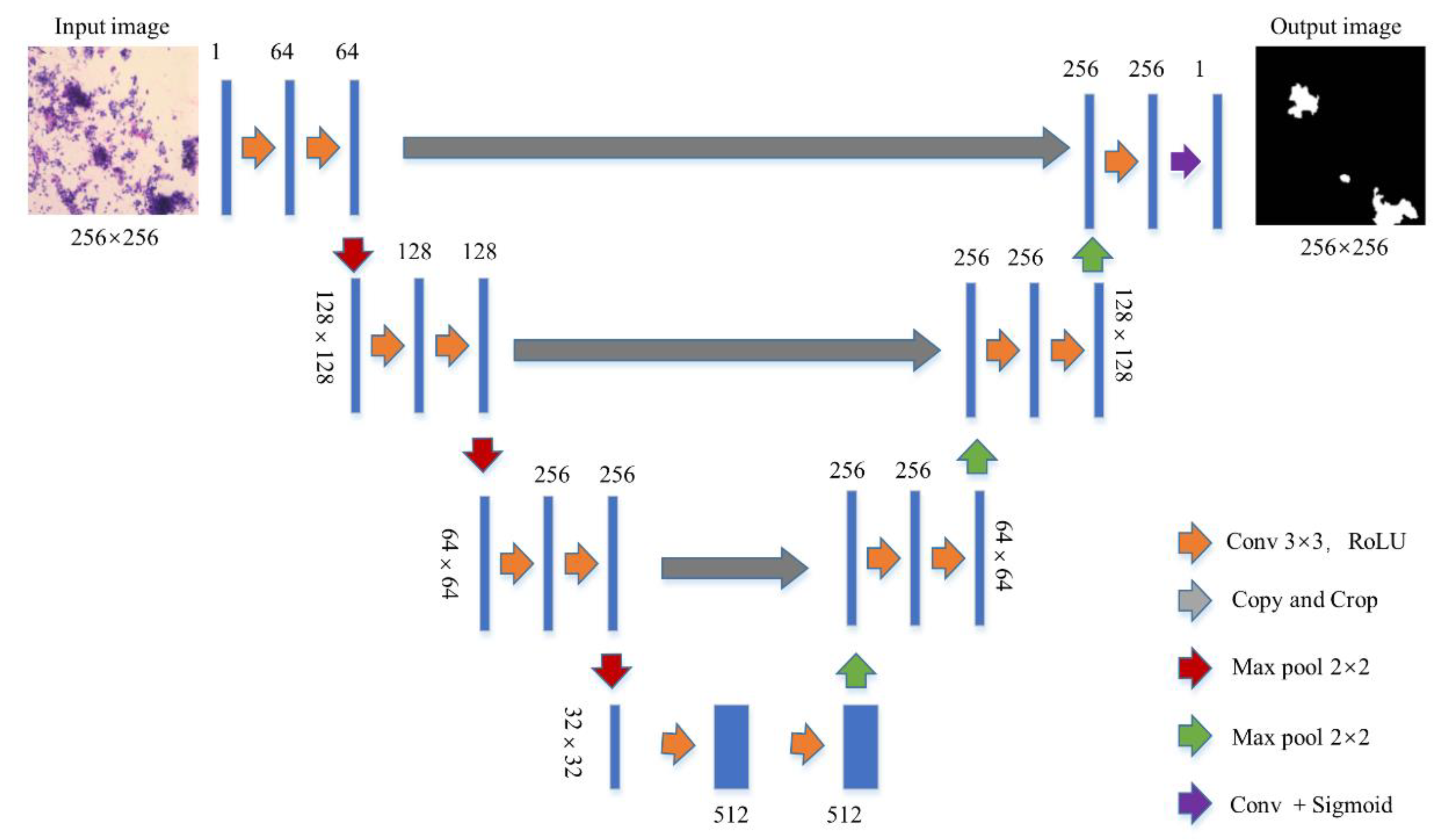
2.5. Segmentation Networks
2.6. Data Preprocessing
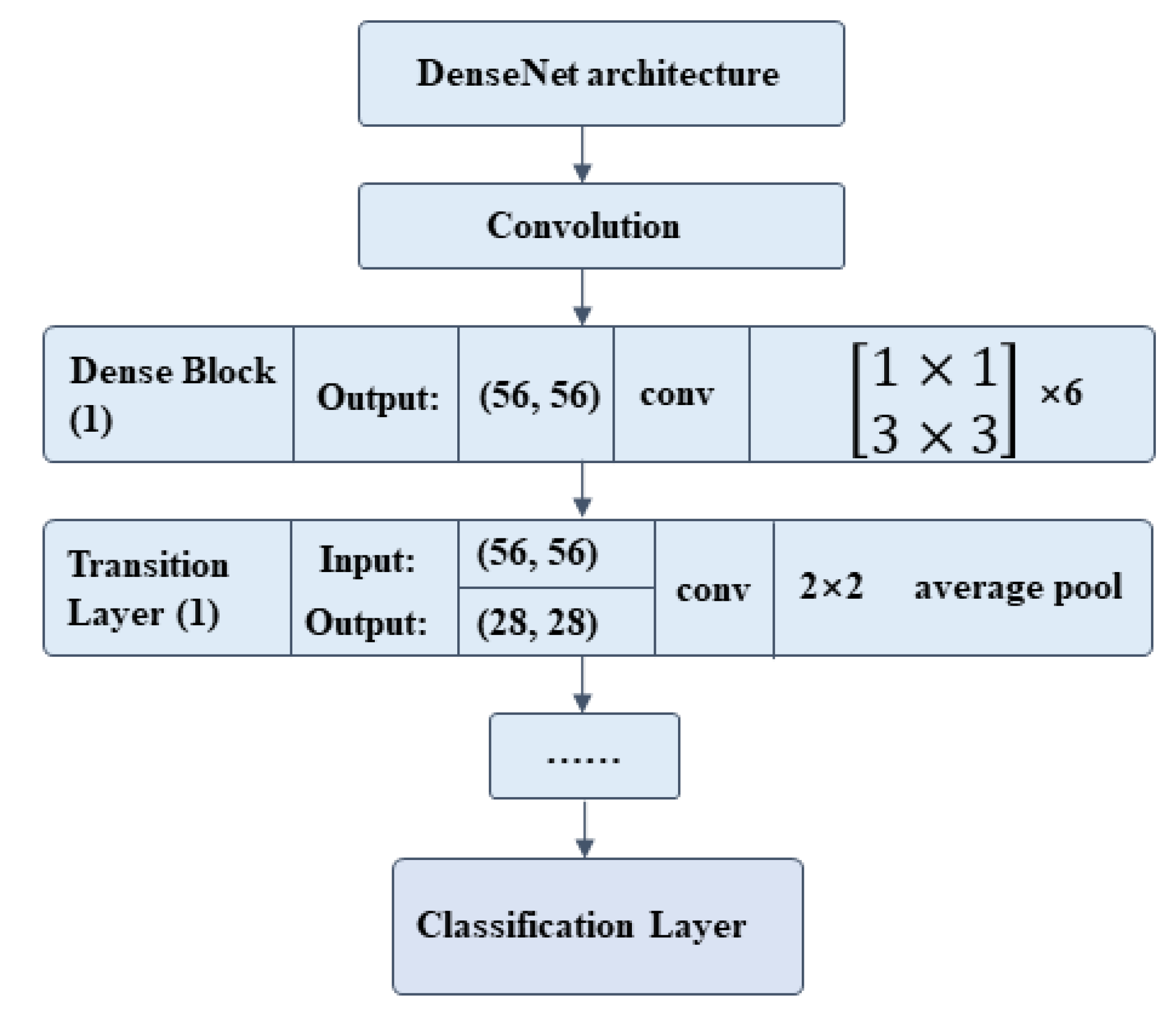
2.7. Classification Network
2.8. Network Evaluation
2.9. Statistical Analysis
2.10. Plots and Charts
3. Results
3.1. Baseline Characteristics
3.2. Dataset
3.3. Verification Set and Test Set
3.4. False Results
3.5. Data Supporting
4. Discussion
Principal Findings
5. Results
5.1. Clinical Implications
5.2. Research Implications
5.3. Strengths and Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef]
- Feng, R.M.; Zong, Y.N.; Cao, S.M.; Xu, R.H. Current cancer situation in China: Good or bad news from the 2018 Global Cancer Statistics? Cancer Commun. 2019, 39, 22. [Google Scholar] [CrossRef]
- Ganz, P.A. Current US Cancer Statistics: Alarming Trends in Young Adults? J. Natl. Cancer Inst. 2019, 111, 1241–1242. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA A Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Amant, F.; Mirza, M.R.; Koskas, M.; Creutzberg, C.L. Cancer of the corpus uteri. Int. J. Gynecol. Obstet. 2018, 143, 37–50. [Google Scholar] [CrossRef]
- Nishida, N.; Murakami, F.; Kuroda, A. Clinical Utility of Endometrial Cell Block Cytology in Postmenopausal Women. Acta Cytol. 2017, 61, 441–446. [Google Scholar] [CrossRef]
- Takeda, T.; Wong, T.F.; Adachi, T. Guidelines for office gynecology in Japan: Japan Society of Obstetrics and Gynecology and Japan Association of Obstetricians and Gynecologists 2011 edition. J. Obstet. Gynaecol. Res. 2012, 38, 615–631. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa-Okamura, C.; Sato, S.; Tsuji, I. Effectiveness of mass screening for endometrial cancer. Acta Cytol. 2002, 46, 277–283. [Google Scholar] [CrossRef]
- Kipp, B.R.; Medeiros, F.; Campion, M.B. Direct uterine sampling with the Tao brush sampler using a liquid-based preparation method for the detection of endometrial cancer and atypical hyperplasia: A feasibility study. Cancer 2008, 114, 228–235. [Google Scholar] [CrossRef]
- Remondi, C.; Sesti, F.; Bonanno, E.; Pietropolli, A.; Piccione, E. Diagnostic accuracy of liquid-based endometrial cytology in the evaluation of endometrial pathology in postmenopausal women. Cytopathology 2013, 24, 365–371. [Google Scholar] [CrossRef] [Green Version]
