The Potential and Emerging Role of Quantitative Imaging Biomarkers for Cancer Characterization
Abstract
:Simple Summary
Abstract
1. Introduction
1.1. Achievements in Personalized Cancer Therapy
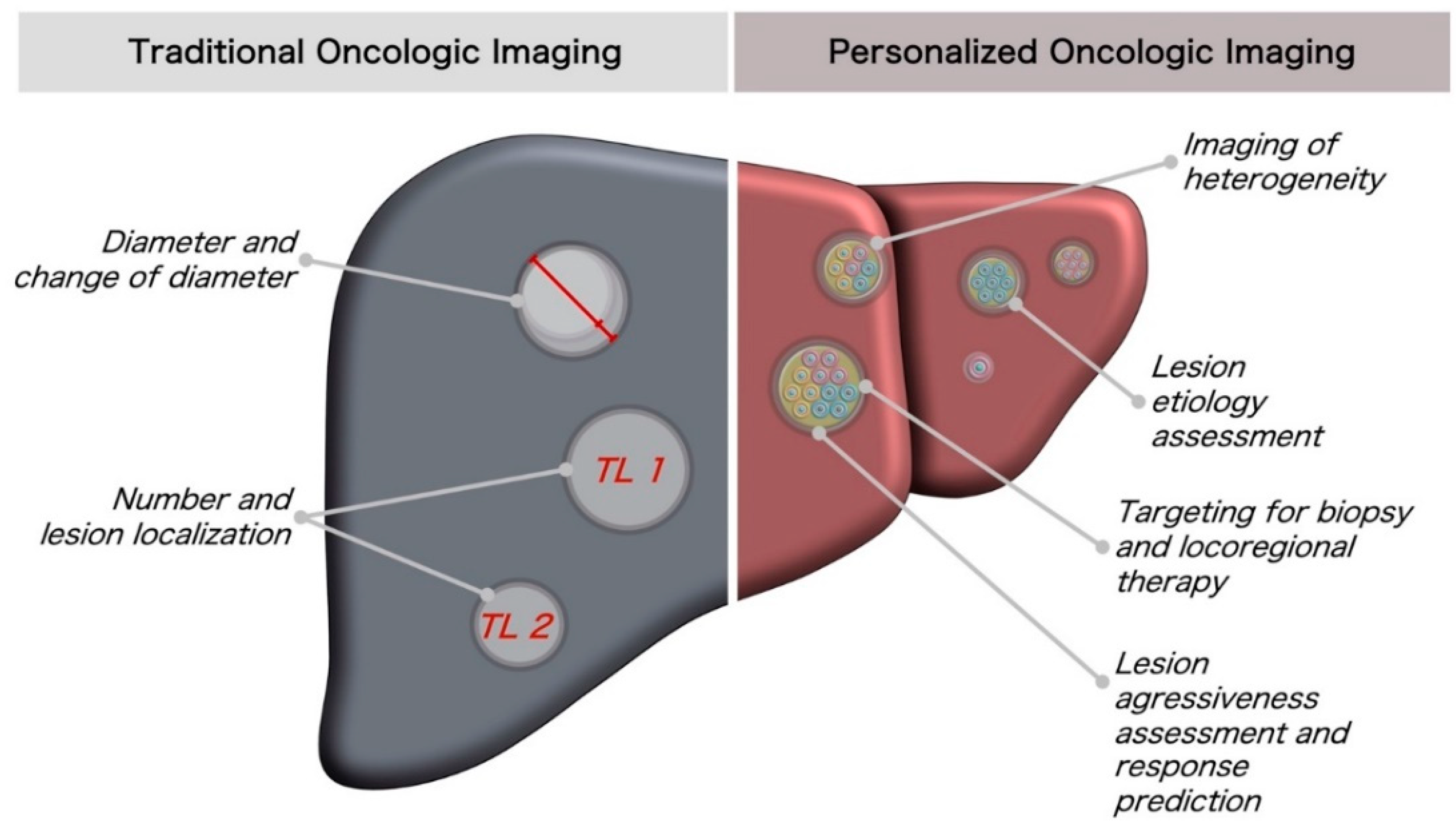
1.2. Current Status of Oncologic Imaging: Response Assessment
1.3. Current Status of Oncologic Imaging: Characterization of Lesions
1.4. Molecular Tumor Diagnostics for Personalized Therapy Stratification
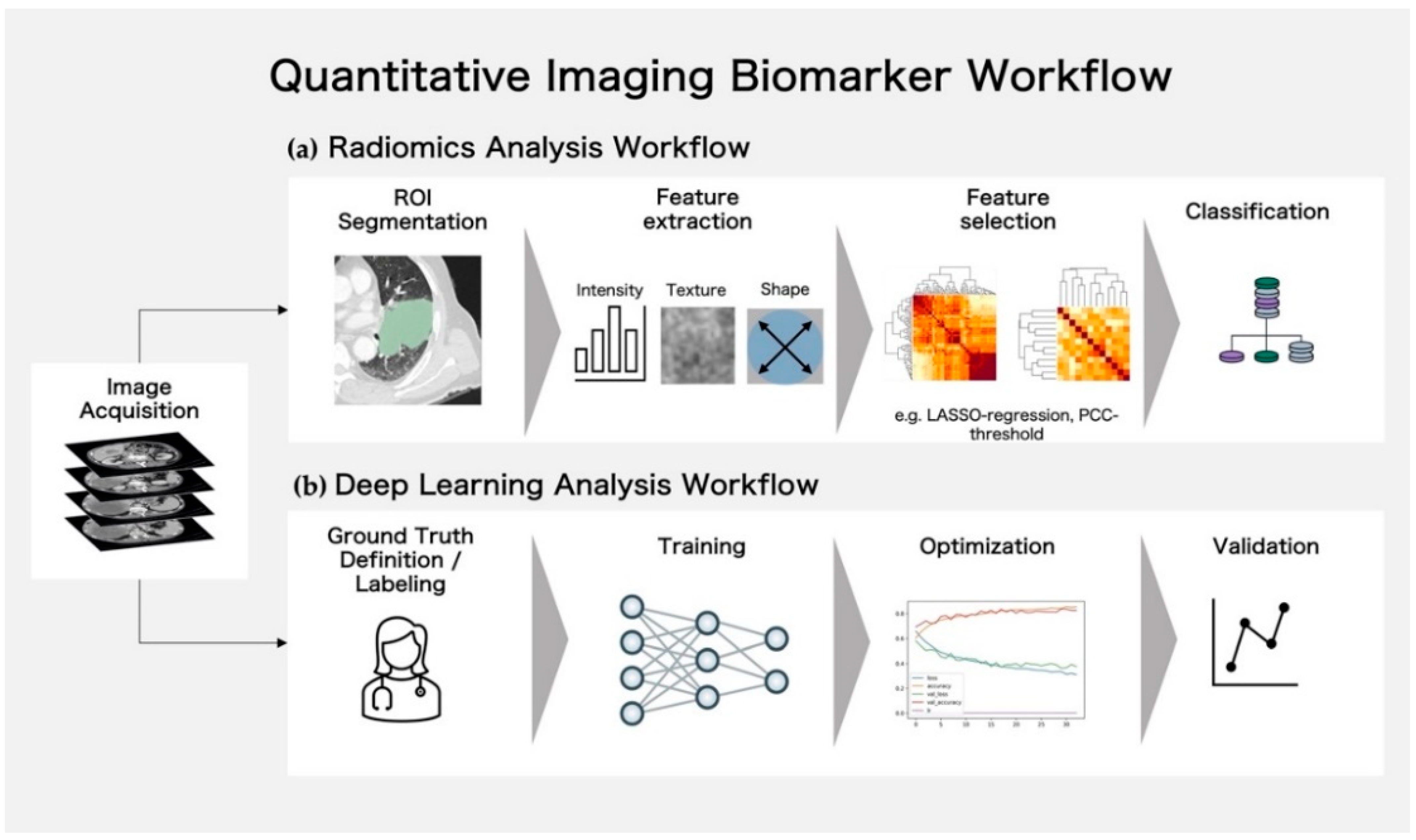
2. Techniques for Lesion Analysis Using Quantitative Imaging Biomarkers
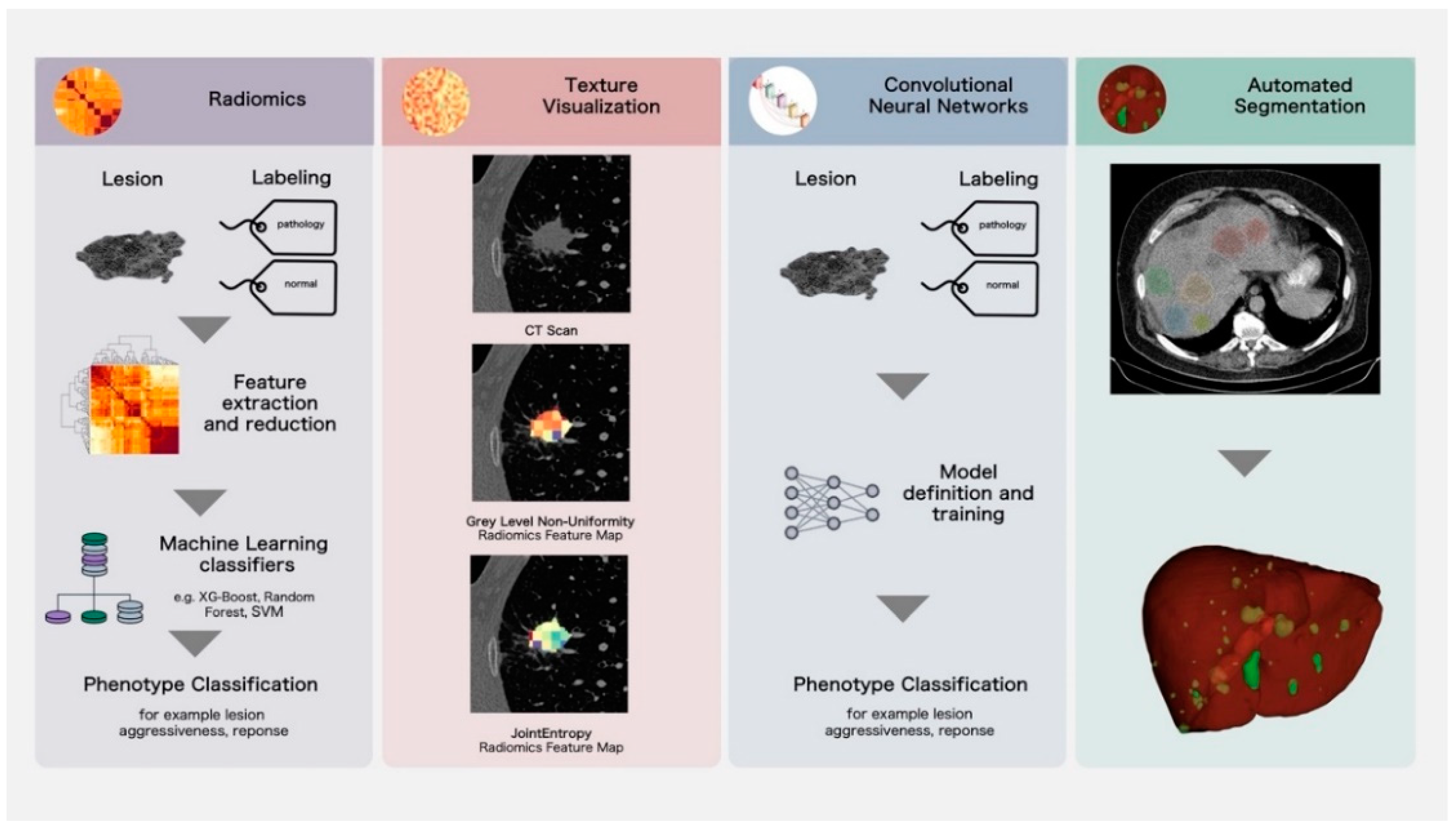
2.1. Classical Radiomics Analysis Workflow and Texture Visualization
2.2. Deep Learning Analysis Workflow
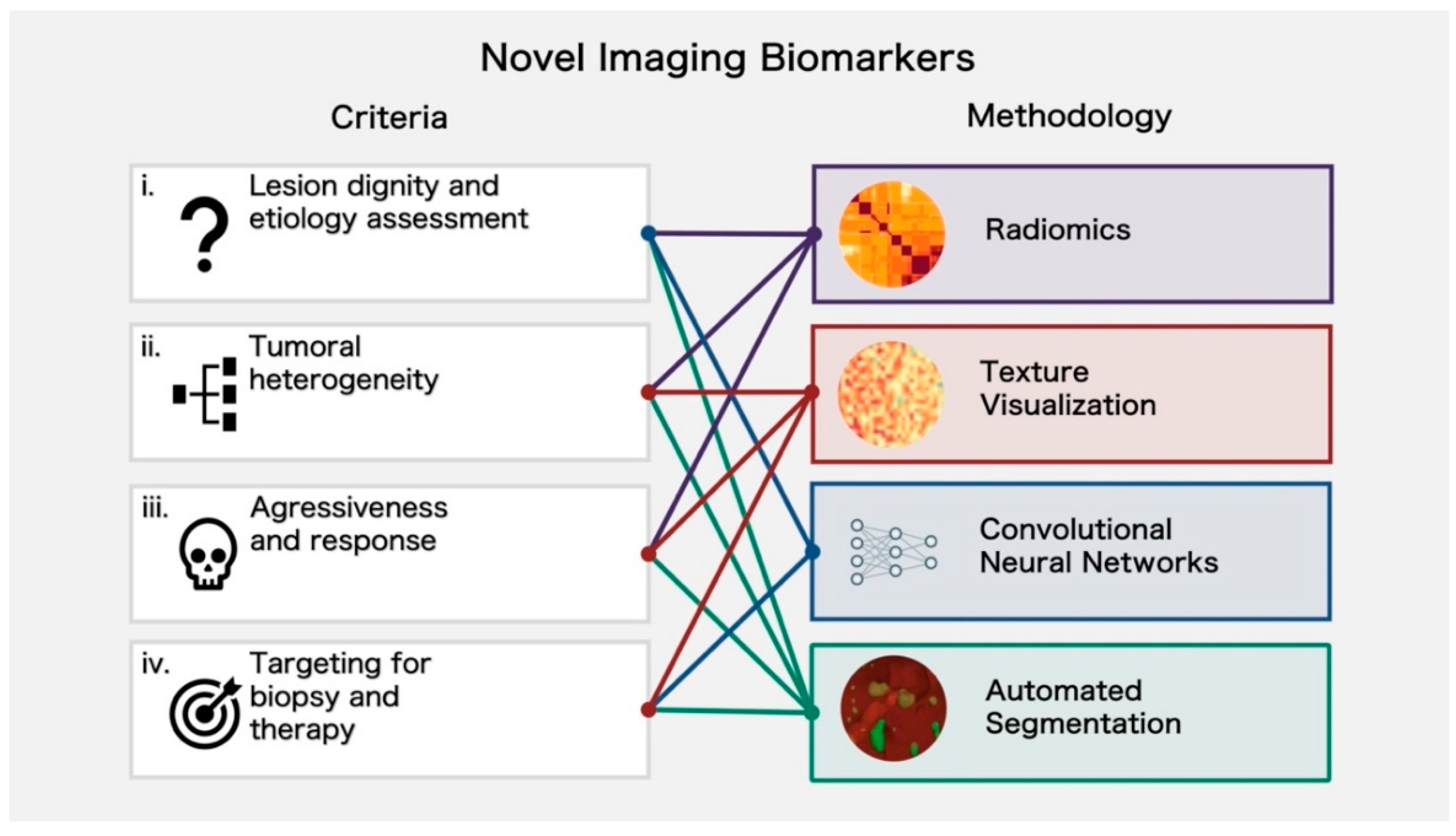
3. Application and Evidence for Novel Imaging Biomarkers for Improved Lesion Characterization and Prognosis
3.1. Lesion Dignity and Etiology Assessment
3.2. Assessment of Tumoral Heterogeneity
3.3. Assessment of Aggressiveness and Response
3.4. Targeting for Biopsy and Therapy
4. Future Perspectives
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hofmarcher, T.; Lindgren, P.; Wilking, N.; Jönsson, B. The Cost of Cancer in Europe 2018. Eur. J. Cancer 2020, 129, 41–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, J.; Ploner, A.; Elfström, K.M.; Wang, J.; Roth, A.; Fang, F.; Sundström, K.; Dillner, J.; Sparén, P. HPV Vaccination and the Risk of Invasive Cervical Cancer. N. Engl. J. Med. 2020, 383, 1340–1348. [Google Scholar] [CrossRef]
- Cardoso, R.; Zhu, A.; Guo, F.; Heisser, T.; Hoffmeister, M.; Brenner, H. Incidence and Mortality of Proximal and Distal Colorectal Cancer in Germany. Dtsch. Ärztebl. Int. 2021, 118, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Brenner, H.; Stock, C.; Hoffmeister, M. Effect of Screening Sigmoidoscopy and Screening Colonoscopy on Colorectal Cancer Incidence and Mortality: Systematic Review and Meta-Analysis of Randomised Controlled Trials and Observational Studies. BMJ 2014, 348, g2467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torre, L.A.; Siegel, R.L.; Ward, E.M.; Jemal, A. Global Cancer Incidence and Mortality Rates and Trends—An Update. Cancer Epidemiol. Biomark. Prev. 2016, 25, 16–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Naghavi, M.; Allen, C.; Barber, R.M.; Bhutta, Z.A.; Carter, A.; Casey, D.C.; Charlson, F.J.; Chen, A.Z.; Coates, M.M.; et al. Global, Regional, and National Life Expectancy, All-Cause Mortality, and Cause-Specific Mortality for 249 Causes of Death, 1980–2015: A Systematic Analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1459–1544. [Google Scholar] [CrossRef] [Green Version]
- Jonker, D.J.; O’Callaghan, C.J.; Karapetis, C.S.; Zalcberg, J.R.; Tu, D.; Au, H.-J.; Berry, S.R.; Krahn, M.; Price, T.; Simes, R.J.; et al. Cetuximab for the Treatment of Colorectal Cancer. N. Engl. J. Med. 2007, 357, 2040–2048. [Google Scholar] [CrossRef] [Green Version]
- Makary, M.S.; Khandpur, U.; Cloyd, J.M.; Mumtaz, K.; Dowell, J.D. Locoregional Therapy Approaches for Hepatocellular Carcinoma: Recent Advances and Management Strategies. Cancers 2020, 12, 1914. [Google Scholar] [CrossRef]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New Response Evaluation Criteria in Solid Tumours: Revised RECIST Guideline (Version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef]
- Schwartz, L.H.; Litière, S.; de Vries, E.; Ford, R.; Gwyther, S.; Mandrekar, S.; Shankar, L.; Bogaerts, J.; Chen, A.; Dancey, J.; et al. RECIST 1.1—Update and Clarification: From the RECIST Committee. Eur. J. Cancer 2016, 62, 132–137. [Google Scholar] [CrossRef] [Green Version]
- Ko, C.-C.; Yeh, L.-R.; Kuo, Y.-T.; Chen, J.-H. Imaging Biomarkers for Evaluating Tumor Response: RECIST and Beyond. Biomark. Res. 2021, 9, 52. [Google Scholar] [CrossRef] [PubMed]
- Birchard, K.R.; Hoang, J.K.; Herndon, J.E.; Patz, E.F. Early Changes in Tumor Size in Patients Treated for Advanced Stage Nonsmall Cell Lung Cancer Do Not Correlate with Survival. Cancer 2009, 115, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Cousin, S.; Taieb, S.; Penel, N. A Paradigm Shift in Tumour Response Evaluation of Targeted Therapy: The Assessment of Novel Drugs in Exploratory Clinical Trials. Curr. Opin. Oncol. 2012, 24, 338–344. [Google Scholar] [CrossRef]
