Radiomics for Discrimination between Early-Stage Nasopharyngeal Carcinoma and Benign Hyperplasia with Stable Feature Selection on MRI
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods and Materials
2.1. Patient Characteristics
- (i)
- (ii)
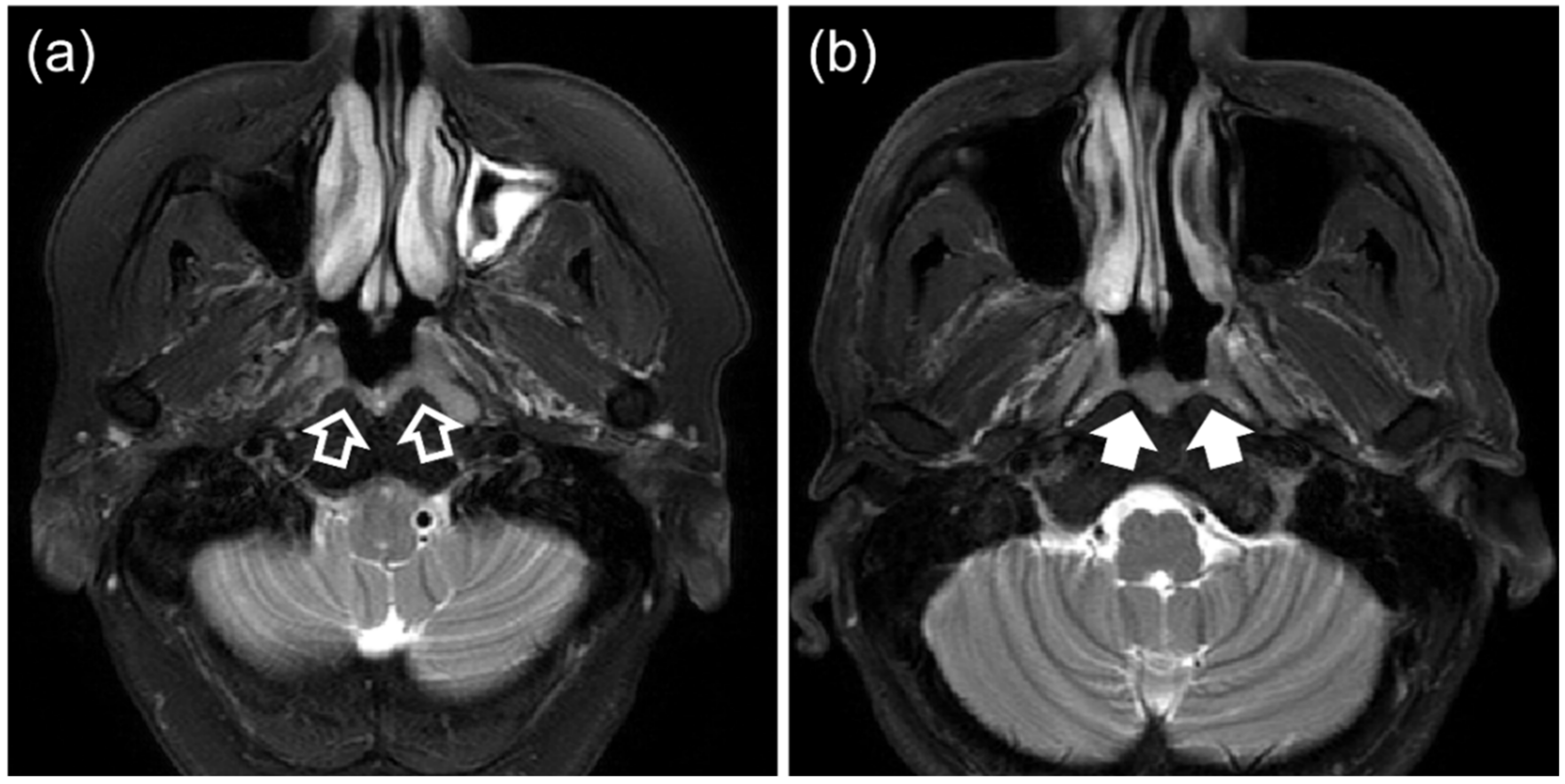
- Those with BH with a segmentable thickness of ≥3 mm on at least one axial slice without any evidence of NPC on MRI or endoscopic examination, followed-up for a minimum of 12 months.