- Han, L.; Du, J.; Zhao, L. An Efficacious Endometrial Sampler for Screening Endometrial Cancer. Front. Oncol. 2019, 9, 67. [Google Scholar] [CrossRef] [PubMed]
- Fourcade, A.; Khonsari, R.H. Deep learning in medical image analysis: A third eye for doctors. J. Stomatol. Oral Maxillofac. Surg. 2019, 120, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Soenksen, L.R.; Kassis, T.; Conover, S.T. Using deep learning for dermatologist-level detection of suspicious pigmented skin lesions from wide-field images. Sci. Transl. Med. 2021, 13, eabb3652. [Google Scholar] [CrossRef] [PubMed]
- Gulshan, V.; Peng, L.; Coram, M. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 2016, 316, 2402–2410. [Google Scholar] [CrossRef]
- Colling, R.; Pitman, H.; Oien, K. Artificial intelligence in digital pathology: A roadmap to routine use in clinical practice. J. Pathol. 2019, 249, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Jha, S.; Topol, E.J. Adapting to Artificial Intelligence: Radiologists and Pathologists as Information Specialists. JAMA 2016, 316, 2353–2354. [Google Scholar] [CrossRef]
- Bera, K.; Schalper, K.A.; Rimm, D.L.; Velcheti, V.; Madabhushi, A. Artificial intelligence in digital pathology—new tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 2019, 16, 703–715. [Google Scholar] [CrossRef]
- Makris, G.M.; Pouliakis, A.; Siristatidis, C. Image analysis and multi-layer perceptron artificial neural networks for the discrimination between benign and malignant endometrial lesions. Diagn. Cytopathol. 2017, 45, 202–211. [Google Scholar] [CrossRef]
- Sun, H.; Zeng, X.; Xu, T.; Peng, G.; Ma, Y. Computer-Aided Diagnosis in Histopathological Images of the Endometrium Using a Convolutional Neural Network and Attention Mechanisms. IEEE J. Biomed. Health Inform. 2020, 24, 1664–1676. [Google Scholar] [CrossRef]
- The American Society for Bone and Mineral Research. Issue Information-Declaration of Helsinki. J. Bone Miner. Res. 2018, 33, BM i–BM ii. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.; Xiang, Y.; Ma, X.X. Advices on standards of endometrial cancer screening. Zhonghua Fu Chan Ke Za Zhi. 2020, 55, 307–311. [Google Scholar] [PubMed]
