Anti-Tumoral Effect of Chemerin on Ovarian Cancer Cell Lines Mediated by Activation of Interferon Alpha Response
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. RNA Preparation and Real-Time RT-PCR
- IFI44:
- 5′-GCGGCCTGTGCAGGGATGAC-3′ and 5′-TGTCCTTCAGCGATGGGGAATCA-3′,
- OAS1:
- 5′-GAGGCAGCTGGCACAAGAGGC-3′ and 5′-CGTCGGTCTCATCGTCTGCAC-3′,
- DDX60:
- 5′-CGCGGGTCTTTGGACACCACC-3′ and 5′-GCTGCCTGTGCCTCCAACCTG-3′,
- MX1:
- 5′-GGCTGTTTACCAGACTCCGACA-3′ and 5′-CACAAAGCCTGGCAGCTCTCTA-3′
- ACTB:
- 5′-CACCATTGGCAATGAGCGGTTC-3′ and 5′-AGGTCTTTGCGGATGTCCACGT-3′
2.3. Cell Culture, Cell Viability, Soft Agar Colony and Spheroid Formation
2.4. Apoptosis Assays
2.5. Invasion and Migration Assay
2.6. Affymetrix Clariom S Microarray Assay
2.7. Western Blot Analysis
2.8. Statistical Analysis
3. Results
3.1. Expression of Chemerin Receptors CMKLR1 and GPR1 in Ovarian Cancer Cell Lines
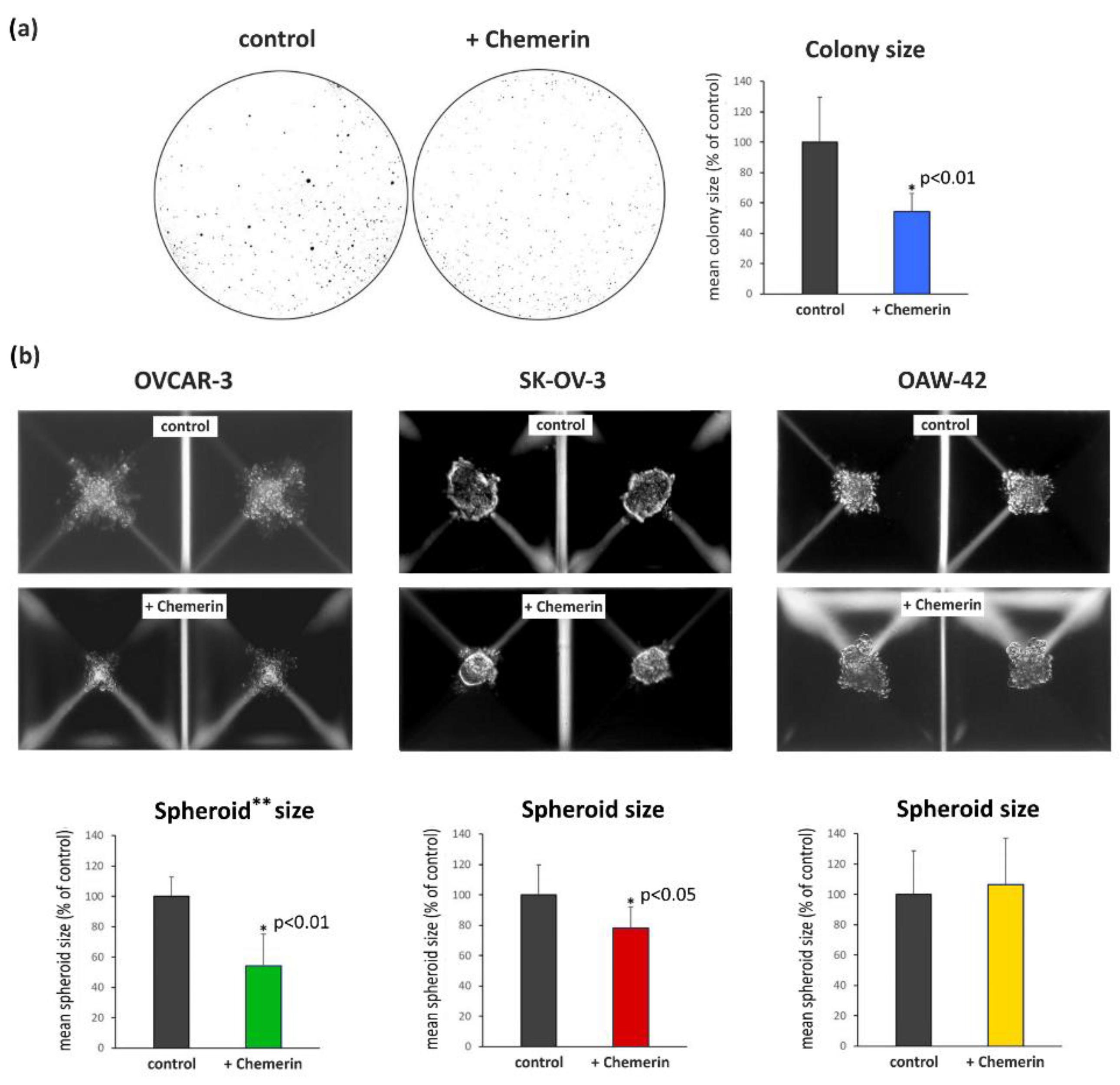
3.2. Effect of Chemerin (huChem-157) on Ovarian Cancer Cell Lines Using 2D and 3D In Vitro Culture Models
3.3. Effect of Chemerin on Apoptosis of Ovarian Cancer Cell Lines
3.4. Effect of Chemerin on Migration and Invasiveness of Ovarian Cancer Cell Lines
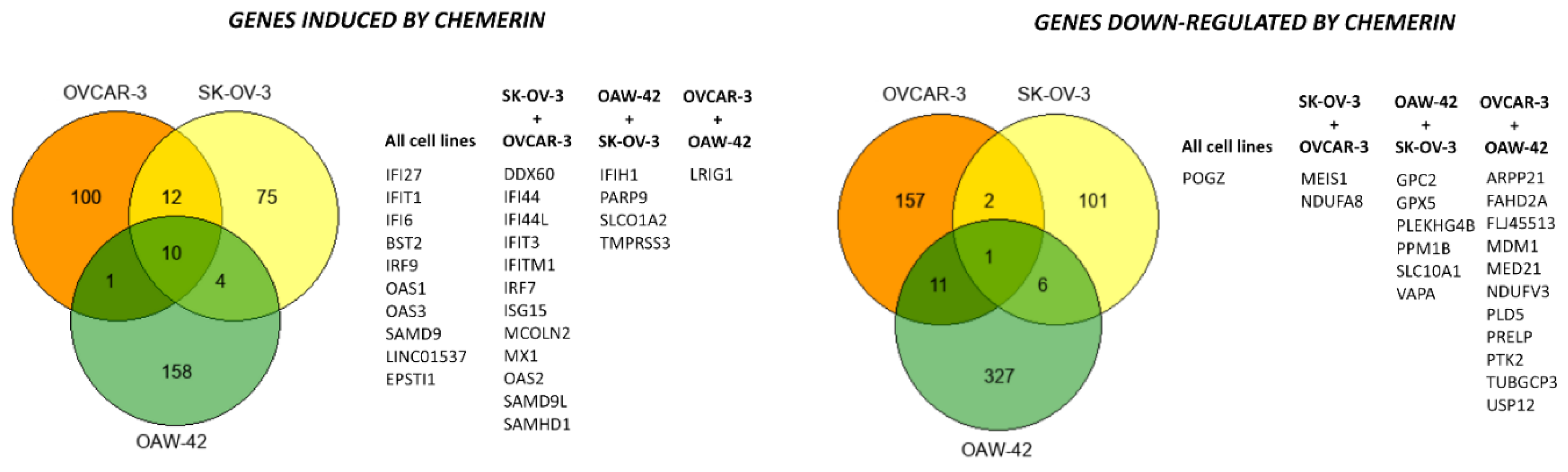
3.5. Chemerin Effects on Transcriptomes of Ovarian Cancer Cell Lines
3.5.1. Verification of the Microarray Data by RT-qPCR and Western Blot Analyses
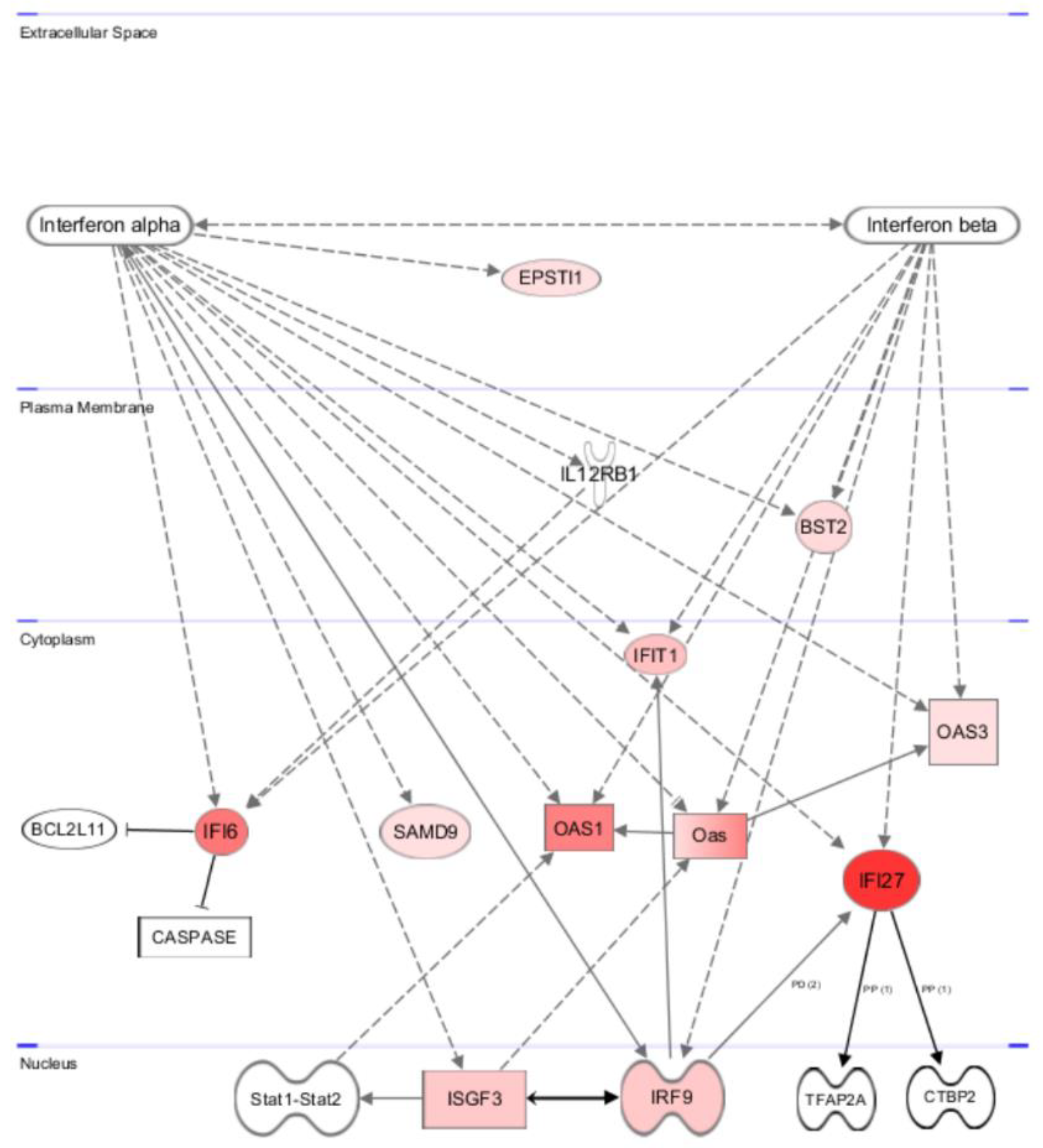
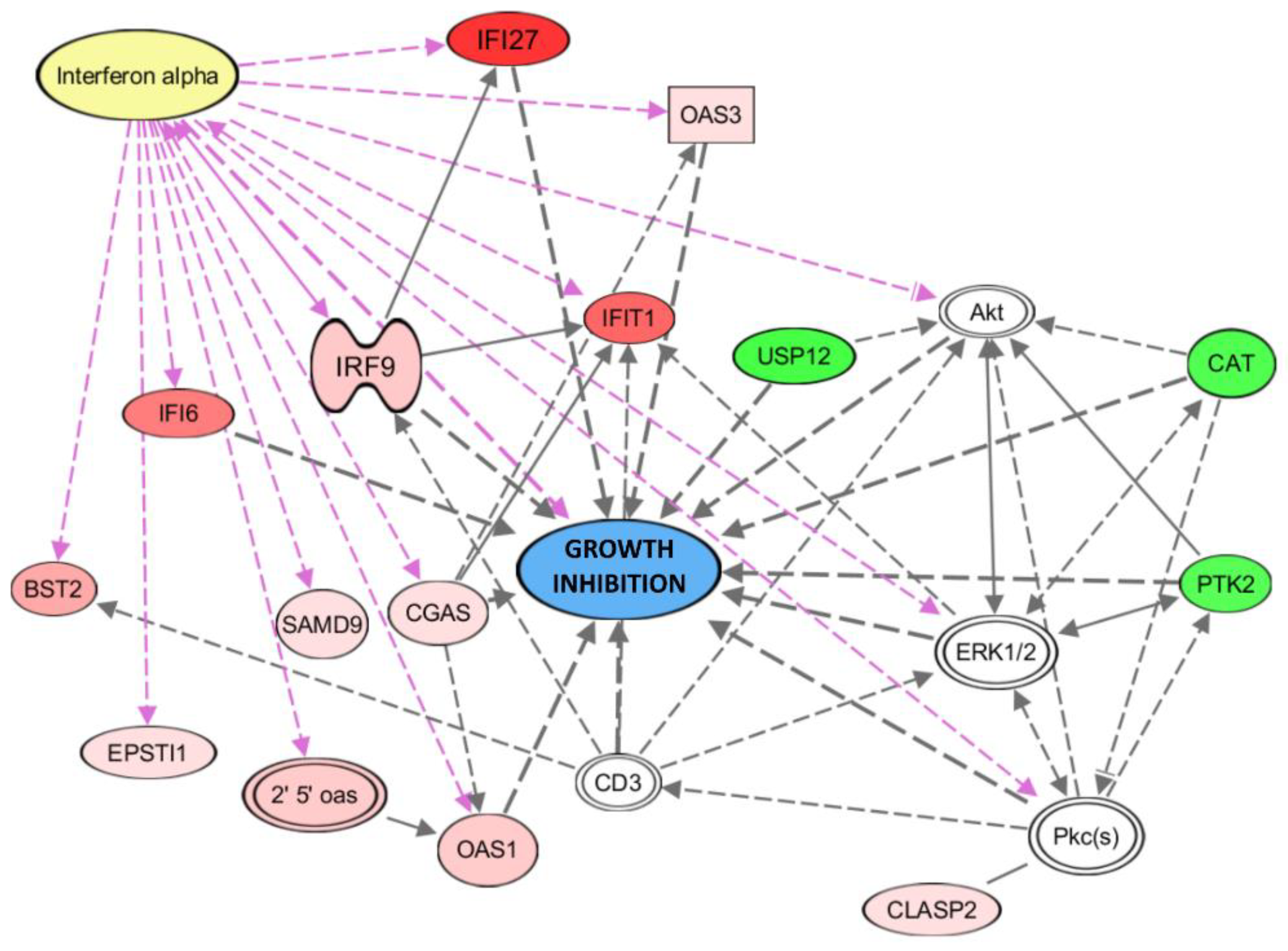
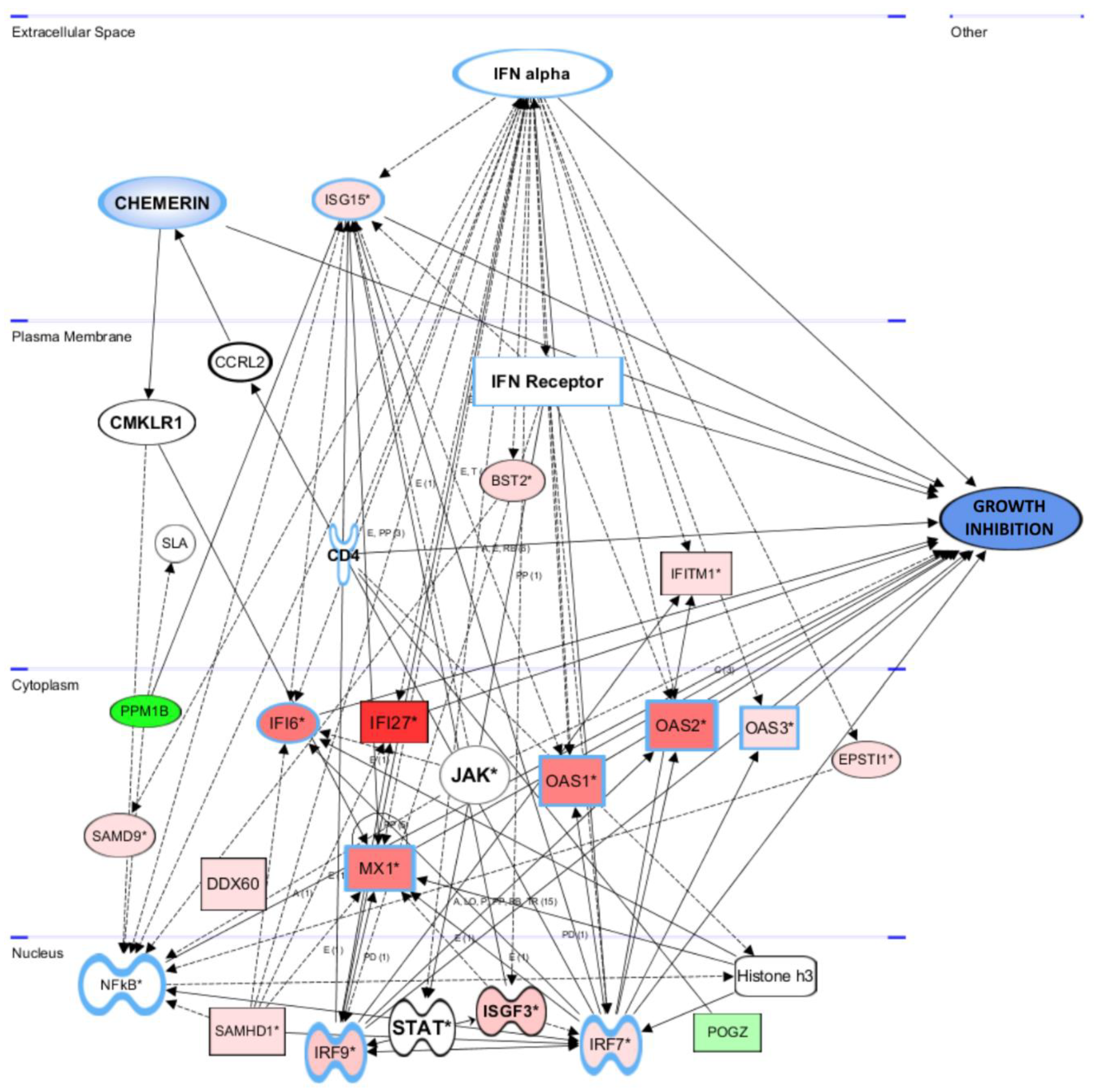
3.5.2. Gene and Pathway Analyses Based on the Chemerin-Triggered Transcriptome Changes
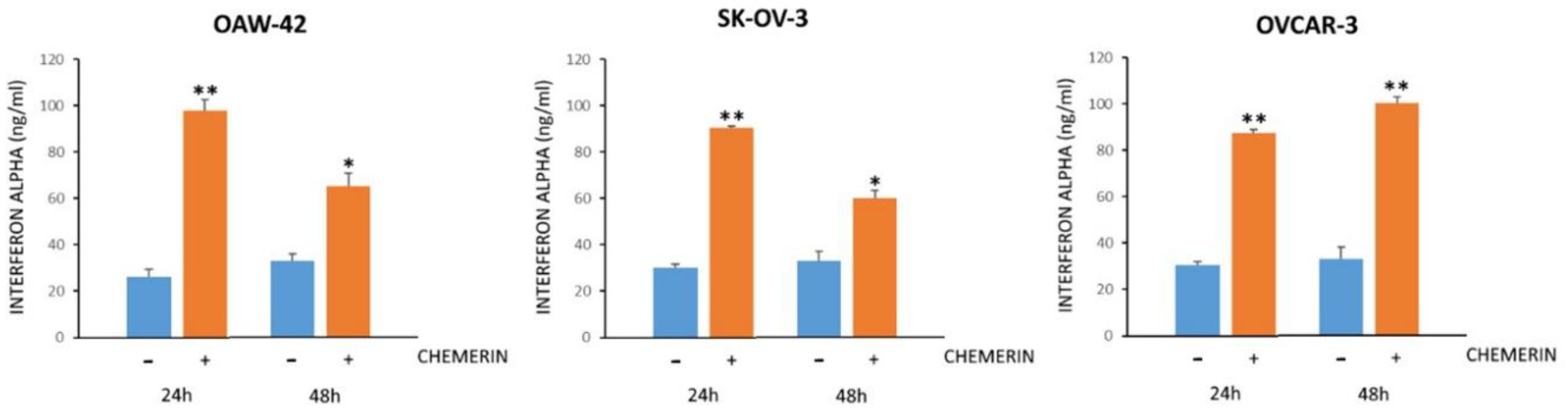
3.6. Chemerin Effect on IFNα Levels
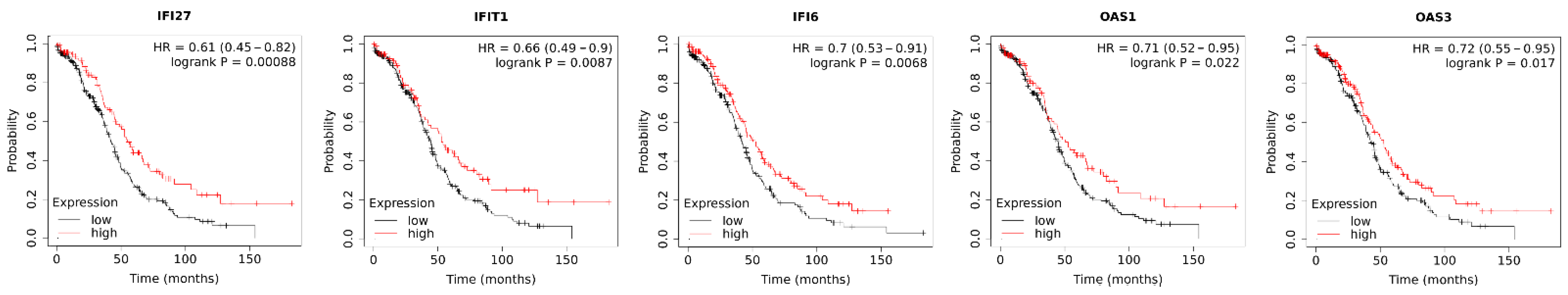
3.7. Association of Chemerin-Regulated Genes with Survival of Ovarian Cancer Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2015. CA Cancer J. Clin. 2015, 65, 5–29. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Trabert, B.; DeSantis, C.E.; Miller, K.D.; Samimi, G.; Runowicz, C.D.; Gaudet, M.M.; Jemal, A.; Siegel, R.L. Ovarian cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Bäumler, M.; Gallant, D.; Druckmann, R.; Kuhn, W. Ultrasound screening of ovarian cancer. Horm. Mol. Biol. Clin. Investig. 2019, 41. [Google Scholar] [CrossRef]
- Karnezis, A.N.; Cho, K.R.; Gilks, C.B.; Pearce, C.L.; Huntsman, D.G. The disparate origins of ovarian cancers: Pathogenesis and prevention strategies. Nat. Rev. Cancer 2017, 17, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Cojocaru, E.; Parkinson, C.A.; Brenton, J.D. Personalising Treatment for High-Grade Serous Ovarian Carcinoma. Clin. Oncol. 2018, 30, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Giusti, I.; Bianchi, S.; Nottola, S.A.; Macchiarelli, G.; Dolo, V. Clinical electron microscopy in the study of human ovarian tissues. Euromediterranean Biomed. J. 2019, 14, 145–151. [Google Scholar]