- Choi, H.; Charnsangavej, C.; Faria, S.C.; Macapinlac, H.A.; Burgess, M.A.; Patel, S.R.; Chen, L.L.; Podoloff, D.A.; Benjamin, R.S. Correlation of Computed Tomography and Positron Emission Tomography in Patients with Metastatic Gastrointestinal Stromal Tumor Treated at a Single Institution with Imatinib Mesylate: Proposal of New Computed Tomography Response Criteria. J. Clin. Oncol. 2007, 25, 1753–1759. [Google Scholar] [CrossRef]
- Weng, Z.; Ertle, J.; Zheng, S.; Lauenstein, T.; Mueller, S.; Bockisch, A.; Gerken, G.; Yang, D.; Schlaak, J.F. Choi Criteria Are Superior in Evaluating Tumor Response in Patients Treated with Transarterial Radioembolization for Hepatocellular Carcinoma. Oncol. Lett. 2013, 6, 1707–1712. [Google Scholar] [CrossRef] [Green Version]
- Heinemann, V.; Stintzing, S.; Modest, D.P.; Giessen-Jung, C.; Michl, M.; Mansmann, U.R. Early Tumour Shrinkage (ETS) and Depth of Response (DpR) in the Treatment of Patients with Metastatic Colorectal Cancer (MCRC). Eur. J. Cancer 2015, 51, 1927–1936. [Google Scholar] [CrossRef] [PubMed]
- Piessevaux, H.; Buyse, M.; Schlichting, M.; Van Cutsem, E.; Bokemeyer, C.; Heeger, S.; Tejpar, S. Use of Early Tumor Shrinkage to Predict Long-Term Outcome in Metastatic Colorectal Cancer Treated with Cetuximab. J. Clin. Oncol. 2013, 31, 3764–3775. [Google Scholar] [CrossRef]
- Winter, K.S.; Hofmann, F.O.; Thierfelder, K.M.; Holch, J.W.; Hesse, N.; Baumann, A.B.; Modest, D.P.; Stintzing, S.; Heinemann, V.; Ricke, J.; et al. Towards Volumetric Thresholds in RECIST 1.1: Therapeutic Response Assessment in Hepatic Metastases. Eur. Radiol. 2018, 28, 4839–4848. [Google Scholar] [CrossRef]
- Froelich, M.F.; Petersen, E.L.; Heinemann, V.; Nörenberg, D.; Hesse, N.; Gesenhues, A.B.; Modest, D.P.; Sommer, W.H.; Hofmann, F.O.; Stintzing, S.; et al. Impact of Size and Location of Metastases on Early Tumor Shrinkage and Depth of Response in Patients with Metastatic Colorectal Cancer: Subgroup Findings of the Randomized, Open-Label Phase 3 Trial FIRE-3/AIO KRK-0306. Clin. Colorectal Cancer 2020, 19, 291–300.e5. [Google Scholar] [CrossRef]
- Froelich, M.F.; Heinemann, V.; Sommer, W.H.; Holch, J.W.; Schoeppe, F.; Hesse, N.; Baumann, A.B.; Kunz, W.G.; Reiser, M.F.; Ricke, J.; et al. CT Attenuation of Liver Metastases before Targeted Therapy Is a Prognostic Factor of Overall Survival in Colorectal Cancer Patients. Results from the Randomised, Open-Label FIRE-3/AIO KRK0306 Trial. Eur. Radiol. 2018, 28, 5284–5292. [Google Scholar] [CrossRef]
- Nadler, A.; Cukier, M.; Rowsell, C.; Kamali, S.; Feinberg, Y.; Singh, S.; Law, C.H.L. Ki-67 Is a Reliable Pathological Grading Marker for Neuroendocrine Tumors. Virchows Arch. 2013, 462, 501–505. [Google Scholar] [CrossRef] [PubMed]
- Yip, S.S.F.; Liu, Y.; Parmar, C.; Li, Q.; Liu, S.; Qu, F.; Ye, Z.; Gillies, R.J.; Aerts, H.J.W.L. Associations between Radiologist-Defined Semantic and Automatically Computed Radiomic Features in Non-Small Cell Lung Cancer. Sci. Rep. 2017, 7, 3519. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhou, B.; Xia, Y.; Zuo, J.; Liu, Y.; Bi, X.; Luo, X.; Zhang, C. A Methylation-Based Nomogram for Predicting Survival in Patients with Lung Adenocarcinoma. BMC Cancer 2021, 21, 801. [Google Scholar] [CrossRef]
- Hayashi, H.; Takiguchi, Y.; Minami, H.; Akiyoshi, K.; Segawa, Y.; Ueda, H.; Iwamoto, Y.; Kondoh, C.; Matsumoto, K.; Takahashi, S.; et al. Site-Specific and Targeted Therapy Based on Molecular Profiling by Next-Generation Sequencing for Cancer of Unknown Primary Site: A Nonrandomized Phase 2 Clinical Trial. JAMA Oncol. 2020, 6, 1931. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Li, X.; Song, M.; Wang, C. Molecular Analysis of Oncogenic Mutations in Resected Margins by Next-Generation Sequencing Predicts Relapse in Non-Small Cell Lung Cancer Patients. OncoTargets Ther. 2020, 13, 9525–9531. [Google Scholar] [CrossRef]