- Margari, N.; Pouliakis, A.; Anoinos, D. A reporting system for endometrial cytology: Cytomorphologic criteria-Implied risk of malignancy. Diagn. Cytopathol. 2016, 44, 888–901. [Google Scholar] [CrossRef]
- Norimatsu, Y.; Shimizu, K.; Kobayashi, T.K. Cellular features of endometrial hyperplasia and well differentiated adenocarcinoma using the Endocyte sampler: Diagnostic criteria based on the cytoarchitecture of tissue fragments. Cancer 2006, 108, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Yanoh, K.; Norimatsu, Y.; Hirai, Y. New diagnostic reporting format for endometrial cytology based on cytoarchitectural criteria. Cytopathology 2009, 20, 388–394. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, M.L.; Seto, M.L.; Hing, A.V.; Bull, M.J.; Hopkin, R.J.; Leppig, K.A. Cleidocranial dysplasia with severe parietal bone dysplasia: C-terminal RUNX2 mutations. Birth Defects Res. Part A Clin. Mol. Teratol. 2006, 76, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Dance, A. AI spots cell structures that humans can’t. Nature 2021, 592, 154–155. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Tsuzuki, T.; Akatsuka, J. Automated acquisition of explainable knowledge from unannotated histopathology images. Nat. Commun. 2019, 10, 5642. [Google Scholar] [CrossRef]
- Araujo, T.; Aresta, G.; Castro, E. Classification of breast cancer histology images using Convolutional Neural Networks. PLoS ONE 2017, 12, e0177544. [Google Scholar] [CrossRef]
- Xu, Y.; Jia, Z.; Wang, L.B. Large scale tissue histopathology image classification, segmentation, and visualization via deep convolutional activation features. BMC Bioinform. 2017, 18, 281. [Google Scholar] [CrossRef]
- Papaefthimiou, M.; Symiakaki, H.; Mentzelopoulou, P. Study on the morphology and reproducibility of the diagnosis of endometrial lesions utilizing liquid-based cytology. Cancer 2005, 105, 56–64. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, Q.; Zhao, L. Endometrial Cytology as a Method to Improve the Accuracy of Diagnosis of Endometrial Cancer: Case Report and Meta-Analysis. Front. Oncol. 2019, 9, 256. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Zou, S.; Zhou, W. Clinically applicable histopathological diagnosis system for gastric cancer detection using deep learning. Nat. Commun. 2020, 11, 4294. [Google Scholar] [CrossRef] [PubMed]