- Alkady, M.M.; Abdel-Messeih, P.L.; Nosseir, N.M. Assessment of Serum Levels of the Adipocytokine Chemerin in Colorectal Cancer Patients. J. Med. Biochem. 2018, 37, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Eichelmann, F.; Schulze, M.B.; Wittenbecher, C.; Menzel, J.; Weikert, C.; Di Giuseppe, R.; Biemann, R.; Isermann, B.; Fritsche, A.; Boeing, H.; et al. Association of Chemerin Plasma Concentration With Risk of Colorectal Cancer. JAMA Netw. Open 2019, 2, e190896. [Google Scholar] [CrossRef]
- El-Sagheer, G.; Gayyed, M.; Ahmad, A.; Abd El-Fattah, A.; Mohamed, M. Expression of chemerin correlates with a poor prognosis in female breast cancer patients. Breast Cancer 2018, 10, 169–176. [Google Scholar] [CrossRef]
- Goralski, K.B.; Jackson, A.E.; McKeown, B.T.; Sinal, C.J. More Than an Adipokine: The Complex Roles of Chemerin Signaling in Cancer. Int. J. Mol. Sci. 2019, 20, 4778. [Google Scholar] [CrossRef]
- Kim, H.; Lee, J.-H.; Lee, S.K.; Song, N.-Y.; Son, S.H.; Kim, K.R.; Chung, W.-Y. Chemerin Treatment Inhibits the Growth and Bone Invasion of Breast Cancer Cells. Int. J. Mol. Sci. 2020, 21, 2871. [Google Scholar] [CrossRef] [PubMed]
- Pachynski, R.K.; Wang, P.; Salazar, N.; Zheng, Y.; Nease, L.; Rosalez, J.; Leong, W.-I.; Virdi, G.; Rennier, K.; Shin, W.J.; et al. Chemerin Suppresses Breast Cancer Growth by Recruiting Immune Effector Cells into the Tumor Microenvironment. Front. Immunol. 2019, 10, 983. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Fan, J.; Zhu, J.; Ling, Y.; Mi, S.; Chen, H.; Fan, C.; Li, Y. Circulating chemerin level and risk of cancer: A systematic review and meta-analysis. Biomark. Med. 2020, 14, 919–928. [Google Scholar] [CrossRef]
- Shin, W.J.; Zabel, B.A.; Pachynski, R.K. Mechanisms and Functions of Chemerin in Cancer: Potential Roles in Therapeutic Intervention. Front. Immunol. 2018, 9, 2772. [Google Scholar] [CrossRef]
- Treeck, O.; Buechler, C.; Ortmann, O. Chemerin and Cancer. Int. J. Mol. Sci. 2019, 20, 3750. [Google Scholar] [CrossRef] [PubMed]
- Ashtary-Larky, D.; Lamuchi-Deli, N.; Kashkooli, S.; Mombaini, D.; Alipour, M.; Khodadadi, F.; Bagheri, R.; Dutheil, F.; Wong, A. The effects of exercise training on serum concentrations of chemerin in individuals with overweight and obesity: A systematic review, meta-analysis, and meta-regression of 43 clinical trials. Arch. Physiol. Biochem. 2021, 1–16. [Google Scholar] [CrossRef]
- Léniz, A.; González, M.; Besné, I.; Carr-Ugarte, H.; Gómez-García, I.; Portillo, M.P. Role of chemerin in the control of glucose homeostasis. Mol. Cell. Endocrinol. 2022, 541, 111504. [Google Scholar] [CrossRef]
- Bondue, B.; Wittamer, V.; Parmentier, M. Chemerin and its receptors in leukocyte trafficking, inflammation and metabolism. Cytokine Growth Factor Rev. 2011, 22, 331–338. [Google Scholar] [CrossRef]
- Ernst, M.C.; Sinal, C.J. Chemerin: At the crossroads of inflammation and obesity. Trends Endocrinol. Metab. TEM 2010, 21, 660–667. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, J.; Wang, H. Correlation of blood glucose, serum chemerin and insulin resistance with NAFLD in patients with type 2 diabetes mellitus. Exp. Ther. Med. 2018, 15, 2936–2940. [Google Scholar] [CrossRef]
- Chang, S.-S.; Eisenberg, D.; Zhao, L.; Adams, C.; Leib, R.; Morser, J.; Leung, L. Chemerin activation in human obesity. Obesity 2016, 24, 1522–1529. [Google Scholar] [CrossRef] [PubMed]
