The Genomic Landscape in Philadelphia-Negative Myeloproliferative Neoplasm Patients with Second Cancers
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Source, WES, and Data Analysis
2.2. Mutation Validation with Sanger Sequencing
2.3. Measurement of Plasma Levels of Inflammatory Cytokines
2.4. Genotyping for JAK2 46/1 Haplotype
3. Results
3.1. MPN Patients with SCs Are Older but Do Not Exhibit Unique Clinical Characteristics
3.2. Patterns of Genomic Variations in MPN Patients with SCs Are Not strikingly Disparate from Those of Control Cases
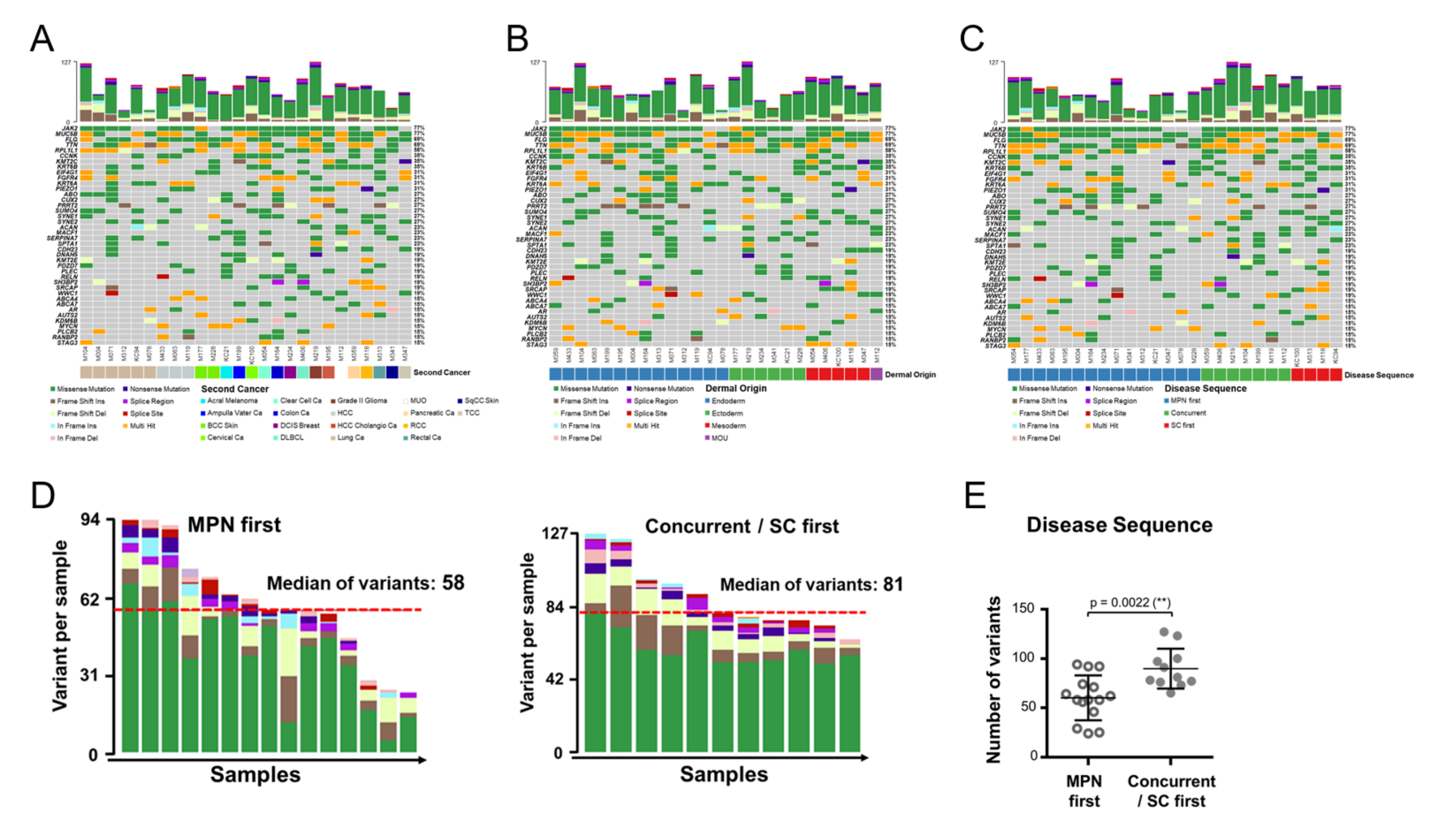
3.3. Genomic Alterations Are Allocated in Distinct Genes and Manifest Unique Co-Occurring Patterns in MPN with an SC
3.4. Critical Variant Replaces JAK2 as the more Prominent Disease Driver in MPN with an SC
3.5. Genomic Variants in MPN with an SC Are Enriched in Inflammation Signaling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tefferi, A.; Pardanani, A. Myeloproliferative Neoplasms: A Contemporary Review. JAMA Oncol. 2015, 1, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Vainchenker, W.; Kralovics, R. Genetic basis and molecular pathophysiology of classical myeloproliferative neoplasms. Blood 2017, 129, 667–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rumi, E.; Passamonti, F.; Elena, C.; Pietra, D.; Arcaini, L.; Astori, C.; Zibellini, S.; Boveri, E.; Pascutto, C.; Lazzarino, M. Increased risk of lymphoid neoplasm in patients with myeloproliferative neoplasm: A study of 1915 patients. Haematologica 2011, 96, 454–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchetti, M.; Carobbio, A.; Capitoni, E.; Barbui, T. Lymphoproliferative disorders in patients with chronic myeloproliferative neoplasms: A systematic review. Am. J. Hematol 2018, 93, 698–703. [Google Scholar] [CrossRef] [Green Version]
- Ghirardi, A.; Carobbio, A.; Masciulli, A.; Barbui, T. Incidence of solid tumors in polycythemia vera treated with phlebotomy with or without hydroxyurea: ECLAP follow-up data. Blood Cancer J. 2018, 8, 5. [Google Scholar] [CrossRef] [Green Version]
- Landtblom, A.R.; Bower, H.; Andersson, T.M.; Dickman, P.W.; Samuelsson, J.; Bjorkholm, M.; Kristinsson, S.Y.; Hultcrantz, M. Second malignancies in patients with myeloproliferative neoplasms: A population-based cohort study of 9379 patients. Leukemia 2018, 32, 2203–2210. [Google Scholar] [CrossRef]
- Frederiksen, H.; Farkas, D.K.; Christiansen, C.F.; Hasselbalch, H.C.; Sorensen, H.T. Chronic myeloproliferative neoplasms and subsequent cancer risk: A Danish population-based cohort study. Blood 2011, 118, 6515–6520. [Google Scholar] [CrossRef] [Green Version]