- Malik, V.; Kalakoti, Y.; Sundar, D. Deep Learning Assisted Multi-Omics Integration for Survival and Drug-Response Prediction in Breast Cancer. BMC Genom. 2021, 22, 214. [Google Scholar] [CrossRef]
- Chaudhary, K.; Poirion, O.B.; Lu, L.; Garmire, L.X. Deep Learning–Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin. Cancer Res. 2018, 24, 1248–1259. [Google Scholar] [CrossRef] [Green Version]
- Snyder, A.; Makarov, V.; Merghoub, T.; Yuan, J.; Zaretsky, J.M.; Desrichard, A.; Walsh, L.A.; Postow, M.A.; Wong, P.; Ho, T.S.; et al. Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. N. Engl. J. Med. 2014, 371, 2189–2199. [Google Scholar] [CrossRef] [Green Version]
- Schumacher, T.N.; Schreiber, R.D. Neoantigens in Cancer Immunotherapy. Science 2015, 348, 69–74. [Google Scholar] [CrossRef] [Green Version]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [Green Version]
- Prayer, F.; Hofmanninger, J.; Weber, M.; Kifjak, D.; Willenpart, A.; Pan, J.; Röhrich, S.; Langs, G.; Prosch, H. Variability of Computed Tomography Radiomics Features of Fibrosing Interstitial Lung Disease: A Test-Retest Study. Methods 2021, 188, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.J.; Kim, D.; Elgeti, T.; Steffen, I.G.; Hamm, B.; Nagel, S.N. Stability of Radiomic Features across Different Region of Interest Sizes—A CT and MR Phantom Study. Tomography 2021, 7, 238–252. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.; Baine, M.; Wisnoskie, S.; Bennion, N.; Zheng, D.; Yu, L.; Dalal, V.; Hollingsworth, M.A.; Lin, C.; Zheng, D. Effects of Interobserver and Interdisciplinary Segmentation Variabilities on CT-Based Radiomics for Pancreatic Cancer. Sci. Rep. 2021, 11, 16328. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Steinmann, A.; Ding, Y.; Lee, H.; Owens, C.; Wang, J.; Yang, J.; Followill, D.; Ger, R.; MacKin, D.; et al. Radiomics Feature Robustness as Measured Using an MRI Phantom. Sci. Rep. 2021, 11, 3973. [Google Scholar] [CrossRef]
- Lu, L.; Liang, Y.; Schwartz, L.H.; Zhao, B. Reliability of Radiomic Features Across Multiple Abdominal CT Image Acquisition Settings: A Pilot Study Using ACR CT Phantom. Tomography 2019, 5, 226–231. [Google Scholar] [CrossRef]
- Korte, J.C.; Cardenas, C.; Hardcastle, N.; Kron, T.; Wang, J.; Bahig, H.; Elgohari, B.; Ger, R.; Court, L.; Fuller, C.D.; et al. Radiomics Feature Stability of Open-Source Software Evaluated on Apparent Diffusion Coefficient Maps in Head and Neck Cancer. Sci. Rep. 2021, 11, 17633. [Google Scholar] [CrossRef]
- van Timmeren, J.E.; Leijenaar, R.T.H.; van Elmpt, W.; Wang, J.; Zhang, Z.; Dekker, A.; Lambin, P. Test–Retest Data for Radiomics Feature Stability Analysis: Generalizable or Study-Specific? Tomography 2016, 2, 361–365. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-Based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Liu, Z.; He, L.; Chen, X.; Pan, D.; Ma, Z.; Liang, C.; Tian, J.; Liang, C. Radiomics Signature: A Potential Biomarker for the Prediction of Disease-Free Survival in Early-Stage (I or II) Non—Small Cell Lung Cancer. Radiology 2016, 281, 947–957. [Google Scholar] [CrossRef]
- Shi, R.; Chen, W.; Yang, B.; Qu, J.; Cheng, Y.; Zhu, Z.; Gao, Y.; Wang, Q.; Liu, Y.; Li, Z.; et al. Prediction of KRAS, NRAS and BRAF Status in Colorectal Cancer Patients with Liver Metastasis Using a Deep Artificial Neural Network Based on Radiomics and Semantic Features. Am. J. Cancer Res. 2020, 10, 4513–4526. [Google Scholar]
- Enke, J.S.; Moltz, J.H.; D’Anastasi, M.; Kunz, W.G.; Schmidt, C.; Maurus, S.; Mühlberg, A.; Katzmann, A.; Sühling, M.; Hahn, H.; et al. Radiomics Features of the Spleen as Surrogates for CT-Based Lymphoma Diagnosis and Subtype Differentiation. Cancers 2022, 14, 713. [Google Scholar] [CrossRef] [PubMed]
- Woźnicki, P.; Westhoff, N.; Huber, T.; Riffel, P.; Froelich, M.F.; Gresser, E.; von Hardenberg, J.; Mühlberg, A.; Michel, M.S.; Schoenberg, S.O.; et al. Multiparametric MRI for Prostate Cancer Characterization: Combined Use of Radiomics Model with PI-RADS and Clinical Parameters. Cancers 2020, 12, 1767. [Google Scholar] [CrossRef] [PubMed]