- Mårtensson, U.E.A.; Fenyö, E.M.; Olde, B.; Owman, C. Characterization of the human chemerin receptor--ChemR23/CMKLR1--as co-receptor for human and simian immunodeficiency virus infection, and identification of virus-binding receptor domains. Virology 2006, 355, 6–17. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rourke, J.L.; Muruganandan, S.; Dranse, H.J.; McMullen, N.M.; Sinal, C.J. Gpr1 is an active chemerin receptor influencing glucose homeostasis in obese mice. J. Endocrinol. 2014, 222, 201–215. [Google Scholar] [CrossRef] [PubMed]
- Monnier, J.; Lewén, S.; O’Hara, E.; Huang, K.; Tu, H.; Butcher, E.C.; Zabel, B.A. Expression, regulation, and function of atypical chemerin receptor CCRL2 on endothelial cells. J. Immunol. 2012, 189, 956–967. [Google Scholar] [CrossRef]
- Kennedy, A.J.; Davenport, A.P. International Union of Basic and Clinical Pharmacology CIII: Chemerin Receptors CMKLR1 (Chemerin1) and GPR1 (Chemerin2) Nomenclature, Pharmacology, and Function. Pharmacol. Rev. 2018, 70, 174–196. [Google Scholar] [CrossRef]
- Zhai, Y.; Pang, Y. Systemic and ovarian inflammation in women with polycystic ovary syndrome. J. Reprod. Immunol. 2022, 151, 103628. [Google Scholar] [CrossRef] [PubMed]
- Bongrani, A.; Plotton, I.; Mellouk, N.; Ramé, C.; Guerif, F.; Froment, P.; Dupont, J. High androgen concentrations in follicular fluid of polycystic ovary syndrome women. Reprod. Biol. Endocrinol. 2022, 20, 88. [Google Scholar] [CrossRef] [PubMed]
- Reverchon, M.; Cornuau, M.; Ramé, C.; Guerif, F.; Royère, D.; Dupont, J. Chemerin inhibits IGF-1-induced progesterone and estradiol secretion in human granulosa cells. Hum. Reprod. 2012, 27, 1790–1800. [Google Scholar] [CrossRef]
- Chen, P.; Jia, R.; Liu, Y.; Cao, M.; Zhou, L.; Zhao, Z. Progress of Adipokines in the Female Reproductive System: A Focus on Polycystic Ovary Syndrome. Front. Endocrinol. 2022, 13, 881684. [Google Scholar] [CrossRef]
- Yao, J.; Li, Z.; Fu, Y.; Wu, R.; Wang, Y.; Liu, C.; Yang, L.; Zhang, H. Involvement of obesity-associated upregulation of chemerin/chemokine-like receptor 1 in oxidative stress and apoptosis in ovaries and granulosa cells. Biochem. Biophys. Res. Commun. 2019, 510, 449–455. [Google Scholar] [CrossRef]
- Bozaoglu, K.; Curran, J.E.; Stocker, C.J.; Zaibi, M.S.; Segal, D.; Konstantopoulos, N.; Morrison, S.; Carless, M.; Dyer, T.D.; Cole, S.A.; et al. Chemerin, a novel adipokine in the regulation of angiogenesis. J. Clin. Endocrinol. Metab. 2010, 95, 2476–2485. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Adya, R.; Tan, B.K.; Chen, J.; Randeva, H.S. Identification of chemerin receptor (ChemR23) in human endothelial cells: Chemerin-induced endothelial angiogenesis. Biochem. Biophys. Res. Commun. 2010, 391, 1762–1768. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, N.; Naruse, K.; Kobayashi, Y.; Miyabe, M.; Saiki, T.; Enomoto, A.; Takahashi, M.; Matsubara, T. Chemerin promotes angiogenesis in vivo. Physiol. Rep. 2018, 6, e13962. [Google Scholar] [CrossRef]
