Tensin2 Is a Novel Diagnostic Marker in GIST, Associated with Gastric Location and Non-Metastatic Tumors
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Clinical Data
2.2. Tissue Microarray Construction and Immunohistochemistry
2.3. RNA Extraction and qPCR
2.4. DNA Extraction, PCR, and Sequencing
2.5. Statistical Analyses
3. Results
3.1. Clinicopathological Data of Patients
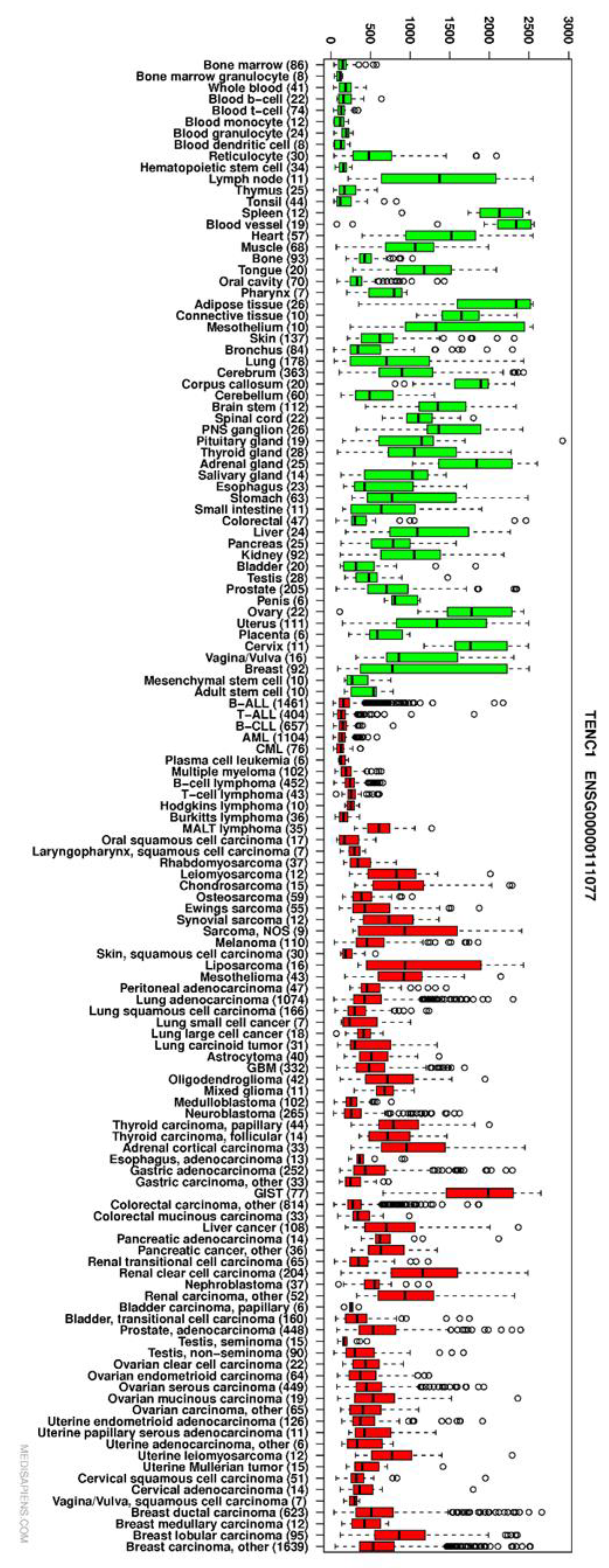
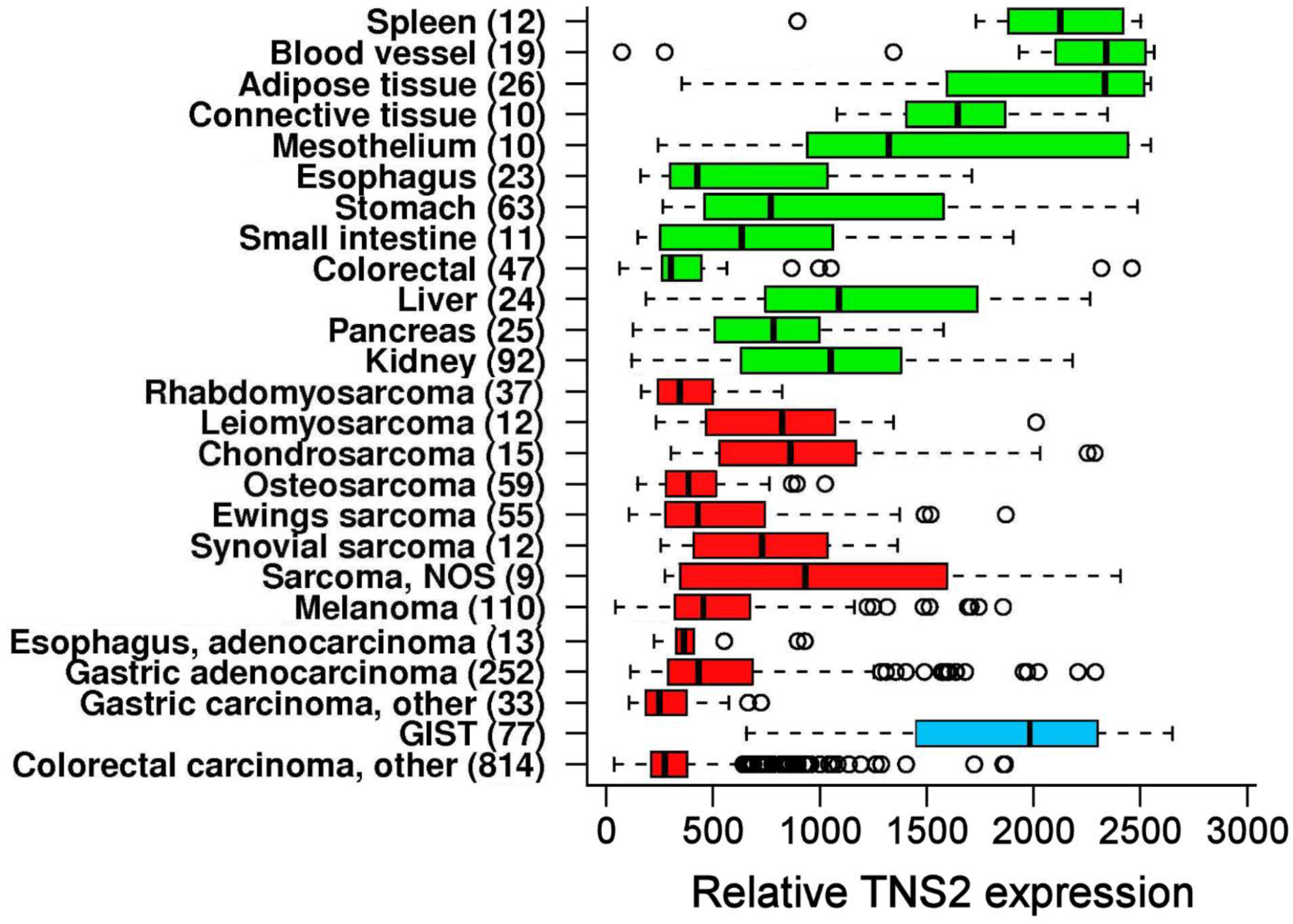
3.2. TNS2 Is Highly Overexpressed in GIST Compared with Other Cancers
3.3. TNS2 Is a Potent Diagnostic Marker for GIST
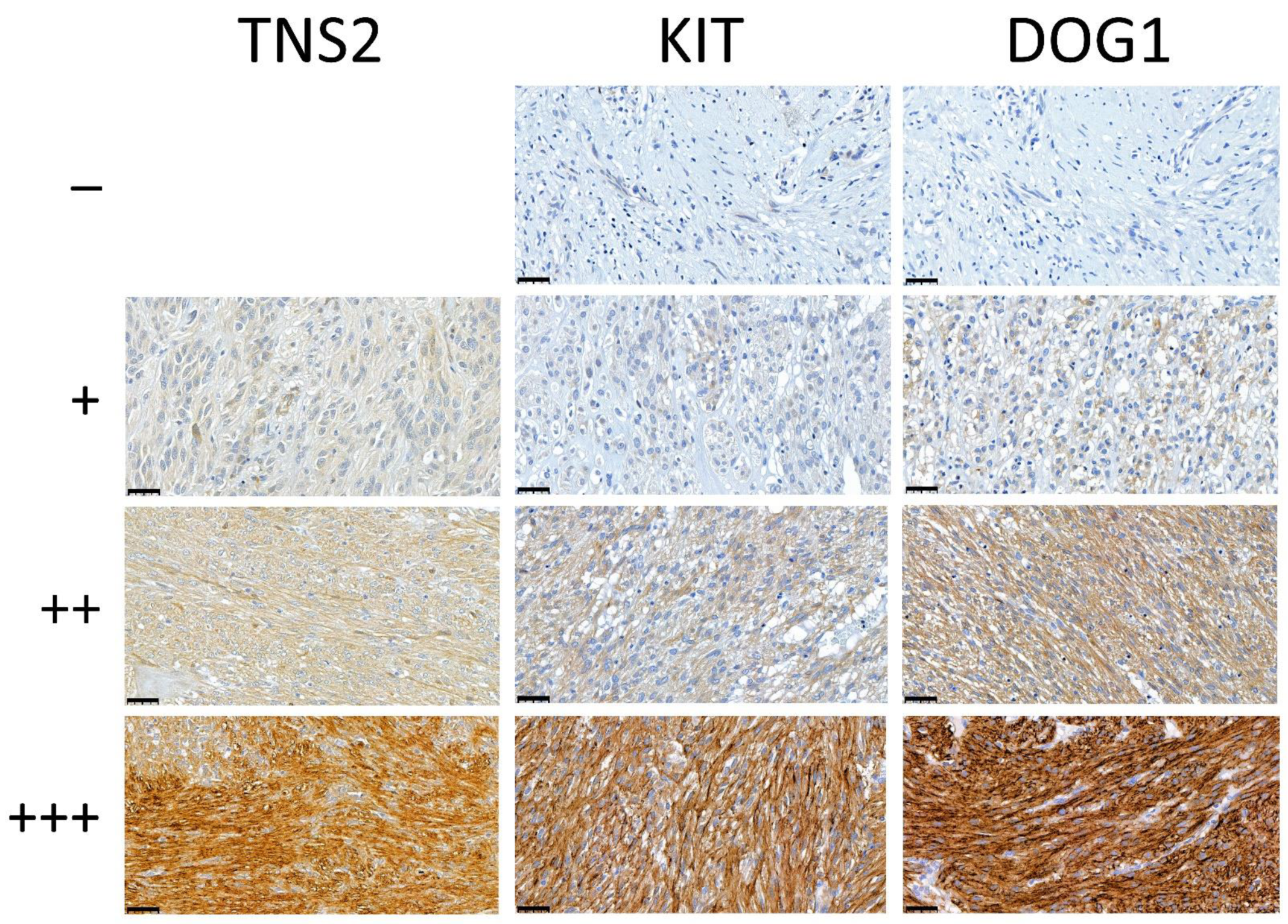
3.4. Sensitivity of TNS2 as a Diagnostic Marker Compared with KIT and DOG1
3.5. Immunohistochemical TNS2 Expression Is Stronger in Gastric and Non-Metastatic GISTs
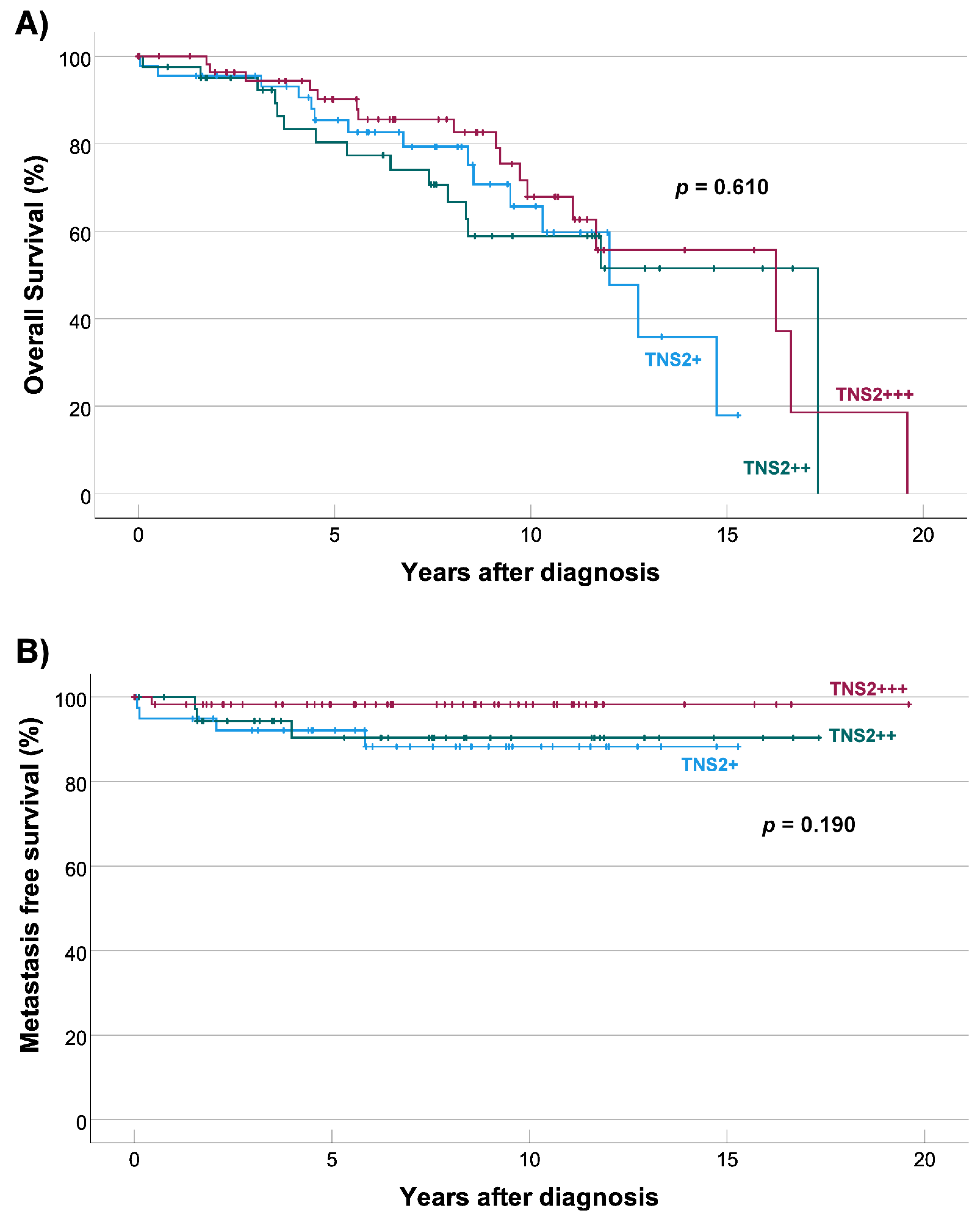
3.6. TNS2 Expression Is Not Associated with Overall or Metastasis-Free Survival
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Soreide, K.; Sandvik, O.M.; Soreide, J.A.; Giljaca, V.; Jureckova, A.; Bulusu, V.R. Global epidemiology of gastrointestinal stromal tumours (GIST): A systematic review of population-based cohort studies. Cancer Epidemiol. 2016, 40, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Sircar, K.; Hewlett, B.R.; Huizinga, J.D.; Chorneyko, K.; Berezin, I.; Riddell, R.H. Interstitial cells of Cajal as precursors of gastrointestinal stromal tumors. Am. J. Surg. Pathol. 1999, 23, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Kondo, J.; Huh, W.J.; Franklin, J.L.; Heinrich, M.C.; Rubin, B.P.; Coffey, R.J. A smooth muscle-derived, Braf-driven mouse model of gastrointestinal stromal tumor (GIST): Evidence for an alternative GIST cell-of-origin. J. Pathol. 2020, 252, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Corless, C.L.; Fletcher, J.A.; Heinrich, M.C. Biology of gastrointestinal stromal tumors. J. Clin. Oncol. 2004, 22, 3813–3825. [Google Scholar] [CrossRef]
- Hostein, I.; Faur, N.; Primois, C.; Boury, F.; Denard, J.; Emile, J.F.; Bringuier, P.P.; Scoazec, J.Y.; Coindre, J.M. BRAF mutation status in gastrointestinal stromal tumors. Am. J. Clin. Pathol. 2010, 133, 141–148. [Google Scholar] [CrossRef]