2.2. Image Acquisition and Preprocessing
2.3. Lesion Delineation and Feature Extraction
2.4. Feature Selection
2.4.1. Preliminary Feature Filtering
2.4.2. Supervised Feature Selection
Bagged-Boosted RENT
2.5. Discrimination of NPC and Benign Hyperplasia Using Selected Features
2.6. Evaluating Discrimination Performance, Feature Selection Stability, and Statistical Analysis
2.6.1. Building the Radiomics Model
2.6.2. Testing the Final Radiomics Model
2.6.3. Evaluating the Stability of the Radiomic Features Selected by the Proposed BB-RENT
2.6.4. Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. Performance of the Radiomic Model in the 3 T MRI Training Cohort
3.2.1. Fivefold Cross Validation
3.2.2. Building the Final Ensemble Radiomic Model
3.3. Performance of the Final Model on the 1.5 T Testing Cohort
3.4. Stability of Radiomic Feature Selection by Different Feature Selection Methods
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chan, K.C.A.; Woo, J.K.S.; King, A.; Zee, B.C.Y.; Lam, W.K.J.; Chan, S.L.; Chu, S.W.I.; Mak, C.; Tse, I.O.L.; Leung, S.Y.M.; et al. Analysis of Plasma Epstein-Barr Virus DNA to Screen for Nasopharyngeal Cancer. N. Engl. J. Med. 2017, 377, 513–522. [Google Scholar] [CrossRef] [PubMed]
- King, A.D.; Woo, J.K.S.; Ai, Q.Y.; Chan, J.S.M.; Lam, W.K.J.; Tse, I.O.L.; Bhatia, K.S.; Zee, B.C.Y.; Hui, E.P.; Ma, B.B.Y.; et al. Complementary roles of MRI and endoscopic examination in the early detection of nasopharyngeal carcinoma. Ann. Oncol. 2019, 30, 977–982. [Google Scholar] [CrossRef] [PubMed]
- Ai, Q.-Y.; King, A.; So, T.; Lam, W.; Mo, F.; Tse, I.; Woo, J.; Chan, K. MRI of benign hyperplasia in the nasopharynx: Is there an association with Epstein–Barr virus? Clin. Radiol. 2020, 75, 711.e713–711.e718. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, K.S.; King, A.D.; Vlantis, A.C.; Ahuja, A.T.; Tse, G.M. Nasopharyngeal mucosa and adenoids: Appearance at MR imaging. Radiology 2012, 263, 437–443. [Google Scholar] [CrossRef]
- King, A.; Wong, L.; Law, B.; Bhatia, K.; Woo, J.; Ai, Q.-Y.; Tan, T.; Goh, J.; Chuah, K.; Mo, F. MR imaging criteria for the detection of nasopharyngeal carcinoma: Discrimination of early-stage primary tumors from benign hyperplasia. Am. J. Neuroradiol. 2018, 39, 515–523. [Google Scholar] [CrossRef] [Green Version]
- King, A.D.; Woo, J.K.S.; Ai, Q.Y.; Mo, F.K.F.; So, T.Y.; Lam, W.K.J.; Tse, I.O.L.; Vlantis, A.C.; Yip, K.W.N.; Hui, E.P.; et al. Early Detection of Cancer: Evaluation of MR Imaging Grading Systems in Patients with Suspected Nasopharyngeal Carcinoma. AJNR Am. J. Neuroradiol. 2020, 41, 515–521. [Google Scholar] [CrossRef] [Green Version]
- Wong, L.M.; King, A.D.; Ai, Q.Y.H.; Lam, W.K.J.; Poon, D.M.C.; Ma, B.B.Y.; Chan, K.C.A.; Mo, F.K.F. Convolutional neural network for discriminating nasopharyngeal carcinoma and benign hyperplasia on MRI. Eur. Radiol. 2021, 31, 3856–3863. [Google Scholar] [CrossRef]
- Deng, Y.; Li, C.; Lv, X.; Xia, W.; Shen, L.; Jing, B.; Li, B.; Guo, X.; Sun, Y.; Xie, C. The contrast-enhanced MRI can be substituted by unenhanced MRI in identifying and automatically segmenting primary nasopharyngeal carcinoma with the aid of deep learning models: An exploratory study in large-scale population of endemic area. Comput. Methods Programs Biomed. 2022, 217, 106702. [Google Scholar] [CrossRef]