- Schutyser, E.; Struyf, S.; Proost, P.; Opdenakker, G.; Laureys, G.; Verhasselt, B.; Peperstraete, L.; van de Putte, I.; Saccani, A.; Allavena, P.; et al. Identification of biologically active chemokine isoforms from ascitic fluid and elevated levels of CCL18/pulmonary and activation-regulated chemokine in ovarian carcinoma. J. Biol. Chem. 2002, 277, 24584–24593. [Google Scholar] [CrossRef]
- Hoffmann, M.; Rak, A.; Ptak, A. Bisphenol A and its derivatives decrease expression of chemerin, which reverses its stimulatory action in ovarian cancer cells. Toxicol. Lett. 2018, 291, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Lánczky, A.; Győrffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Kulkarni, N.S.; Vaidya, B.; Gupta, V. Nano-synergistic combination of Erlotinib and Quinacrine for non-small cell lung cancer (NSCLC) therapeutics-Evaluation in biologically relevant in-vitro models. Mater. Sci. Eng. C Mater. Biol. Appl. 2021, 121, 111891. [Google Scholar] [CrossRef]
- Schmitz, C.; Potekhina, E.; Belousov, V.V.; Lavrentieva, A. Hypoxia Onset in Mesenchymal Stem Cell Spheroids: Monitoring With Hypoxia Reporter Cells. Front. Bioeng. Biotechnol. 2021, 9, 611837. [Google Scholar] [CrossRef]
- Zuppinger, C. 3D Cardiac Cell Culture: A Critical Review of Current Technologies and Applications. Front. Cardiovasc. Med. 2019, 6, 87. [Google Scholar] [CrossRef] [PubMed]
- Schmidhauser, M.; Renz, P.F.; Tsikrika, P.; Freimann, R.; Wutz, A.; Wrana, J.L.; Beyer, T.A. Gaining Insights into the Function of Post-Translational Protein Modification Using Genome Engineering and Molecular Cell Biology. J. Mol. Biol. 2019, 431, 3920–3932. [Google Scholar] [CrossRef] [PubMed]
- Treeck, O.; Belgutay, D.; Häring, J.; Schüler, S.; Lattrich, C.; Ortmann, O. Network analysis of icb-1 gene function in human breast cancer cells. J. Cell. Biochem. 2012, 113, 2979–2988. [Google Scholar] [CrossRef] [PubMed]
- Pohl, R.; Eichelberger, L.; Feder, S.; Haberl, E.M.; Rein-Fischboeck, L.; McMullen, N.; Sinal, C.J.; Bruckmann, A.; Weiss, T.S.; Beck, M.; et al. Hepatocyte expressed chemerin-156 does not protect from experimental non-alcoholic steatohepatitis. Mol. Cell. Biochem. 2022, 477, 2059–2071. [Google Scholar] [CrossRef]
- Sell, H.; Divoux, A.; Poitou, C.; Basdevant, A.; Bouillot, J.-L.; Bedossa, P.; Tordjman, J.; Eckel, J.; Clément, K. Chemerin correlates with markers for fatty liver in morbidly obese patients and strongly decreases after weight loss induced by bariatric surgery. J. Clin. Endocrinol. Metab. 2010, 95, 2892–2896. [Google Scholar] [CrossRef]
- Chen, C.-L.; Cheung, L.W.T.; Lau, M.-T.; Choi, J.-H.; Auersperg, N.; Wang, H.-S.; Wong, A.S.T.; Leung, P.C.K. Differential role of gonadotropin-releasing hormone on human ovarian epithelial cancer cell invasion. Endocrine 2007, 31, 311–320. [Google Scholar] [CrossRef]
- Roy, S.; Ghosh, S.; Mallick, P.; Maity, P. Acivicin with glutaminase regulates proliferation and invasion of human MCF-7 and OAW-42 cells—An in vitro study. Indian J. Exp. Biol. 2008, 46, 22–26. [Google Scholar]