- Janeway, K.A.; Kim, S.Y.; Lodish, M.; Nose, V.; Rustin, P.; Gaal, J.; Dahia, P.L.; Liegl, B.; Ball, E.R.; Raygada, M.; et al. Defects in succinate dehydrogenase in gastrointestinal stromal tumors lacking KIT and PDGFRA mutations. Proc. Natl. Acad. Sci. USA 2011, 108, 314–318. [Google Scholar] [CrossRef]
- Miettinen, M.; Lasota, J. Gastrointestinal stromal tumors: Review on morphology, molecular pathology, prognosis, and differential diagnosis. Arch. Pathol. Lab. Med. 2006, 130, 1466–1478. [Google Scholar] [CrossRef]
- Hassan, H.T. c-Kit expression in human normal and malignant stem cells prognostic and therapeutic implications. Leuk. Res. 2009, 33, 5–10. [Google Scholar] [CrossRef]
- Liegl, B.; Hornick, J.L.; Corless, C.L.; Fletcher, C.D. Monoclonal antibody DOG1.1 shows higher sensitivity than KIT in the diagnosis of gastrointestinal stromal tumors, including unusual subtypes. Am. J. Surg. Pathol. 2009, 33, 437–446. [Google Scholar] [CrossRef]
- Miettinen, M.; Wang, Z.F.; Lasota, J. DOG1 antibody in the differential diagnosis of gastrointestinal stromal tumors: A study of 1840 cases. Am. J. Surg. Pathol. 2009, 33, 1401–1408. [Google Scholar] [CrossRef]
- Miettinen, M.; Virolainen, M.; Sarlomo-Rikala, M. Gastrointestinal stromal tumors-value of CD34 antigen in their identification and separation from true leiomyomas and schwannomas. Am. J. Surg. Pathol. 1995, 19, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.E.; Kim, M.A.; Lee, H.S.; Lee, B.L.; Kim, W.H. Characteristics of KIT-negative gastrointestinal stromal tumours and diagnostic utility of protein kinase C theta immunostaining. J. Clin. Pathol. 2008, 61, 722–729. [Google Scholar] [CrossRef] [PubMed]
- Parkkila, S.; Lasota, J.; Fletcher, J.A.; Ou, W.B.; Kivela, A.J.; Nuorva, K.; Parkkila, A.K.; Ollikainen, J.; Sly, W.S.; Waheed, A.; et al. Carbonic anhydrase II. A novel biomarker for gastrointestinal stromal tumors. Mod. Pathol. 2010, 23, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Lasota, J.; Wang, Z.; Kim, S.Y.; Helman, L.; Miettinen, M. Expression of the receptor for type i insulin-like growth factor (IGF1R) in gastrointestinal stromal tumors: An immunohistochemical study of 1078 cases with diagnostic and therapeutic implications. Am. J. Surg. Pathol. 2013, 37, 114–119. [Google Scholar] [CrossRef]