- Dunbar, A.J.; Rampal, R.K.; Levine, R. Leukemia secondary to myeloproliferative neoplasms. Blood 2020, 136, 61–70. [Google Scholar] [CrossRef]
- Grinfeld, J.; Nangalia, J.; Baxter, E.J.; Wedge, D.C.; Angelopoulos, N.; Cantrill, R.; Godfrey, A.L.; Papaemmanuil, E.; Gundem, G.; MacLean, C.; et al. Classification and Personalized Prognosis in Myeloproliferative Neoplasms. N. Engl. J. Med. 2018, 379, 1416–1430. [Google Scholar] [CrossRef]
- Guglielmelli, P.; Lasho, T.L.; Rotunno, G.; Mudireddy, M.; Mannarelli, C.; Nicolosi, M.; Pacilli, A.; Pardanani, A.; Rumi, E.; Rosti, V.; et al. MIPSS70: Mutation-Enhanced International Prognostic Score System for Transplantation-Age Patients with Primary Myelofibrosis. J. Clin. Oncol. 2018, 36, 310–318. [Google Scholar] [CrossRef]
- Tefferi, A.; Guglielmelli, P.; Nicolosi, M.; Mannelli, F.; Mudireddy, M.; Bartalucci, N.; Finke, C.M.; Lasho, T.L.; Hanson, C.A.; Ketterling, R.P.; et al. GIPSS: Genetically inspired prognostic scoring system for primary myelofibrosis. Leukemia 2018, 32, 1631–1642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tefferi, A.; Guglielmelli, P.; Pardanani, A.; Vannucchi, A.M. Myelofibrosis Treatment Algorithm 2018. Blood Cancer J. 2018, 8, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mora, B.; Rumi, E.; Guglielmelli, P.; Barraco, D.; Maffioli, M.; Rambaldi, A.; Caramella, M.; Komrokji, R.; Gotlib, J.; Kiladjian, J.J.; et al. Second primary malignancies in postpolycythemia vera and postessential thrombocythemia myelofibrosis: A study on 2233 patients. Cancer Med. 2019, 8, 4089–4092. [Google Scholar] [CrossRef] [PubMed]
- Barbui, T.; Ghirardi, A.; Masciulli, A.; Carobbio, A.; Palandri, F.; Vianelli, N.; De Stefano, V.; Betti, S.; Di Veroli, A.; Iurlo, A.; et al. Second cancer in Philadelphia negative myeloproliferative neoplasms (MPN-K). A nested case-control study. Leukemia 2019, 33, 1996–2005. [Google Scholar] [CrossRef] [Green Version]
- Hasselbalch, H.C.; Bjorn, M.E. MPNs as Inflammatory Diseases: The Evidence, Consequences, and Perspectives. Mediat. Inflamm. 2015, 2015, 102476. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.C.; You, J.Y.; Lung, J.; Huang, C.E.; Chen, Y.Y.; Leu, Y.W.; Ho, H.Y.; Li, C.P.; Lu, C.H.; Lee, K.D.; et al. Aberrant let7a/HMGA2 signaling activity with unique clinical phenotype in JAK2-mutated myeloproliferative neoplasms. Haematologica 2017, 102, 509–518. [Google Scholar] [CrossRef] [Green Version]
- Hsu, C.C.; Huang, C.E.; Wu, Y.Y.; Chen, Y.Y.; Lung, J.; Leu, Y.W.; Li, C.P.; Tsou, H.Y.; Chuang, W.H.; Lu, C.H.; et al. Quantitative competitive allele-specific TaqMan duplex PCR (qCAST-Duplex PCR) assay: A refined method for highly sensitive and specific detection of JAK2V617F mutant allele burdens. Haematologica 2018, 103, e450–e454. [Google Scholar] [CrossRef] [Green Version]