- Majoros, A.; Platanitis, E.; Kernbauer-Hölzl, E.; Rosebrock, F.; Müller, M.; Decker, T. Canonical and Non-Canonical Aspects of JAK-STAT Signaling: Lessons from Interferons for Cytokine Responses. Front. Immunol. 2017, 8, 29. [Google Scholar] [CrossRef]
- Tsuno, T.; Mejido, J.; Zhao, T.; Schmeisser, H.; Morrow, A.; Zoon, K.C. IRF9 is a key factor for eliciting the antiproliferative activity of IFN-alpha. J. Immunother. 2009, 32, 803–816. [Google Scholar] [CrossRef]
- Michalska, A.; Blaszczyk, K.; Wesoly, J.; Bluyssen, H.A.R. A Positive Feedback Amplifier Circuit That Regulates Interferon (IFN)-Stimulated Gene Expression and Controls Type I and Type II IFN Responses. Front. Immunol. 2018, 9, 1135. [Google Scholar] [CrossRef]
- Liu-Chittenden, Y.; Patel, D.; Gaskins, K.; Giordano, T.J.; Assie, G.; Bertherat, J.; Kebebew, E. Serum RARRES2 Is a Prognostic Marker in Patients with Adrenocortical Carcinoma. J. Clin. Endocrinol. Metab. 2016, 101, 3345–3352. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhou, J.; Tang, X.; Zhou, L.-Y.; Zhai, L.-L.; Vanessa, M.E.-D.; Yi, J.; Yi, Y.-Y.; Lin, J.; Qian, J.; et al. Reduced expression of chemerin is associated with poor clinical outcome in acute myeloid leukemia. Oncotarget 2017, 8, 92536–92544. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pachynski, R.K.; Zabel, B.A.; Kohrt, H.E.; Tejeda, N.M.; Monnier, J.; Swanson, C.D.; Holzer, A.K.; Gentles, A.J.; Sperinde, G.V.; Edalati, A.; et al. The chemoattractant chemerin suppresses melanoma by recruiting natural killer cell antitumor defenses. J. Exp. Med. 2012, 209, 1427–1435. [Google Scholar] [CrossRef] [PubMed]
- Tümmler, C.; Snapkov, I.; Wickström, M.; Moens, U.; Ljungblad, L.; Maria Elfman, L.H.; Winberg, J.-O.; Kogner, P.; Johnsen, J.I.; Sveinbjørnsson, B. Inhibition of chemerin/CMKLR1 axis in neuroblastoma cells reduces clonogenicity and cell viability in vitro and impairs tumor growth in vivo. Oncotarget 2017, 8, 95135–95151. [Google Scholar] [CrossRef]
- Lu, Z.; Liang, J.; He, Q.; Wan, Q.; Hou, J.; Lian, K.; Wang, A. The serum biomarker chemerin promotes tumorigenesis and metastasis in oral squamous cell carcinoma. Clin. Sci. 2019, 133, 681–695. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Sun, P.; Dong, K.; Xin, Y.; TaiLulu, A.; Li, Q.; Sun, J.; Peng, M.; Shi, P. Chemerin reverses the malignant phenotype and induces differentiation of human hepatoma SMMC7721 cells. Arch. Pharm. Res. 2021, 44, 194–204. [Google Scholar] [CrossRef]
- Gao, C.; Shi, J.; Zhang, J.; Li, Y.; Zhang, Y. Chemerin promotes proliferation and migration of ovarian cancer cells by upregulating expression of PD-L1. J. Zhejiang Univ. Sci. B 2022, 23, 164–170. [Google Scholar] [CrossRef]
- Kumar, J.D.; Aolymat, I.; Tiszlavicz, L.; Reisz, Z.; Garalla, H.M.; Beynon, R.; Simpson, D.; Dockray, G.J.; Varro, A. Chemerin acts via CMKLR1 and GPR1 to stimulate migration and invasion of gastric cancer cells: Putative role of decreased TIMP-1 and TIMP-2. Oncotarget 2019, 10, 98–112. [Google Scholar] [CrossRef][Green Version]
- Kumar, J.D.; Kandola, S.; Tiszlavicz, L.; Reisz, Z.; Dockray, G.J.; Varro, A. The role of chemerin and ChemR23 in stimulating the invasion of squamous oesophageal cancer cells. Br. J. Cancer 2016, 114, 1152–1159. [Google Scholar] [CrossRef]