- Doyle, L.A.; Nelson, D.; Heinrich, M.C.; Corless, C.L.; Hornick, J.L. Loss of succinate dehydrogenase subunit B (SDHB) expression is limited to a distinctive subset of gastric wild-type gastrointestinal stromal tumours: A comprehensive genotype-phenotype correlation study. Histopathology 2012, 61, 801–809. [Google Scholar] [CrossRef]
- Lo, S.H.; Janmey, P.A.; Hartwig, J.H.; Chen, L.B. Interactions of tensin with actin and identification of its three distinct actin-binding domains. J. Cell. Biol. 1994, 125, 1067–1075. [Google Scholar] [CrossRef]
- Hong, S.Y.; Shih, Y.P.; Sun, P.; Hsieh, W.J.; Lin, W.C.; Lo, S.H. Down-regulation of tensin2 enhances tumorigenicity and is associated with a variety of cancers. Oncotarget 2016, 7, 38143–38153. [Google Scholar] [CrossRef]
- Calderwood, D.A.; Fujioka, Y.; de Pereda, J.M.; Garcia-Alvarez, B.; Nakamoto, T.; Margolis, B.; McGlade, C.J.; Liddington, R.C.; Ginsberg, M.H. Integrin beta cytoplasmic domain interactions with phosphotyrosine-binding domains: A structural prototype for diversity in integrin signaling. Proc. Natl. Acad. Sci. USA 2003, 100, 2272–2277. [Google Scholar] [CrossRef]
- Hafizi, S.; Ibraimi, F.; Dahlbäck, B. C1-TEN is a negative regulator of the Akt/PKB signal transduction pathway and inhibits cell survival, proliferation, and migration. FASEB J. 2005, 19, 971–973. [Google Scholar] [CrossRef]
- Koh, A.; Lee, M.N.; Yang, Y.R.; Jeong, H.; Ghim, J.; Noh, J.; Kim, J.; Ryu, D.; Park, S.; Song, P.; et al. C1-Ten Is a Protein Tyrosine Phosphatase of Insulin Receptor Substrate 1 (IRS-1), Regulating IRS-1 Stability and Muscle Atrophy. Mol. Cell. Biol. 2013, 33, 1608–1620. [Google Scholar] [CrossRef]
- Cheng, L.C.; Chen, Y.L.; Cheng, A.N.; Lee, A.Y.; Cho, C.Y.; Huang, J.S.; Chuang, S.E. AXL phosphorylates and up-regulates TNS2 and its implications in IRS-1-associated metabolism in cancer cells. J. Biomed. Sci. 2018, 25, 80. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Duncan, I.C.; Bozorgchami, H.; Lo, S.H. Tensin1 and a previously undocumented family member, tensin2, positively regulate cell migration. Proc. Natl. Acad. Sci. USA 2002, 99, 733–738. [Google Scholar] [CrossRef]