- Shan, Q.; Hu, H.; Feng, S.; Peng, Z.; Chen, S.; Zhou, Q.; Li, X.; Xie, X.; Lu, M.; Wang, W.; et al. CT-Based Peritumoral Radiomics Signatures to Predict Early Recurrence in Hepatocellular Carcinoma after Curative Tumor Resection or Ablation. Cancer Imaging 2019, 19, 11. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Cao, K.; Jin, S.; Zhu, X.; Ding, J.; Peng, W. Differentiation of Renal Cell Carcinoma Subtypes through MRI-Based Radiomics Analysis. Eur. Radiol. 2020, 30, 5738–5747. [Google Scholar] [CrossRef]
- Zheng, X.; Yao, Z.; Huang, Y.; Yu, Y.; Wang, Y.; Liu, Y.; Mao, R.; Li, F.; Xiao, Y.; Wang, Y.; et al. Deep Learning Radiomics Can Predict Axillary Lymph Node Status in Early-Stage Breast Cancer. Nat. Commun. 2020, 11, 1236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shu, Z.-Y.; Mao, D.-W.; Xu, Y.; Shao, Y.; Pang, P.-P.; Gong, X.-Y. Prediction of the Progression from Mild Cognitive Impairment to Alzheimer’s Disease Using a Radiomics-Integrated Model. Ther. Adv. Neurol. Disord. 2021, 14, 175628642110295. [Google Scholar] [CrossRef]
- De Perrot, T.; Hofmeister, J.; Burgermeister, S.; Martin, S.P.; Feutry, G.; Klein, J.; Montet, X. Differentiating Kidney Stones from Phleboliths in Unenhanced Low-Dose Computed Tomography Using Radiomics and Machine Learning. Eur. Radiol. 2019, 29, 4776–4782. [Google Scholar] [CrossRef]
- Liang, C.-H.; Liu, Y.-C.; Wan, Y.-L.; Yun, C.-H.; Wu, W.-J.; López-González, R.; Huang, W.-M. Quantification of Cancer-Developing Idiopathic Pulmonary Fibrosis Using Whole-Lung Texture Analysis of HRCT Images. Cancers 2021, 13, 5600. [Google Scholar] [CrossRef]
- Lin, A.; Kolossváry, M.; Cadet, S.; McElhinney, P.; Goeller, M.; Han, D.; Yuvaraj, J.; Nerlekar, N.; Slomka, P.J.; Marwan, M.; et al. Radiomics-Based Precision Phenotyping Identifies Unstable Coronary Plaques from Computed Tomography Angiography. JACC Cardiovasc. Imaging 2022, 15, 859–871. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Huang, M.; Lai, Q.; Huang, J.; Chen, J. Feasibility of Constructing an Automatic Meniscus Injury Detection Model Based on Dual-Mode Magnetic Resonance Imaging (MRI) Radiomics of the Knee Joint. Comput. Math. Methods Med. 2022, 2022, 2155132. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Proceedings of the 18th International Conference, Munich, Germany, 5–9 October2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Depeursinge, A.; Andrearczyk, V.; Whybra, P.; van Griethuysen, J.; Müller, H.; Schaer, R.; Vallières, M.; Zwanenburg, A. Standardised Convolutional Filtering for Radiomics. arXiv 2021, arXiv:2006.05470. [Google Scholar]
- Parmar, C.; Grossmann, P.; Bussink, J.; Lambin, P.; Aerts, H.J.W.L. Machine Learning Methods for Quantitative Radiomic Biomarkers. Sci. Rep. 2015, 5, 13087. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Wang, D.; Mok, V.C.; Shi, L. Comparison of Feature Selection Methods and Machine Learning Classifiers for Radiomics Analysis in Glioma Grading. IEEE Access 2019, 7, 102010–102020. [Google Scholar] [CrossRef]
- Vuong, D.; Tanadini-Lang, S.; Wu, Z.; Marks, R.; Unkelbach, J.; Hillinger, S.; Eboulet, E.I.; Thierstein, S.; Peters, S.; Pless, M.; et al. Radiomics Feature Activation Maps as a New Tool for Signature Interpretability. Front. Oncol. 2020, 10, 578895. [Google Scholar] [CrossRef]
- McCulloch, W.S.; Pitts, W. A logical calculus of the ideas immanent in nervous activity. Bull. Math. Biophys. 1943, 5, 115–133. [Google Scholar] [CrossRef]
- Rajpurkar, P.; Chen, E.; Banerjee, O.; Topol, E.J. AI in Health and Medicine. Nat. Med. 2022, 28, 31–38. [Google Scholar] [CrossRef]
- Chlap, P.; Min, H.; Vandenberg, N.; Dowling, J.; Holloway, L.; Haworth, A. A Review of Medical Image Data Augmentation Techniques for Deep Learning Applications. J. Med. Imaging Radiat. Oncol. 2021, 65, 545–563. [Google Scholar] [CrossRef]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J.W.L. Artificial Intelligence in Radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef]