- Ke, L.; Deng, Y.; Xia, W.; Qiang, M.; Chen, X.; Liu, K.; Jing, B.; He, C.; Xie, C.; Guo, X. Development of a self-constrained 3D DenseNet model in automatic detection and segmentation of nasopharyngeal carcinoma using magnetic resonance images. Oral Oncol. 2020, 110, 104862. [Google Scholar] [CrossRef]
- Duan, W.; Xiong, B.; Tian, T.; Zou, X.; He, Z.; Zhang, L. Radiomics in Nasopharyngeal Carcinoma. Clin. Med. Insights Oncol. 2022, 16, 1–10. [Google Scholar] [CrossRef]
- Zhang, Y.M.; Gong, G.Z.; Qiu, Q.T.; Han, Y.W.; Lu, H.M.; Yin, Y. Radiomics for Diagnosis and Radiotherapy of Nasopharyngeal Carcinoma. Front. Oncol. 2021, 11, 767134. [Google Scholar] [CrossRef] [PubMed]
- Traverso, A.; Wee, L.; Dekker, A.; Gillies, R. Repeatability and reproducibility of radiomic features: A systematic review. Int. J. Radiat. Oncol. Biol. Phys. 2018, 102, 1143–1158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jha, A.; Mithun, S.; Jaiswar, V.; Sherkhane, U.; Purandare, N.; Prabhash, K.; Rangarajan, V.; Dekker, A.; Wee, L.; Traverso, A. Repeatability and reproducibility study of radiomic features on a phantom and human cohort. Sci. Rep. 2021, 11, 2055. [Google Scholar] [CrossRef] [PubMed]
- Pfaehler, E.; Zhovannik, I.; Wei, L.; Boellaard, R.; Dekker, A.; Monshouwer, R.; El Naqa, I.; Bussink, J.; Gillies, R.; Wee, L. A systematic review and quality of reporting checklist for repeatability and reproducibility of radiomic features. Phys. Imaging Radiat. Oncol. 2021, 20, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Yuan, J.; Zhou, Y.; Wong, O.L.; Cheung, K.Y.; Yu, S.K. Acquisition repeatability of MRI radiomics features in the head and neck: A dual-3D-sequence multi-scan study. Vis. Comput. Ind. Biomed. Art 2022, 5, 10. [Google Scholar] [CrossRef]
- Alelyani, S. Stable bagging feature selection on medical data. J. Big Data 2021, 8, 11. [Google Scholar] [CrossRef]
- Jenul, A.; Schrunner, S.; Liland, K.H.; Indahl, U.G.; Futsaether, C.M.; Tomic, O. RENT—Repeated Elastic Net Technique for Feature Selection. IEEE Access 2021, 9, 152333–152346. [Google Scholar] [CrossRef]
- Salman, R.; Alzaatreh, A.; Sulieman, H.; Faisal, S. A bootstrap framework for aggregating within and between feature selection methods. Entropy 2021, 23, 200. [Google Scholar] [CrossRef]
- Wang, A.; Liu, H.; Yang, J.; Chen, G. Ensemble feature selection for stable biomarker identification and cancer classification from microarray expression data. Comput. Biol. Med. 2022, 142, 105208. [Google Scholar] [CrossRef]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The eighth edition AJCC cancer staging manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef]
- Brierley, J.D.; Gospodarowicz, M.K.; Wittekind, C. TNM Classification of Malignant Tumours; John Wiley & Sons: Hoboken, NJ, USA, 2017. [Google Scholar]
- Tustison, N.J.; Avants, B.B.; Cook, P.A.; Zheng, Y.; Egan, A.; Yushkevich, P.A.; Gee, J.C. N4ITK: Improved N3 bias correction. IEEE Trans. Med. Imaging 2010, 29, 1310–1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nyúl, L.G.; Udupa, J.K.; Zhang, X. New variants of a method of MRI scale standardization. IEEE Trans. Med. Imaging 2000, 19, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.-K.; Wang, M.-J.J. Image thresholding by minimizing the measures of fuzziness. Pattern Recognit. 1995, 28, 41–51. [Google Scholar] [CrossRef]
- Van Griethuysen, J.J.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koo, T.K.; Li, M.Y. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef] [Green Version]