- Ewels, P.; Magnusson, M.; Lundin, S.; Kaller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [Green Version]
- Okonechnikov, K.; Conesa, A.; Garcia-Alcalde, F. Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 2016, 32, 292–294. [Google Scholar] [CrossRef] [PubMed]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang le, L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, K. InterVar: Clinical Interpretation of Genetic Variants by the 2015 ACMG-AMP Guidelines. Am. J. Hum. Genet. 2017, 100, 267–280. [Google Scholar] [CrossRef] [Green Version]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef] [Green Version]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013, 76, 7–20. [Google Scholar] [CrossRef] [Green Version]
- Mayakonda, A.; Lin, D.C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef] [Green Version]
- Tamborero, D.; Gonzalez-Perez, A.; Lopez-Bigas, N. OncodriveCLUST: Exploiting the positional clustering of somatic mutations to identify cancer genes. Bioinformatics 2013, 29, 2238–2244. [Google Scholar] [CrossRef]
- Jones, A.V.; Chase, A.; Silver, R.T.; Oscier, D.; Zoi, K.; Wang, Y.L.; Cario, H.; Pahl, H.L.; Collins, A.; Reiter, A.; et al. JAK2 haplotype is a major risk factor for the development of myeloproliferative neoplasms. Nat. Genet. 2009, 41, 446–449. [Google Scholar] [CrossRef] [Green Version]
- Jaiswal, S.; Natarajan, P.; Silver, A.J.; Gibson, C.J.; Bick, A.G.; Shvartz, E.; McConkey, M.; Gupta, N.; Gabriel, S.; Ardissino, D.; et al. Clonal Hematopoiesis and Risk of Atherosclerotic Cardiovascular Disease. N. Engl. J. Med. 2017, 377, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Kilpivaara, O.; Mukherjee, S.; Schram, A.M.; Wadleigh, M.; Mullally, A.; Ebert, B.L.; Bass, A.; Marubayashi, S.; Heguy, A.; Garcia-Manero, G.; et al. A germline JAK2 SNP is associated with predisposition to the development of JAK2(V617F)-positive myeloproliferative neoplasms. Nat. Genet. 2009, 41, 455–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olcaydu, D.; Harutyunyan, A.; Jager, R.; Berg, T.; Gisslinger, B.; Pabinger, I.; Gisslinger, H.; Kralovics, R. A common JAK2 haplotype confers susceptibility to myeloproliferative neoplasms. Nat. Genet. 2009, 41, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, M.; Ghirardi, A.; Masciulli, A.; Carobbio, A.; Palandri, F.; Vianelli, N.; Rossi, E.; Betti, S.; Di Veroli, A.; Iurlo, A.; et al. Second cancers in MPN: Survival analysis from an international study. Am. J. Hematol. 2020, 95, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Strickland, M.; Quek, L.; Psaila, B. The immune land.dscape in BCR-ABL negative myeloproliferative neoplasms: Inflammation, infections and opportunities for immunotherapy. Br. J. Haematol. 2022, 196, 1149–1158. [Google Scholar] [CrossRef]