- McBee, M.P.; Awan, O.A.; Colucci, A.T.; Ghobadi, C.W.; Kadom, N.; Kansagra, A.P.; Tridandapani, S.; Auffermann, W.F. Deep Learning in Radiology. Acad. Radiol. 2018, 25, 1472–1480. [Google Scholar] [CrossRef] [Green Version]
- Esteva, A. A Guide to Deep Learning in Healthcare. Nat. Med. 2019, 25, 6. [Google Scholar] [CrossRef] [PubMed]
- Cirillo, D.; Catuara-Solarz, S.; Morey, C.; Guney, E.; Subirats, L.; Mellino, S.; Gigante, A.; Valencia, A.; Rementeria, M.J.; Chadha, A.S.; et al. Sex and Gender Differences and Biases in Artificial Intelligence for Biomedicine and Healthcare. NPJ Digit. Med. 2020, 3, 81. [Google Scholar] [CrossRef] [PubMed]
- Naik, N.; Hameed, B.M.Z.; Shetty, D.K.; Swain, D.; Shah, M.; Paul, R.; Aggarwal, K.; Ibrahim, S.; Patil, V.; Smriti, K.; et al. Legal and Ethical Consideration in Artificial Intelligence in Healthcare: Who Takes Responsibility? Front. Surg. 2022, 9, 862322. [Google Scholar] [CrossRef]
- Dankwa-Mullan, I.; Scheufele, E.L.; Matheny, M.E.; Quintana, Y.; Chapman, W.W.; Jackson, G.; South, B.R. A Proposed Framework on Integrating Health Equity and Racial Justice into the Artificial Intelligence Development Lifecycle. J. Health Care Poor Underserved 2021, 32, 300–317. [Google Scholar] [CrossRef]
- Touvron, H.; Cord, M.; Douze, M.; Massa, F.; Sablayrolles, A.; Jégou, H. Training Data-Efficient Image Transformers & Distillation through Attention 2021. In Proceedings of the 38th International Conference on Machine Learning, PMLR Virtual Conference, Online, 18–24 July 2021. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image Is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. arXiv 2021, arXiv:physics/2010.11929. Available online: http://arxiv.org/abs/physics/2010.11929 (accessed on 4 June 2022).
- Nie, P.; Yang, G.; Guo, J.; Chen, J.; Li, X.; Ji, Q.; Wu, J.; Cui, J.; Xu, W. A CT-Based Radiomics Nomogram for Differentiation of Focal Nodular Hyperplasia from Hepatocellular Carcinoma in the Non-Cirrhotic Liver. Cancer Imaging 2020, 20, 20. [Google Scholar] [CrossRef]
- Lang, N.; Zhang, Y.; Zhang, E.; Zhang, J.; Chow, D.; Chang, P.; Yu, H.J.; Yuan, H.; Su, M.-Y. Differentiation of Spinal Metastases Originated from Lung and Other Cancers Using Radiomics and Deep Learning Based on DCE-MRI. Magn. Reson. Imaging 2019, 64, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Kirienko, M.; Cozzi, L.; Rossi, A.; Voulaz, E.; Antunovic, L.; Fogliata, A.; Chiti, A.; Sollini, M. Ability of FDG PET and CT Radiomics Features to Differentiate between Primary and Metastatic Lung Lesions. Eur. J. Nucl. Med. Mol. Imaging 2018, 45, 1649–1660. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, J.; Liu, S.; Cui, J.; Zhan, J.; Pang, J.; Zhou, R.; Li, X.; Dong, C. MRI-Based Radiomics Nomogram for Differentiation of Benign and Malignant Lesions of the Parotid Gland. Eur. Radiol. 2021, 31, 4042–4052. [Google Scholar] [CrossRef]
- Hamm, C.A.; Wang, C.J.; Savic, L.J.; Ferrante, M.; Schobert, I.; Schlachter, T.; Lin, M.; Duncan, J.S.; Weinreb, J.C.; Chapiro, J.; et al. Deep Learning for Liver Tumor Diagnosis Part I: Development of a Convolutional Neural Network Classifier for Multi-Phasic MRI. Eur. Radiol. 2019, 29, 3338–3347. [Google Scholar] [CrossRef]
- Wang, C.J.; Hamm, C.A.; Savic, L.J.; Ferrante, M.; Schobert, I.; Schlachter, T.; Lin, M.; Weinreb, J.C.; Duncan, J.S.; Chapiro, J.; et al. Deep Learning for Liver Tumor Diagnosis Part II: Convolutional Neural Network Interpretation Using Radiologic Imaging Features. Eur. Radiol. 2019, 29, 3348–3357. [Google Scholar] [CrossRef] [PubMed]
- Venkadesh, K.V.; Setio, A.A.A.; Schreuder, A.; Scholten, E.T.; Chung, K.; Wille, M.M.W.; Saghir, Z.; van Ginneken, B.; Prokop, M.; Jacobs, C. Deep Learning for Malignancy Risk Estimation of Pulmonary Nodules Detected at Low-Dose Screening CT. Radiology 2021, 300, 438–447. [Google Scholar] [CrossRef] [PubMed]