- Jung, A.S.; Kaushansky, A.; Macbeath, G.; Kaushansky, K. Tensin2 is a novel mediator in thrombopoietin (TPO)-induced cellular proliferation by promoting Akt signaling. Cell Cycle 2011, 10, 1838–1844. [Google Scholar] [CrossRef] [PubMed]
- Chan, L.-K.; Ko, F.C.F.; Ng, I.O.-L.; Yam, J.W.P. Deleted in liver cancer 1 (DLC1) utilizes a novel binding site for Tensin2 PTB domain interaction and is required for tumor-suppressive function. PLoS ONE 2009, 4, e5572. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Joensuu, H. Risk stratification of patients diagnosed with gastrointestinal stromal tumor. Hum. Pathol. 2008, 39, 1411–1419. [Google Scholar] [CrossRef]
- Kilpinen, S.; Autio, R.; Ojala, K.; Iljin, K.; Bucher, E.; Sara, H.; Pisto, T.; Saarela, M.; Skotheim, R.I.; Bjorkman, M.; et al. Systematic bioinformatic analysis of expression levels of 17,330 human genes across 9,783 samples from 175 types of healthy and pathological tissues. Genome Biol. 2008, 9, R139. [Google Scholar] [CrossRef]
- Kang, H.C.; Menias, C.O.; Gaballah, A.H.; Shroff, S.; Taggart, M.W.; Garg, N.; Elsayes, K.M. Beyond the GIST: Mesenchymal tumors of the stomach. Radiographics 2013, 33, 1673–1690. [Google Scholar] [CrossRef]
- Swalchick, W.; Shamekh, R.; Bui, M.M. Is DOG1 Immunoreactivity Specific to Gastrointestinal Stromal Tumor? Cancer Control 2015, 22, 498–504. [Google Scholar] [CrossRef]
- Kocsmar, E.; Kocsmar, I.; Szalai, L.; Lendvai, G.; Szijarto, A.; Schaff, Z.; Kiss, A.; Kovalszky, I.; Papp, G.; Lotz, G. Cross-testing of major molecular markers indicates distinct pathways of tumorigenesis in gastric adenocarcinomas and synchronous gastrointestinal stromal tumors. Sci. Rep. 2020, 10, 22212. [Google Scholar] [CrossRef]
- Lasota, J.; Dansonka-Mieszkowska, A.; Sobin, L.H.; Miettinen, M. A great majority of GISTs with PDGFRA mutations represent gastric tumors of low or no malignant potential. Lab. Invest. 2004, 84, 874–883. [Google Scholar] [CrossRef]