- Yang, P.; Hwa Yang, Y.; Zhou, B.B.; Zomaya, A.Y. A review of ensemble methods in bioinformatics. Curr. Bioinform. 2010, 5, 296–308. [Google Scholar] [CrossRef] [Green Version]
- Freund, Y.; Schapire, R.E. A decision-theoretic generalization of on-line learning and an application to boosting. J. Comput. Syst. Sci. 1997, 55, 119–139. [Google Scholar] [CrossRef] [Green Version]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Youden, W.J. Index for rating diagnostic tests. Cancer 1950, 3, 32–35. [Google Scholar] [CrossRef]
- Nogueira, S.; Sechidis, K.; Brown, G. On the stability of feature selection algorithms. J. Mach. Learn. Res. 2017, 18, 6345–6398. [Google Scholar]
- Shen, H.; Yuan, X.; Liu, D.; Tu, C.; Wang, X.; Liu, R.; Wang, X.; Lan, X.; Fu, K.; Zhang, J. Multiparametric dual-energy CT to differentiate stage T1 nasopharyngeal carcinoma from benign hyperplasia. Quant. Imaging Med. Surg. 2021, 11, 4004. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.M.; Ai, Q.Y.H.; Poon, D.M.C.; Tong, M.; Ma, B.B.Y.; Hui, E.P.; Shi, L.; King, A.D. A convolutional neural network combined with positional and textural attention for the fully automatic delineation of primary nasopharyngeal carcinoma on non-contrast-enhanced MRI. Quant. Imaging Med. Surg. 2021, 11, 3932–3944. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.M.; Ai, Q.Y.H.; Mo, F.K.F.; Poon, D.M.C.; King, A.D. Convolutional neural network in nasopharyngeal carcinoma: How good is automatic delineation for primary tumor on a non-contrast-enhanced fat-suppressed T2-weighted MRI? Jpn. J. Radiol. 2021, 39, 571–579. [Google Scholar] [CrossRef] [PubMed]





| Ref. | Year | Cohort | MRI Sequence | AUC | Strengths |
|---|---|---|---|---|---|
| Ke et al. [9] | 2020 | 3142 NPC; 958 BH | T1w-ce | 0.95 | Large cohort; Performed segmentation and differentiation of NPC and BH simultaneously; |
| Wong et al. [7] | 2021 | 203 stage T1 NPC; 209 BH | T2w-fs | 0.96 | Study designed to include only stage T1 NPC, which is more challenging than including all T stages to discriminate from BH; Non-contrast-enhanced MRI investigated, which could be used for screening; |
| Deng et al. [8] | 2022 | 3582 NPC; 825 BH & 71 IM | T1w-ce, T1w, T2w | 1.0 | Large cohort from the same institution as Ke et al. [9] but also including non-contrast-enhanced MRI; Segmentation and differentiation of NPC and BH performed simultaneously High AUC performance of nearly 1 achieved; |
| Variable | All 1.5 T (n = 213) | All 3 T (n = 442) | Fold 1 (n = 89) | Fold 2 (n = 89) | Fold 3 (n = 88) | Fold 4 (n = 88) | Fold 5 (n = 88) | p |
|---|---|---|---|---|---|---|---|---|
| Age (years) | 48.2 ± 12.7 18–83 | 54.3 ± 9.5 25–90 | 54.6 ± 10.2 35–90 | 53.7 ± 8.2 32–76 | 55.1 ± 10.1 32–80 | 52.9 ± 10.2 25–86 | 55.2 ± 8.34 34–71 | 0.22 |
| Sex | 0.42 | |||||||
| Men | 139 | 392 | 83 | 77 | 80 | 77 | 75 | - |
| Women | 74 | 50 | 6 | 12 | 8 | 11 | 13 | - |
| Pathology | 1.00 | |||||||
| T1 NPC | 99 | 220 | 44 | 44 | 45 | 44 | 44 | - |
| BH | 114 | 222 | 45 | 45 | 43 | 44 | 44 | - |
| Method | AUC | Accuracy | Sensitivity | Specificity | Threshold (≥) |
|---|---|---|---|---|---|
| SVR | 0.85 ± 0.04 | 80.3% ± 1.8% | 79.6% ± 9.7% | 80.8% ± 10.0% | 0.51 ± 0.10 |
| Logistic Regression | 0.77 ± 0.01 | 77.2% ± 1.2% | 74.2% ± 3.9% | 80.1% ± 3.3% | 1 |
| RF | 0.82 ± 0.02 | 76.7% ± 1.4% | 70.1% ± 8.4% | 83.2% ± 7.4% | 0.54 ± 0.10 |