- Ardila, D.; Kiraly, A.P.; Bharadwaj, S.; Choi, B.; Reicher, J.J.; Peng, L.; Tse, D.; Etemadi, M.; Ye, W.; Corrado, G.; et al. End-to-End Lung Cancer Screening with Three-Dimensional Deep Learning on Low-Dose Chest Computed Tomography. Nat. Med. 2019, 25, 954–961. [Google Scholar] [CrossRef]
- Al-masni, M.A.; Al-antari, M.A.; Park, J.-M.; Gi, G.; Kim, T.-Y.; Rivera, P.; Valarezo, E.; Choi, M.-T.; Han, S.-M.; Kim, T.-S. Simultaneous Detection and Classification of Breast Masses in Digital Mammograms via a Deep Learning YOLO-Based CAD System. Comput. Methods Programs Biomed. 2018, 157, 85–94. [Google Scholar] [CrossRef]
- Ehteshami Bejnordi, B.; Veta, M.; Johannes van Diest, P.; van Ginneken, B.; Karssemeijer, N.; Litjens, G.; van der Laak, J.A.W.M.; CAMELYON16 Consortium; Hermsen, M.; Manson, Q.F.; et al. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women with Breast Cancer. JAMA 2017, 318, 2199. [Google Scholar] [CrossRef]
- Daimiel Naranjo, I.; Gibbs, P.; Reiner, J.S.; Lo Gullo, R.; Thakur, S.B.; Jochelson, M.S.; Thakur, N.; Baltzer, P.A.T.; Helbich, T.H.; Pinker, K. Breast Lesion Classification with Multiparametric Breast MRI Using Radiomics and Machine Learning: A Comparison with Radiologists’ Performance. Cancers 2022, 14, 1743. [Google Scholar] [CrossRef]
- Wang, G. Radiomics Based on Digital Mammography Helps to Identify Mammographic Masses Suspicious for Cancer. Front. Oncol. 2022, 12, 10. [Google Scholar] [CrossRef] [PubMed]
- Ristow, I.; Madesta, F.; Well, L.; Shenas, F.; Wright, F.; Molwitz, I.; Farschtschi, S.; Bannas, P.; Adam, G.; Mautner, V.F.; et al. Evaluation of Magnetic Resonance Imaging-Based Radiomics Characteristics for Differentiation of Benign and Malignant Peripheral Nerve Sheath Tumors in Neurofibromatosis Type 1. Neuro-Oncology 2022, noac100. [Google Scholar] [CrossRef]
- Schwarz, R.F.; Ng, C.K.Y.; Cooke, S.L.; Newman, S.; Temple, J.; Piskorz, A.M.; Gale, D.; Sayal, K.; Murtaza, M.; Baldwin, P.J.; et al. Spatial and Temporal Heterogeneity in High-Grade Serous Ovarian Cancer: A Phylogenetic Analysis. PLoS Med. 2015, 12, e1001789. [Google Scholar] [CrossRef] [Green Version]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; Tarpey, P.; et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dagogo-Jack, I.; Shaw, A.T. Tumour Heterogeneity and Resistance to Cancer Therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Tharmaseelan, H.; Hertel, A.; Tollens, F.; Rink, J.; Woźnicki, P.; Haselmann, V.; Ayx, I.; Nörenberg, D.; Schoenberg, S.O.; Froelich, M.F. Identification of CT Imaging Phenotypes of Colorectal Liver Metastases from Radiomics Signatures—Towards Assessment of Interlesional Tumor Heterogeneity. Cancers 2022, 14, 1646. [Google Scholar] [CrossRef] [PubMed]
- Bailly, C.; Bodet-Milin, C.; Bourgeois, M.; Gouard, S.; Ansquer, C.; Barbaud, M.; Sébille, J.-C.; Chérel, M.; Kraeber-Bodéré, F.; Carlier, T. Exploring Tumor Heterogeneity Using PET Imaging: The Big Picture. Cancers 2019, 11, 1282. [Google Scholar] [CrossRef] [Green Version]
- Meignan, M.; Cottereau, A.S.; Versari, A.; Chartier, L.; Dupuis, J.; Boussetta, S.; Grassi, I.; Casasnovas, R.-O.; Haioun, C.; Tilly, H.; et al. Baseline Metabolic Tumor Volume Predicts Outcome in High–Tumor-Burden Follicular Lymphoma: A Pooled Analysis of Three Multicenter Studies. J. Clin. Oncol. 2016, 34, 3618–3626. [Google Scholar] [CrossRef]
- Mühlberg, A.; Holch, J.W.; Heinemann, V.; Huber, T.; Moltz, J.; Maurus, S.; Jäger, N.; Liu, L.; Froelich, M.F.; Katzmann, A.; et al. The Relevance of CT-Based Geometric and Radiomics Analysis of Whole Liver Tumor Burden to Predict Survival of Patients with Metastatic Colorectal Cancer. Eur. Radiol. 2021, 31, 834–846. [Google Scholar] [CrossRef] [PubMed]
- Ytre-Hauge, S.; Husby, J.A.; Magnussen, I.J.; Werner, H.M.J.; Salvesen, Ø.O.; Bjørge, L.; Trovik, J.; Stefansson, I.M.; Salvesen, H.B.; Haldorsen, I.S. Preoperative Tumor Size at MRI Predicts Deep Myometrial Invasion, Lymph Node Metastases, and Patient Outcome in Endometrial Carcinomas. Int. J. Gynecol. Cancer 2015, 25, 459–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cottereau, A.-S.; Hapdey, S.; Chartier, L.; Modzelewski, R.; Casasnovas, O.; Itti, E.; Tilly, H.; Vera, P.; Meignan, M.A.; Becker, S. Baseline Total Metabolic Tumor Volume Measured with Fixed or Different Adaptive Thresholding Methods Equally Predicts Outcome in Peripheral T Cell Lymphoma. J. Nucl. Med. 2017, 58, 276–281. [Google Scholar] [CrossRef] [Green Version]