- Hoffmann, M.; Gogola, J.; Ptak, A. Apelin abrogates the stimulatory effects of 17β-estradiol and insulin-like growth factor-1 on proliferation of epithelial and granulosa ovarian cancer cell lines via crosstalk between APLNR and ERα/IGF1R. Mol. Biol. Rep. 2019, 46, 6325–6338. [Google Scholar] [CrossRef]
- Tagliaferri, P.; Caraglia, M.; Budillon, A.; Marra, M.; Vitale, G.; Viscomi, C.; Masciari, S.; Tassone, P.; Abbruzzese, A.; Venuta, S. New pharmacokinetic and pharmacodynamic tools for interferon-alpha (IFN-alpha) treatment of human cancer. Cancer Immunol. Immunother. 2005, 54, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Gytz, H.; Hansen, M.F.; Skovbjerg, S.; Kristensen, A.C.M.; Hørlyck, S.; Jensen, M.B.; Fredborg, M.; Markert, L.D.; McMillan, N.A.; Christensen, E.I.; et al. Apoptotic properties of the type 1 interferon induced family of human mitochondrial membrane ISG12 proteins. Biol. Cell 2017, 109, 94–112. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zu, T.; Li, T.; Li, M.; Mi, J.; Bai, F.; Liu, G.; Wen, J.; Li, H.; Brakebusch, C.; et al. ATF3 downmodulates its new targets IFI6 and IFI27 to suppress the growth and migration of tongue squamous cell carcinoma cells. PLoS Genet. 2021, 17, e1009283. [Google Scholar] [CrossRef]
- Hsieh, W.-L.; Huang, Y.-H.; Wang, T.-M.; Ming, Y.-C.; Tsai, C.-N.; Pang, J.-H.S. IFI27, a novel epidermal growth factor-stabilized protein, is functionally involved in proliferation and cell cycling of human epidermal keratinocytes. Cell Prolif. 2015, 48, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Zhao, J.; Song, J.; Li, Y.; Zuo, R.; Sa, Y.; Ma, Z.; OuYang, H. Interferon alpha-inducible protein 27 (IFI27) is a prognostic marker for pancreatic cancer based on comprehensive bioinformatics analysis. Bioengineered 2021, 12, 8515–8528. [Google Scholar] [CrossRef] [PubMed]
- Cervantes-Badillo, M.G.; Paredes-Villa, A.; Gómez-Romero, V.; Cervantes-Roldán, R.; Arias-Romero, L.E.; Villamar-Cruz, O.; González-Montiel, M.; Barrios-García, T.; Cabrera-Quintero, A.J.; Rodríguez-Gómez, G.; et al. IFI27/ISG12 Downregulates Estrogen Receptor α Transactivation by Facilitating Its Interaction with CRM1/XPO1 in Breast Cancer Cells. Front. Endocrinol. 2020, 11, 568375. [Google Scholar] [CrossRef]
- Wang, Y.; Jia, W.; Ma, Q.; Li, F.; Ma, Z.; Yang, M.; Pu, J.; Huo, Z.; Dang, J. Identification of IFI44L as a new candidate molecular marker for systemic lupus erythematosus. Clin. Exp. Rheumatol. 2022. [Google Scholar] [CrossRef]
- DeDiego, M.L.; Martinez-Sobrido, L.; Topham, D.J. Novel Functions of IFI44L as a Feedback Regulator of Host Antiviral Responses. J. Virol. 2019, 93, e01159-19. [Google Scholar] [CrossRef]
- Harada, H.; Taniguchi, T.; Tanaka, N. The role of interferon regulatory factors in the interferon system and cell growth control. Biochimie 1998, 80, 641–650. [Google Scholar] [CrossRef]