- Crusz, S.M.; Balkwill, F.R. Inflammation and cancer: Advances and new agents. Nat. Rev. Clin. Oncol. 2015, 12, 584–596. [Google Scholar] [CrossRef]
- Dong, M.; Blobe, G.C. Role of transforming growth factor-beta in hematologic malignancies. Blood 2006, 107, 4589–4596. [Google Scholar] [CrossRef] [Green Version]
- Johnson, B.F.; Clay, T.M.; Hobeika, A.C.; Lyerly, H.K.; Morse, M.A. Vascular endothelial growth factor and immunosuppression in cancer: Current knowledge and potential for new therapy. Expert Opin. Biol. Ther. 2007, 7, 449–460. [Google Scholar] [CrossRef]
- Pettersson, H.; Knutsen, H.; Holmberg, E.; Andreasson, B. Increased incidence of another cancer in myeloproliferative neoplasms patients at the time of diagnosis. Eur. J. Haematol. 2015, 94, 152–156. [Google Scholar] [CrossRef]
- Zhang, J.; Sun, H.; Liu, S.; Huang, W.; Gu, J.; Zhao, Z.; Qin, H.; Luo, L.; Yang, J.; Fang, Y.; et al. Alteration of tumor-associated macrophage subtypes mediated by KRT6A in pancreatic ductal adenocarcinoma. Aging (Albany NY) 2020, 12, 23217–23232. [Google Scholar] [CrossRef]
- Che, D.; Wang, M.; Sun, J.; Li, B.; Xu, T.; Lu, Y.; Pan, H.; Lu, Z.; Gu, X. KRT6A Promotes Lung Cancer Cell Growth and Invasion Through MYC-Regulated Pentose Phosphate Pathway. Front. Cell Dev. Biol. 2021, 9, 694071. [Google Scholar] [CrossRef] [PubMed]
- Hermouet, S.; Vilaine, M. The JAK2 46/1 haplotype: A marker of inappropriate myelomonocytic response to cytokine stimulation, leading to increased risk of inflammation, myeloid neoplasm, and impaired defense against infection? Haematologica 2011, 96, 1575–1579. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, C.; Birgens, H.S.; Nordestgaard, B.G.; Kjaer, L.; Bojesen, S.E. The JAK2 V617F somatic mutation, mortality and cancer risk in the general population. Haematologica 2011, 96, 450–453. [Google Scholar] [CrossRef] [PubMed]




| Variables | MPN without SC (n = 193) | MPN with SC (n = 27) | p-Value |
|---|---|---|---|
| Age # (mean ± SD) | 60.8 ± 16.8 | 70.2 ± 14.6 | 0.006 |
| Male gender | 88 (45.6%) | 16 (59.3%) | 0.183 |
| Diagnosis † | 0.774 | ||
| PV | 61 | 6 | |
| ET | 94 | 16 | |
| PrePMF | 7 | 1 | |
| PMF | 31 | 4 | |
| Driver mutation | 0.448 | ||
| JAK2V617F | 143 | 19 | |
| JAK2 Exon 12 | 1 | 1 | |
| CALR | 23 | 4 | |
| MPL | 6 | 0 | |
| Triple negative | 20 | 3 | |
| Tumor origin | NA+ | ||
| Ectoderm | - | 6 | |
| Mesoderm | - | 6 | |
| Endoderm | - | 14 | |
| MUO ^ | - | 1 | |
| Secondary MF | 21 (13%) * | 3 (13%) * | 1.000 |