- Rong, J.; Chen, S.; Song, C.; Wang, H.; Zhao, Q.; Zhao, R.; He, Y.; Yan, L.; Song, Y.; Wang, F.; et al. The prognostic value of gender in gastric gastrointestinal stromal tumors: A propensity score matching analysis. Biol. Sex Differ. 2020, 11, 43. [Google Scholar] [CrossRef] [PubMed]
- Kramer, K.; Knippschild, U.; Mayer, B.; Bogelspacher, K.; Spatz, H.; Henne-Bruns, D.; Agaimy, A.; Schwab, M.; Schmieder, M. Impact of age and gender on tumor related prognosis in gastrointestinal stromal tumors (GIST). BMC Cancer 2015, 15, 57. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, Y.; Kong, L.; Kan, Y. Study on Clinicopathological Features of Gastrointestinal Stromal Tumor and Relevant Prognostic Factors. Cell Biochem. Biophys. 2015, 73, 743–747. [Google Scholar] [CrossRef] [PubMed]
- Houghton, A.M.; Rzymkiewicz, D.M.; Ji, H.; Gregory, A.D.; Egea, E.E.; Metz, H.E.; Stolz, D.B.; Land, S.R.; Marconcini, L.A.; Kliment, C.R.; et al. Neutrophil elastase-mediated degradation of IRS-1 accelerates lung tumor growth. Nat. Med. 2010, 16, 219–223. [Google Scholar] [CrossRef]
- Tu, Y.; Zuo, R.; Ni, N.; Eilers, G.; Wu, D.; Pei, Y.; Nie, Z.; Wu, Y.; Wu, Y.; Ou, W.B. Activated tyrosine kinases in gastrointestinal stromal tumor with loss of KIT oncoprotein expression. Cell Cycle 2018, 17, 2577–2592. [Google Scholar] [CrossRef]
- Tarn, C.; Rink, L.; Merkel, E.; Flieder, D.; Pathak, H.; Koumbi, D.; Testa, J.R.; Eisenberg, B.; von Mehren, M.; Godwin, A.K. Insulin-like growth factor 1 receptor is a potential therapeutic target for gastrointestinal stromal tumors. Proc. Natl. Acad. Sci. USA 2008, 105, 8387–8392. [Google Scholar] [CrossRef] [PubMed]
- Martuszewska, D.; Ljungberg, B.; Johansson, M.; Landberg, G.; Oslakovic, C.; Dahlback, B.; Hafizi, S. Tensin3 is a negative regulator of cell migration and all four Tensin family members are downregulated in human kidney cancer. PLoS ONE 2009, 4, e4350. [Google Scholar] [CrossRef]
- Nizioł, M.; Pryczynicz, A. The role of tensins in malignant neoplasms. Arch. Med. Sci. 2021. [Google Scholar] [CrossRef]
- Niziol, M.; Zinczuk, J.; Zareba, K.; Guzinska-Ustymowicz, K.; Pryczynicz, A. Immunohistochemical Analysis of the Expression of Adhesion Proteins: TNS1, TNS2 and TNS3 in Correlation with Clinicopathological Parameters in Gastric Cancer. Biomolecules 2021, 11, 640. [Google Scholar] [CrossRef]