- Platanitis, E.; Demiroz, D.; Schneller, A.; Fischer, K.; Capelle, C.; Hartl, M.; Gossenreiter, T.; Müller, M.; Novatchkova, M.; Decker, T. A molecular switch from STAT2-IRF9 to ISGF3 underlies interferon-induced gene transcription. Nat. Commun. 2019, 10, 2921. [Google Scholar] [CrossRef]
- Lee, W.-H.; Liu, F.-H.; Lee, Y.-L.; Huang, H.-M. Interferon-alpha induces the growth inhibition of human T-cell leukaemia line Jurkat through p38alpha and p38beta. J. Biochem. 2010, 147, 645–650. [Google Scholar] [CrossRef] [PubMed]
- Krejcová, D.; Procházková, J.; Kubala, L.; Pacherník, J. Modulation of cell proliferation and differentiation of human lung carcinoma cells by the interferon-alpha. Gen. Physiol. Biophys. 2009, 28, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Grebenová, D.; Kuzelová, K.; Fuchs, O.; Halada, P.; Havlícek, V.; Marinov, I.; Hrkal, Z. Interferon-alpha suppresses proliferation of chronic myelogenous leukemia cells K562 by extending cell cycle S-phase without inducing apoptosis. Blood Cells Mol. Dis. 2004, 32, 262–269. [Google Scholar] [CrossRef]
- Liu, C.; Li, X.; Feng, G.; Cao, M.; Liu, F.; Zhang, G.; Lu, Y. Downregulation of USP12 inhibits tumor growth via the p38/MAPK pathway in hepatocellular carcinoma. Mol. Med. Rep. 2020, 22, 4899–4908. [Google Scholar] [CrossRef]
- Hachiya, M.; Akashi, M. Catalase regulates cell growth in HL60 human promyelocytic cells: Evidence for growth regulation by H2O2. Radiat. Res. 2005, 163, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Cox, B.D.; Natarajan, M.; Stettner, M.R.; Gladson, C.L. New concepts regarding focal adhesion kinase promotion of cell migration and proliferation. J. Cell. Biochem. 2006, 99, 35–52. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.F.; Chen, Q.; Hua, S.H.; Yip, G.W. Roles of Interferon Induced Protein with Tetratricopeptide Repeats (IFIT) Family in Cancer. Curr. Med. Chem. 2021, 28, 5034–5047. [Google Scholar] [CrossRef]
- Zhang, J.-F.; Chen, Y.; Lin, G.-S.; Zhang, J.-D.; Tang, W.-L.; Huang, J.-H.; Chen, J.-S.; Wang, X.-F.; Lin, Z.-X. High IFIT1 expression predicts improved clinical outcome, and IFIT1 along with MGMT more accurately predicts prognosis in newly diagnosed glioblastoma. Hum. Pathol. 2016, 52, 136–144. [Google Scholar] [CrossRef]
- Zhang, X.; Choi, P.S.; Francis, J.M.; Imielinski, M.; Watanabe, H.; Cherniack, A.D.; Meyerson, M. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat. Genet. 2016, 48, 176–182. [Google Scholar] [CrossRef]
- Carragher, N.O.; Fincham, V.J.; Riley, D.; Frame, M.C. Cleavage of focal adhesion kinase by different proteases during SRC-regulated transformation and apoptosis. Distinct roles for calpain and caspases. J. Biol. Chem. 2001, 276, 4270–4275. [Google Scholar] [CrossRef]
- Zhou, J.; Yi, Q.; Tang, L. The roles of nuclear focal adhesion kinase (FAK) on Cancer: A focused review. J. Exp. Clin. Cancer Res. 2019, 38, 250. [Google Scholar] [CrossRef]
- Levy, A.; Alhazzani, K.; Dondapati, P.; Alaseem, A.; Cheema, K.; Thallapureddy, K.; Kaur, P.; Alobid, S.; Rathinavelu, A. Focal Adhesion Kinase in Ovarian Cancer: A Potential Therapeutic Target for Platinum and Taxane-Resistant Tumors. Curr. Cancer Drug Targets 2019, 19, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Mehlen, P.; Puisieux, A. Metastasis: A question of life or death. Nat. Rev. Cancer 2006, 6, 449–458. [Google Scholar] [CrossRef]