- Dai, Z.; Wei, R.; Wang, H.; Hu, W.; Sun, X.; Zhu, J.; Li, H.; Ge, Y.; Song, B. Multimodality MRI-Based Radiomics for Aggressiveness Prediction in Papillary Thyroid Cancer. BMC Med. Imaging 2022, 22, 54. [Google Scholar] [CrossRef]
- Rodrigues, A.; Santinha, J.; Galvão, B.; Matos, C.; Couto, F.M.; Papanikolaou, N. Prediction of Prostate Cancer Disease Aggressiveness Using Bi-Parametric Mri Radiomics. Cancers 2021, 13, 6065. [Google Scholar] [CrossRef]
- Kim, M.; Jung, S.Y.; Park, J.E.; Jo, Y.; Park, S.Y.; Nam, S.J.; Kim, J.H.; Kim, H.S. Diffusion- and Perfusion-Weighted MRI Radiomics Model May Predict Isocitrate Dehydrogenase (IDH) Mutation and Tumor Aggressiveness in Diffuse Lower Grade Glioma. Eur. Radiol. 2020, 30, 2142–2151. [Google Scholar] [CrossRef]
- Chen, T.; Li, M.; Gu, Y.; Zhang, Y.; Yang, S.; Wei, C.; Wu, J.; Li, X.; Zhao, W.; Shen, J. Prostate Cancer Differentiation and Aggressiveness: Assessment with a Radiomic-Based Model vs. PI-RADS V2. J. Magn. Reson. Imaging 2019, 49, 875–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.; Kobayashi, Y.; Urayama, K.Y.; Yamaura, H.; Yatabe, Y.; Hida, T. Imaging Characteristics of Driver Mutations in EGFR, KRAS, and ALK among Treatment-Naïve Patients with Advanced Lung Adenocarcinoma. PLoS ONE 2016, 11, e0161081. [Google Scholar] [CrossRef] [Green Version]
- Pinker, K.; Shitano, F.; Sala, E.; Do, R.K.; Young, R.J.; Wibmer, A.G.; Hricak, H.; Sutton, E.J.; Morris, E.A. Background, Current Role, and Potential Applications of Radiogenomics: Role and Applications of Radiogenomics. J. Magn. Reson. Imaging 2018, 47, 604–620. [Google Scholar] [CrossRef] [PubMed]
- Mingzhu, L.; Yaqiong, G.; Mengru, L.; Wei, W. Prediction of BRCA Gene Mutation Status in Epithelial Ovarian Cancer by Radiomics Models Based on 2D and 3D CT Images. BMC Med. Imaging 2021, 21, 180. [Google Scholar] [CrossRef]
- Iwatate, Y.; Hoshino, I.; Yokota, H.; Ishige, F.; Itami, M.; Mori, Y.; Chiba, S.; Arimitsu, H.; Yanagibashi, H.; Nagase, H.; et al. Radiogenomics for Predicting P53 Status, PD-L1 Expression, and Prognosis with Machine Learning in Pancreatic Cancer. Br. J. Cancer 2020, 123, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Manikis, G.C.; Ioannidis, G.S.; Siakallis, L.; Nikiforaki, K.; Iv, M.; Vozlic, D.; Surlan-Popovic, K.; Wintermark, M.; Bisdas, S.; Marias, K. Multicenter DSC–MRI-Based Radiomics Predict IDH Mutation in Gliomas. Cancers 2021, 13, 3965. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Qian, Z.; Xu, K.; Wang, K.; Fan, X.; Li, S.; Jiang, T.; Liu, X.; Wang, Y. MRI Features Predict P53 Status in Lower-Grade Gliomas via a Machine-Learning Approach. NeuroImage Clin. 2018, 17, 306–311. [Google Scholar] [CrossRef]
- Wang, G.; Wang, B.; Wang, Z.; Li, W.; Xiu, J.; Liu, Z.; Han, M. Radiomics Signature of Brain Metastasis: Prediction of EGFR Mutation Status. Eur. Radiol. 2021, 31, 4538–4547. [Google Scholar] [CrossRef]
- Cui, Y.; Liu, H.; Ren, J.; Du, X.; Xin, L.; Li, D.; Yang, X.; Wang, D. Development and Validation of a MRI-Based Radiomics Signature for Prediction of KRAS Mutation in Rectal Cancer. Eur. Radiol. 2020, 30, 1948–1958. [Google Scholar] [CrossRef]
- Liang, S.; Zhang, R.; Liang, D.; Song, T.; Ai, T.; Xia, C.; Xia, L.; Wang, Y. Multimodal 3D DenseNet for IDH Genotype Prediction in Gliomas. Genes 2018, 9, 382. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Lin, F.; Zhang, J.; Lv, X.; Zhou, J.; Li, Z.-C.; Chen, Y. Deep Learning Radiomics to Predict PTEN Mutation Status from Magnetic Resonance Imaging in Patients with Glioma. Front. Oncol. 2021, 11, 734433. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Liu, X.; Li, M.; Li, X.; Yang, H.; Zhang, H. Noninvasive KRAS Mutation Estimation in Colorectal Cancer Using a Deep Learning Method Based on CT Imaging. BMC Med. Imaging 2020, 20, 59. [Google Scholar] [CrossRef] [PubMed]