- Oppelt, P.J.; Hirbe, A.C.; Van Tine, B.A. Gastrointestinal stromal tumors (GISTs): Point mutations matter in management, a review. J. Gastrointest. Oncol. 2017, 8, 466–473. [Google Scholar] [CrossRef]
- Kaderbhai, C.G.; Boidot, R.; Beltjens, F.; Chevrier, S.; Arnould, L.; Favier, L.; Lagrange, A.; Coudert, B.; Ghiringhelli, F. Use of dedicated gene panel sequencing using next generation sequencing to improve the personalized care of lung cancer. Oncotarget 2016, 7, 24860–24870. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.A.; Chun, S.M.; Choi, Y.D.; Kweon, S.S.; Jung, S.T.; Shim, H.J.; Yun, S.J. BRAF mutations and KIT aberrations and their clinicopathological correlation in 202 Korean melanomas. J. Invest. Dermatol. 2013, 133, 579–582. [Google Scholar] [CrossRef] [PubMed]

| Target | Forward Primer | Reverse Primer | qPCR Probe |
|---|---|---|---|
| TNS2 | 5′ -AC AGA AAA TGT GAA GCA AAG GT- 3′ | 5′ -GTG CTC TAT GCG CCT GAC T- 3′ | Universal Probe Library #41 |
| GAPDH | 5′ -AGC CAC ATC GCT CAG ACA C- 3′ | 5′ -GCC CAA TAC GAC CAA ATC C- 3′ | Universal Probe Library #60 |
| KIT exon 9 | 5’-AGC CAG GGC TTT TGT TTT CT-3’ | 5’-CAG AGC CTA AAC ATC CCC TTA-3’ | |
| KIT exon 11 | 5’-TTT CCC TTT CTC CCC ACA G-3’ | 5’-CCC AAA AAG GTG ACA TGG A-3 | |
| KIT exon 13 | 5’-TAC TGC ATG CGC TTG ACA TC-3 | 5’-CAT GTT TTG ATA ACC TGA CAG ACA-3 | |
| KIT exon 17 | 5’-TGG TTT TCT TTT CTC CTC CAA-3 | 5’-TCA CAG GAA ACA ATT TTT ATC GAA-3 | |
| PDGFRA exon 12 | 5’-TCC AGT CAC TGT GCT GCT TC-3’ | 5’-GGA GGT TAC CCC ATG GAA CT-3 | |
| PDGFRA exon 18 | 5’-CTT GCA GGG GTG ATG CTA TT-3’ | 5’-TGA AGG AGG ATG AGC CTG AC-3 |
| Characteristic | n = 148 (%) |
|---|---|
| Sex | |
| Male | 74 (50.0) |
| Female | 74 (50.0) |
| Age | Range 10–91 |
| ≤50 | 19 (12.8) |
| 51–60 | 37 (25.0) |
| 61–70 | 46 (31.1) |
| 71–80 | 39 (26.4) |
| ≥81 | 7 (4.7) |
| Primary tumor location | |
| Gastric | 106 (71.6) |
| Small intestinal | 33 (22.3) |
| Large intestinal | 2 (1.4) |
| Extragastrointestinal | 7 (4.7) |
| Tumor diameter in cm | Range 0.9–29.0 |
| ≤2.0 | 6 (4.1) |