| Perceptron | 0.84 ± 0.03 | 79.4% ± 1.6% | 73.8% ± 4.8% | 85.1% ± 4.4% | 0.51 ± 0.06 |
| kNN | 0.82 ± 0.03 | 77.2% ± 1.4% | 71.5% ± 7.9% | 82.9% ± 9.2% | 0.55 ± 0.10 |
| Imaging Filter | Feature Type | Feature Name | SVR 1 | SVR 2 | SVR 3 | SVR 4 | SVR 5 |
|---|---|---|---|---|---|---|---|
| original | shape | SurfaceVolumeRatio | −0.34350 | −0.41914 | −0.35446 | −0.41196 | −0.35104 |
| lbp-3D-k | glrlm | LongRunHighGrayLevelEmphasis | −0.23201 | −0.23496 | −0.19496 | −0.23516 | −0.18680 |
| original | shape | SurfaceArea | −0.15209 | −0.12242 | −0.13933 | −0.05791 | −0.10701 |
| lbp-3D-m2 | first-order | Kurtosis | −0.07727 | −0.16856 | −0.15919 | −0.17252 | - |
| log-sigma−0-4492-mm-3D | first-order | Mean | 0.08879 | - | 0.05899 | −0.06722 | 0.15951 |
| original | shape | LeastAxisLength | 0.10404 | 0.11893 | 0.10616 | - | - |
| exponential | glcm | SumEntropy | 0.04001 | - | - | −0.00654 | 0.13320 |
| lbp-3D-m1 | first-order | Kurtosis | −0.08219 | - | - | - | −0.14464 |
| gradient | first-order | Energy | 0.05771 | 0.01792 | - | - | - |
| lbp-2D | glcm | DifferenceVariance | −0.02907 | - | - | - | - |
| exponential | first-order | Energy | - | −0.05008 | - | - | 0.02571 |
| exponential | first-order | Variance | - | - | - | - | −0.12470 |
| exponential | glrlm | RunVariance | - | −0.04025 | - | - | - |
| lbp-3D-m1 | glcm | ClusterShade | - | - | −0.06451 | - | - |
| lbp-3D-m2 | glrlm | ShortRunHighGrayLevelEmphasis | - | 0.00498 | - | - | - |
| log-sigma−0-4492-mm-3D | first-order | RobustMeanAbsoluteDeviation | - | - | - | −0.10530 | - |
| original | glrlm | RunEntropy | - | - | - | - | 0.05171 |
| Intercepts | 0.48176 | 0.49719 | 0.49814 | 0.47659 | 0.47752 | ||
| 0.33508 | 0.36677 | 0.62281 | 0.42553 | 0.33341 | |||
| Variable | Single EN | RENT | Boosted RENT | Bagged RENT | BB-RENT |
|---|---|---|---|---|---|
| Mean # features (n = 100) | 30.6 ± 4.0 | 5.7 ± 1.5 | 24.9 ± 0.7 | 5.2 ± 1.6 | 6.6 ± 1.7 |
| Nogueira score [31] | 0.34 | 0.37 | 0.20 | 0.34 | 0.54 |
| = 4950) | 0.24 ± 0.06 | 0.25 ± 0.13 | 0.14 ± 0.05 | 0.23 ± 0.15 | 0.39 ± 0.14 |
| t-test p-values [31] | |||||
| RENT | 0.047 | - | - | - | - |
| Boosted RENT | <0.001 | <0.001 | - | - | - |
| Bagged RENT | 0.962 | 0.162 | <0.001 | - | - |
| BB-RENT | <0.001 | <0.001 | <0.001 | <0.001 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wong, L.M.; Ai, Q.Y.H.; Zhang, R.; Mo, F.; King, A.D. Radiomics for Discrimination between Early-Stage Nasopharyngeal Carcinoma and Benign Hyperplasia with Stable Feature Selection on MRI. Cancers 2022, 14, 3433. https://doi.org/10.3390/cancers14143433
Wong LM, Ai QYH, Zhang R, Mo F, King AD. Radiomics for Discrimination between Early-Stage Nasopharyngeal Carcinoma and Benign Hyperplasia with Stable Feature Selection on MRI. Cancers. 2022; 14(14):3433. https://doi.org/10.3390/cancers14143433
Chicago/Turabian StyleWong, Lun M., Qi Yong H. Ai, Rongli Zhang, Frankie Mo, and Ann D. King. 2022. "Radiomics for Discrimination between Early-Stage Nasopharyngeal Carcinoma and Benign Hyperplasia with Stable Feature Selection on MRI" Cancers 14, no. 14: 3433. https://doi.org/10.3390/cancers14143433
APA StyleWong, L. M., Ai, Q. Y. H., Zhang, R., Mo, F., & King, A. D. (2022). Radiomics for Discrimination between Early-Stage Nasopharyngeal Carcinoma and Benign Hyperplasia with Stable Feature Selection on MRI. Cancers, 14(14), 3433. https://doi.org/10.3390/cancers14143433