- Yin, G.; Wang, Z.; Song, Y.; Li, X.; Chen, Y.; Zhu, L.; Su, Q.; Dai, D.; Xu, W. Prediction of EGFR Mutation Status Based on 18F-FDG PET/CT Imaging Using Deep Learning-Based Model in Lung Adenocarcinoma. Front. Oncol. 2021, 11, 709137. [Google Scholar] [CrossRef] [PubMed]
- Nagano, T.; Tachihara, M.; Nishimura, Y. Mechanism of Resistance to Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitors and a Potential Treatment Strategy. Cells 2018, 7, 212. [Google Scholar] [CrossRef] [Green Version]
- Chiu, F.-Y.; Yen, Y. Efficient Radiomics-Based Classification of Multi-Parametric MR Images to Identify Volumetric Habitats and Signatures in Glioblastoma: A Machine Learning Approach. Cancers 2022, 14, 1475. [Google Scholar] [CrossRef]
- Iacobuzio-Donahue, C.A.; Michael, C.; Baez, P.; Kappagantula, R.; Hooper, J.E.; Hollman, T.J. Cancer Biology as Revealed by the Research Autopsy. Nat. Rev. Cancer 2019, 19, 686–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, C.; Ahdoot, M.; Daneshvar, M.A.; Hague, C.; Wilbur, A.R.; Gomella, P.T.; Shih, J.; Khondakar, N.; Yerram, N.; Mehralivand, S.; et al. Why Does Magnetic Resonance Imaging-Targeted Biopsy Miss Clinically Significant Cancer? J. Urol. 2022, 207, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Falagario, U.G.; Jambor, I.; Lantz, A.; Ettala, O.; Stabile, A.; Taimen, P.; Aronen, H.J.; Knaapila, J.; Perez, I.M.; Gandaglia, G.; et al. Combined Use of Prostate-Specific Antigen Density and Magnetic Resonance Imaging for Prostate Biopsy Decision Planning: A Retrospective Multi-Institutional Study Using the Prostate Magnetic Resonance Imaging Outcome Database (PROMOD). Eur. Urol. Oncol. 2021, 4, 971–979. [Google Scholar] [CrossRef]
- Interlenghi, M.; Salvatore, C.; Magni, V.; Caldara, G.; Schiavon, E.; Cozzi, A.; Schiaffino, S.; Carbonaro, L.A.; Castiglioni, I.; Sardanelli, F. A Machine Learning Ensemble Based on Radiomics to Predict BI-RADS Category and Reduce the Biopsy Rate of Ultrasound-Detected Suspicious Breast Masses. Diagnostics 2022, 12, 187. [Google Scholar] [CrossRef] [PubMed]
- Robb, T.J.; Tse, R.; Blenkiron, C. Reviving the Autopsy for Modern Cancer Evolution Research. Cancers 2021, 13, 409. [Google Scholar] [CrossRef]
- Haselmann, V.; Hedtke, M.; Neumaier, M. Liquid Profiling for Cancer Patient Stratification in Precision Medicine—Current Status and Challenges for Successful Implementation in Standard Care. Diagnostics 2022, 12, 748. [Google Scholar] [CrossRef] [PubMed]
- Cucchiara, F.; Del Re, M.; Valleggi, S.; Romei, C.; Petrini, I.; Lucchesi, M.; Crucitta, S.; Rofi, E.; De Liperi, A.; Chella, A.; et al. Integrating Liquid Biopsy and Radiomics to Monitor Clonal Heterogeneity of EGFR-Positive Non-Small Cell Lung Cancer. Front. Oncol. 2020, 10, 593831. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tharmaseelan, H.; Hertel, A.; Rennebaum, S.; Nörenberg, D.; Haselmann, V.; Schoenberg, S.O.; Froelich, M.F. The Potential and Emerging Role of Quantitative Imaging Biomarkers for Cancer Characterization. Cancers 2022, 14, 3349. https://doi.org/10.3390/cancers14143349
Tharmaseelan H, Hertel A, Rennebaum S, Nörenberg D, Haselmann V, Schoenberg SO, Froelich MF. The Potential and Emerging Role of Quantitative Imaging Biomarkers for Cancer Characterization. Cancers. 2022; 14(14):3349. https://doi.org/10.3390/cancers14143349
Chicago/Turabian StyleTharmaseelan, Hishan, Alexander Hertel, Shereen Rennebaum, Dominik Nörenberg, Verena Haselmann, Stefan O. Schoenberg, and Matthias F. Froelich. 2022. "The Potential and Emerging Role of Quantitative Imaging Biomarkers for Cancer Characterization" Cancers 14, no. 14: 3349. https://doi.org/10.3390/cancers14143349
APA StyleTharmaseelan, H., Hertel, A., Rennebaum, S., Nörenberg, D., Haselmann, V., Schoenberg, S. O., & Froelich, M. F. (2022). The Potential and Emerging Role of Quantitative Imaging Biomarkers for Cancer Characterization. Cancers, 14(14), 3349. https://doi.org/10.3390/cancers14143349