| 2.1–5.0 | 61 (42.1) |
| 5.1–10.0 | 54 (37.2) |
| >10.0 | 24 (16.6) |
| Not available | 3 |
| Cell mitoses per 50 HPF | Range 0–100 |
| ≤5 | 90 (65.7) |
| 6–10 | 21 (15.3) |
| >10 | 26 (19.0) |
| Not available | 11 |
| Risk category | |
| Very low | 4 (2.9) |
| Low | 45 (32.1) |
| Intermediate | 29 (20.7) |
| High | 62 (44.3) |
| Not available | 8 |
| Metastasis status | |
| Metastatic at diagnosis | 9 (6.1) |
| Metastatic after diagnosis | 8 (5.4) |
| Total metastatic | 17 (11.5) |
| Not metastatic | 131 (88.5) |
| Not available | 8 |
| Mutated gene and exon | |
| KIT exon 9 | 11 (7.6) |
| KIT exon 11 | 86 (59.7) |
| KIT exon 13 | 6 (4.2) |
| PDGFRA exon 12 | 2 (1.4) |
| PDGFRA exon 18 | 17 (11.8) |
| KIT/PDGFRA WT | 22 (15.3) |
| Not available | 4 |
| Sarcoma | n | Negative (%) | Weak (%) | Intermediate (%) | Strong (%) |
|---|---|---|---|---|---|
| Fibrosarcoma | 19 | 19 (100) | 0 (0) | 0 (0) | 0 (0) |
| Liposarcoma | 89 | 77 (86.5) | 7 (7.9) | 5 (5.6) | 0 (0) |
| Leiomyosarcoma | 57 | 50 (87.7) | 5 (8.8) | 2 (3.5) | 0 (0) |
| Malignant fibrous histiocytoma | 220 | 200 (90.9) | 15 (6.8) | 5 (2.3) | 0 (0) |
| Malignant peripheral nerve sheath tumor | 19 | 17 (89.5) | 2 (10.5) | 0 (0) | 0 (0) |
| Myxofibrosarcoma | 45 | 37 (82.2) | 6 (13.3) | 2 (4.5) | 0 (0) |
| Sarcoma of unknown primary (NUD) | 31 | 28 (90.3) | 3 (9.7) | 0 (0) | 0 (0) |
| Synovial sarcoma | 41 | 40 (97.6) | 0 (0) | 1 (2.4) | 0 (0) |
| Total other sarcomas | 521 | 468 (89.8) | 38 (7.3) | 15 (2.9) | 0 (0) |
| Sarcoma series; GIST | 27 | 0 (0) | 5 (18.6) | 11 (40.7) | 11 (40.7) |
| GIST series | 148 | 0 (0) | 45 (30.4) | 44 (29.7) | 59 (39.9) |
| Total GISTs | 175 | 0 (0) | 50 (28.6) | 55 (31.4) | 70 (40.0) |
| Characteristic | TNS2 Expression | p-Value | ||
|---|---|---|---|---|
| Weak (n = 45) | Intermediate (n = 44) | Strong (n = 59) | ||
| Sex, n (%) | 0.029 | |||
| Male (n = 74) | 30 (40.5) | 19 (25.7) | 25 (33.8) | |
| Female (n = 74) | 15 (20.3) | 25 (33.8) | 34 (45.9) | |
| Median age at diagnosis (range), years | 59.0 (10–91) | 65.5 (44–87) | 66.0 (35–89) | 0.131 |
| Primary tumor location, n (%) | <0.001 | |||
| Gastric (n = 106) | 14 (13.2) | 36 (34.0) | 56 (52.8) | |