| Thromboembolism | 52 (26.9%) | 9 (33.3%) | 0.487 |
| Major bleeding history | 31 (16.1%) | 8 (29.6%) | 0.084 |
| White cell count, ×109/L | 14.88 ± 10.95 | 14.90 ± 8.11 | 0.992 |
| Hemoglobin, g/dL | 14.2 ± 3.7 | 12.8 ± 3.3 | 0.062 |
| Platelet, ×109/L | 619 ± 375 | 725 ± 361 | 0.172 |
| Splenomegaly | 103 (58.5%) * | 12 (50%) * | 0.343 |
| Coding Sequence Mutation | Amino Acid Mutation | Case Number | Cancer Type in Enrolled MPN Patients # |
|---|---|---|---|
| c.721_722delGGinsAA, c.745T>C |
p.Gly241Asn (G241N), p.Phe249Leu (F249L) | 3 | MUO, HCC, Pancreas |
| c.745T>C | p.Phe249Leu (F249L) | 2 | Lung, Lung |
| c.745T>C, c.721_722insAGAGA, c.716_717insAAGACAGAAGACAGACACACACAGTGAGAGAGACAGA | p.Phe249Leu (F249L), p.Gly241fs *29, p.Gly241fs *25 | 1 | HCC |
| c.721_722delGGinsAA | p.Gly241Asn (G241N) | 1 | Lung |
| c.418G>A | p.Val140Ile (V140I) | 1 | Cervix |
| Coding Sequence Mutation | Amino Acid Mutation | COSMIC | FATHMM Prediction | Cancer Type in COSMIC |
|---|---|---|---|---|
| c.745T>C | p.Phe249Leu (F249L) | COSM1739982 |
Pathogenic (score 0.86) |
14 Carcinoma, 1 Lymphoma |
| c.418G>A | p.Val140Ile (V140I) | COSM1239293 |
Neutral (score 0.09) |
4 Carcinoma, 1 Lymphoma |
| c.721_722delGGinsAA | p.Gly241Asn (G241N) | Not found | ||
| c.721_722insAGAGA | p.Gly241fs *29 | Not found | ||
| c.716_717insAAGACAGAAGACAGACACACACAGTGAGAGAGACAGA | p.Gly241fs *25 | Not found |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsu, C.-C.; Wang, Y.-H.; Chen, Y.-Y.; Chen, Y.-J.; Lu, C.-H.; Wu, Y.-Y.; Yang, Y.-R.; Tsou, H.-Y.; Li, C.-P.; Huang, C.-E.; et al. The Genomic Landscape in Philadelphia-Negative Myeloproliferative Neoplasm Patients with Second Cancers. Cancers 2022, 14, 3435. https://doi.org/10.3390/cancers14143435
Hsu C-C, Wang Y-H, Chen Y-Y, Chen Y-J, Lu C-H, Wu Y-Y, Yang Y-R, Tsou H-Y, Li C-P, Huang C-E, et al. The Genomic Landscape in Philadelphia-Negative Myeloproliferative Neoplasm Patients with Second Cancers. Cancers. 2022; 14(14):3435. https://doi.org/10.3390/cancers14143435
Chicago/Turabian StyleHsu, Chia-Chen, Ying-Hsuan Wang, Yi-Yang Chen, Ying-Ju Chen, Chang-Hsien Lu, Yu-Ying Wu, Yao-Ren Yang, Hsing-Yi Tsou, Chian-Pei Li, Cih-En Huang, and et al. 2022. "The Genomic Landscape in Philadelphia-Negative Myeloproliferative Neoplasm Patients with Second Cancers" Cancers 14, no. 14: 3435. https://doi.org/10.3390/cancers14143435
APA StyleHsu, C.-C., Wang, Y.-H., Chen, Y.-Y., Chen, Y.-J., Lu, C.-H., Wu, Y.-Y., Yang, Y.-R., Tsou, H.-Y., Li, C.-P., Huang, C.-E., & Chen, C.-C. (2022). The Genomic Landscape in Philadelphia-Negative Myeloproliferative Neoplasm Patients with Second Cancers. Cancers, 14(14), 3435. https://doi.org/10.3390/cancers14143435