- Ward, K.K.; Tancioni, I.; Lawson, C.; Miller, N.L.G.; Jean, C.; Chen, X.L.; Uryu, S.; Kim, J.; Tarin, D.; Stupack, D.G.; et al. Inhibition of focal adhesion kinase (FAK) activity prevents anchorage-independent ovarian carcinoma cell growth and tumor progression. Clin. Exp. Metastasis 2013, 30, 579–594. [Google Scholar] [CrossRef] [PubMed]
- Reid, P.E.; Brown, N.J.; Holen, I. Breast cancer cells stimulate osteoprotegerin (OPG) production by endothelial cells through direct cell contact. Mol. Cancer 2009, 8, 49. [Google Scholar] [CrossRef] [PubMed]
- Mancini, L.; Moradi-Bidhendi, N.; Brandi, M.L.; Perretti, M.; MacIntyre, I. Modulation of the effects of osteoprotegerin (OPG) ligand in a human leukemic cell line by OPG and calcitonin. Biochem. Biophys. Res. Commun. 2000, 279, 391–397. [Google Scholar] [CrossRef]
- Zauli, G.; Melloni, E.; Capitani, S.; Secchiero, P. Role of full-length osteoprotegerin in tumor cell biology. Cell. Mol. Life Sci. 2009, 66, 841–851. [Google Scholar] [CrossRef]
- Harada, H.; Matsumoto, M.; Sato, M.; Kashiwazaki, Y.; Kimura, T.; Kitagawa, M.; Yokochi, T.; Tan, R.S.; Takasugi, T.; Kadokawa, Y.; et al. Regulation of IFN-alpha/beta genes: Evidence for a dual function of the transcription factor complex ISGF3 in the production and action of IFN-alpha/beta. Genes Cells 1996, 1, 995–1005. [Google Scholar] [CrossRef]
- Honda, K.; Takaoka, A.; Taniguchi, T. Type I interferon corrected gene induction by the interferon regulatory factor family of transcription factors. Immunity 2006, 25, 349–360. [Google Scholar] [CrossRef]
- Sato, M.; Hata, N.; Asagiri, M.; Nakaya, T.; Taniguchi, T.; Tanaka, N. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 1998, 441, 106–110. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schmitt, M.; Gallistl, J.; Schüler-Toprak, S.; Fritsch, J.; Buechler, C.; Ortmann, O.; Treeck, O. Anti-Tumoral Effect of Chemerin on Ovarian Cancer Cell Lines Mediated by Activation of Interferon Alpha Response. Cancers 2022, 14, 4108. https://doi.org/10.3390/cancers14174108
Schmitt M, Gallistl J, Schüler-Toprak S, Fritsch J, Buechler C, Ortmann O, Treeck O. Anti-Tumoral Effect of Chemerin on Ovarian Cancer Cell Lines Mediated by Activation of Interferon Alpha Response. Cancers. 2022; 14(17):4108. https://doi.org/10.3390/cancers14174108
Chicago/Turabian StyleSchmitt, Meike, Johanna Gallistl, Susanne Schüler-Toprak, Jürgen Fritsch, Christa Buechler, Olaf Ortmann, and Oliver Treeck. 2022. "Anti-Tumoral Effect of Chemerin on Ovarian Cancer Cell Lines Mediated by Activation of Interferon Alpha Response" Cancers 14, no. 17: 4108. https://doi.org/10.3390/cancers14174108
APA StyleSchmitt, M., Gallistl, J., Schüler-Toprak, S., Fritsch, J., Buechler, C., Ortmann, O., & Treeck, O. (2022). Anti-Tumoral Effect of Chemerin on Ovarian Cancer Cell Lines Mediated by Activation of Interferon Alpha Response. Cancers, 14(17), 4108. https://doi.org/10.3390/cancers14174108