| Non-gastric (n = 42) | 31 (73.8) | 8 (19.0) | 3 (7.2) | |
| Median diameter of primary tumor (range), cm | 6.0 (2.0–25.0) | 5.8 (2.0–29.0) | 5.0 (0.9–19.0) | 0.197 |
| Not available | 1 | 0 | 2 | |
| Median amount of cell mitoses per 50 HPF (range) | 5.0 (1–80) | 4.0 (0–100) | 4.0 (0–45) | 0.260 |
| Not available | 4 | 2 | 5 | |
| Metastatic GIST tumor, n (%) | 0.004 | |||
| Not metastatic (n = 131) | 36 (27.5) | 37 (28.2) | 58 (44.3) | |
| Metastatic (n = 17) | 9 (52.9) | 7 (41.2) | 1 (5.9) | |
| Risk category, n (%) | 0.020 | |||
| Very low/Low (n = 59) | 13 (26.5) | 13 (26.5) | 23 (46.9) | |
| Intermediate (n = 29) | 3 (10.3) | 11 (37.9) | 15 (51.7) | |
| High (n = 62) | 26 (41.9) | 18 (29.0) | 18 (29.0) | |
| Not available | 3 | 2 | 3 | |
| Mutated gene, n (%) | 0.442 | |||
| KIT (n = 103) | 33 (32.0) | 26 (25.2) | 44 (42.7) | |
| PDGFRA (n = 19) | 4 (21.1) | 9 (47.4) | 6 (31.6) | |
| KIT/PDGFRA WT (n = 22) | 7 (31.8) | 7 (31.8) | 8 (36.4) | |
| Not available | 1 | 2 | 1 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salmikangas, S.; Böhling, T.; Merikoski, N.; Jagdeo, J.; Sampo, M.; Vesterinen, T.; Sihto, H. Tensin2 Is a Novel Diagnostic Marker in GIST, Associated with Gastric Location and Non-Metastatic Tumors. Cancers 2022, 14, 3212. https://doi.org/10.3390/cancers14133212
Salmikangas S, Böhling T, Merikoski N, Jagdeo J, Sampo M, Vesterinen T, Sihto H. Tensin2 Is a Novel Diagnostic Marker in GIST, Associated with Gastric Location and Non-Metastatic Tumors. Cancers. 2022; 14(13):3212. https://doi.org/10.3390/cancers14133212
Chicago/Turabian StyleSalmikangas, Sami, Tom Böhling, Nanna Merikoski, Joanna Jagdeo, Mika Sampo, Tiina Vesterinen, and Harri Sihto. 2022. "Tensin2 Is a Novel Diagnostic Marker in GIST, Associated with Gastric Location and Non-Metastatic Tumors" Cancers 14, no. 13: 3212. https://doi.org/10.3390/cancers14133212
APA StyleSalmikangas, S., Böhling, T., Merikoski, N., Jagdeo, J., Sampo, M., Vesterinen, T., & Sihto, H. (2022). Tensin2 Is a Novel Diagnostic Marker in GIST, Associated with Gastric Location and Non-Metastatic Tumors. Cancers, 14(13), 3212. https://doi.org/10.3390/cancers14133212