- Denny, J.C.; Collins, F.S. Precision medicine in 2030-seven ways to transform healthcare. Cell 2021, 184, 1415–1419. [Google Scholar] [CrossRef] [PubMed]



| Characteristics | n |
|---|---|
| Source | |
| Inpatient Department | 66 |
| Outpatient Department | 47 |
| Age | |
| <40 years old | 13 |
| ≥40 years old | 100 |
| Menstrual Status | |
| Premenopausal | 51 |
| Postmenopausal | 66 |
| Abnormal uterine bleeding | 35 |
| Other Disease | |
| Ovarian cancer | 0 |
| Hypertension | 10 |
| Diabetes | 4 |
| Hormone replacement therapy | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Wang, R.; Xie, Z.; Zhao, L.; Wang, Y.; Sun, C.; Han, L.; Liu, Y.; Hou, H.; Liu, C.; et al. Clinically Applicable Pathological Diagnosis System for Cell Clumps in Endometrial Cancer Screening via Deep Convolutional Neural Networks. Cancers 2022, 14, 4109. https://doi.org/10.3390/cancers14174109
Li Q, Wang R, Xie Z, Zhao L, Wang Y, Sun C, Han L, Liu Y, Hou H, Liu C, et al. Clinically Applicable Pathological Diagnosis System for Cell Clumps in Endometrial Cancer Screening via Deep Convolutional Neural Networks. Cancers. 2022; 14(17):4109. https://doi.org/10.3390/cancers14174109
Chicago/Turabian StyleLi, Qing, Ruijie Wang, Zhonglin Xie, Lanbo Zhao, Yiran Wang, Chao Sun, Lu Han, Yu Liu, Huilian Hou, Chen Liu, and et al. 2022. "Clinically Applicable Pathological Diagnosis System for Cell Clumps in Endometrial Cancer Screening via Deep Convolutional Neural Networks" Cancers 14, no. 17: 4109. https://doi.org/10.3390/cancers14174109
APA StyleLi, Q., Wang, R., Xie, Z., Zhao, L., Wang, Y., Sun, C., Han, L., Liu, Y., Hou, H., Liu, C., Zhang, G., Shi, G., Zhong, D., & Li, Q. (2022). Clinically Applicable Pathological Diagnosis System for Cell Clumps in Endometrial Cancer Screening via Deep Convolutional Neural Networks. Cancers, 14(17), 4109. https://doi.org/10.3390/cancers14174109






