A Systematic Review of the Current Status and Quality of Radiomics for Glioma Differential Diagnosis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
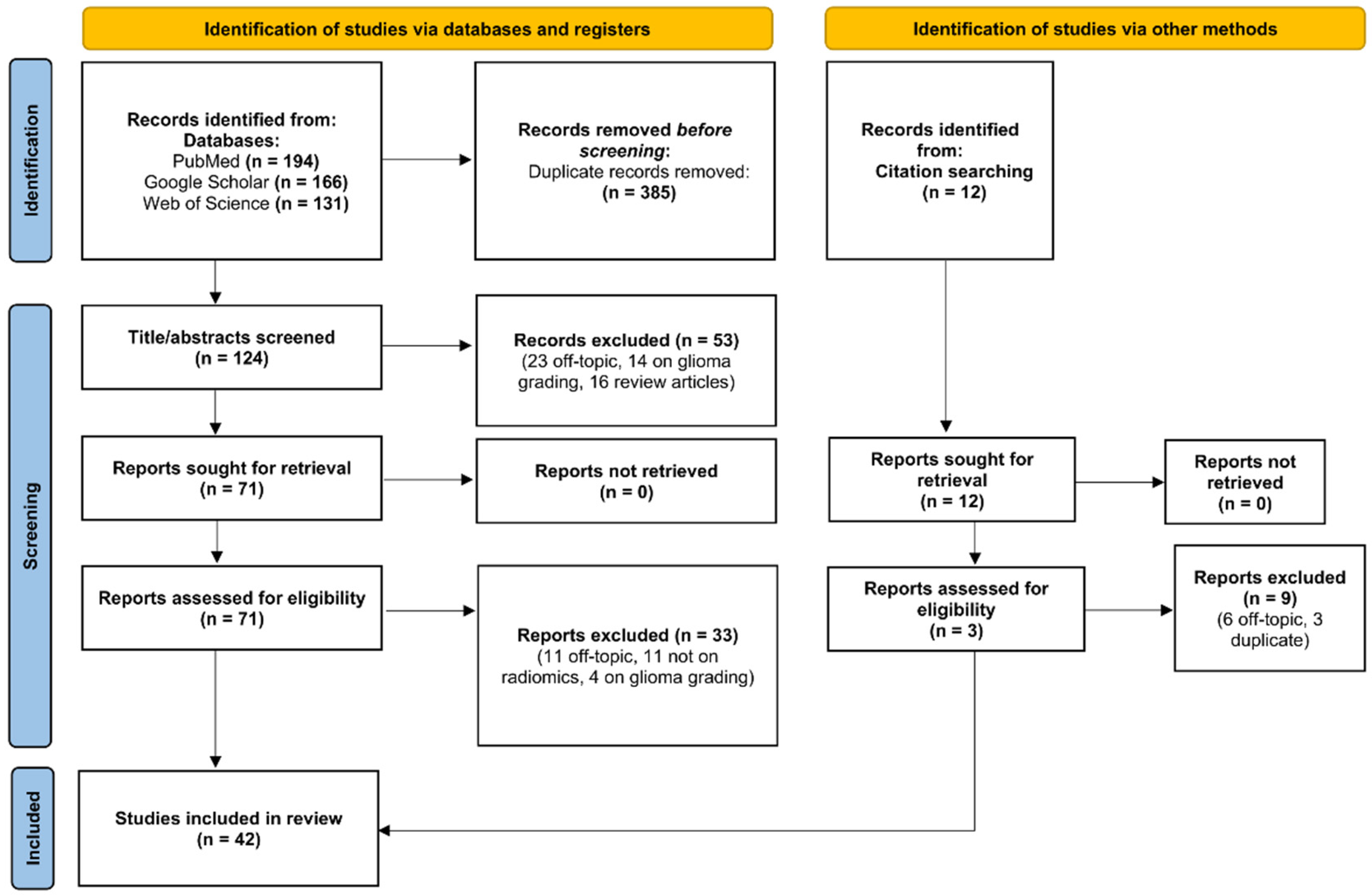
2.1. Search Strategy and Selection Criteria
2.2. Planning and Conducting the Review
2.3. Quality Assessment Using RQS Evaluation
2.4. Statistical Analysis
3. Results
3.1. Study Selection
3.2. Characteristics of Included Studies
| Authors, Year | ST | Diseases | NP (Type) | Modalities Used for Feature Extraction | Seg | Region for Feature Extraction | Software Used for Feature Extraction | Features Number (Type) | FS | CM | VM | Model Applied to a Separate Dataset? | Most Important Result | Main Findings |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Choi et al., 2016 [27] | R | PCNSL, GBM | 42 (19 GBM, 23 PCNSL) | CE-T1WI (IAUC), ADC | S, 3D | CE tumor (no necrosis) | MIPAV | 3 (histogram) | no | multivariate model | LOOCV | no | AUC = 0.886 | The IAUC may be a useful parameter together with ADC for differentiating between PCNSL and atypical GBM. |
| Alcaide-Leon et al., 2017 [28] | R | PCNSL, glioma | 106 (35 PCNSL, 71 glioma) | CE-T1WI | M, 3D | CE tumor | NR | 153 (first-order, second-order texture metrics) | SVM— F-statistic | SVM | nested 10-fold CV | no | AUC = 0.87 | SVM based on textural features of CE-T1WI is not inferior to expert human evaluation in PCNSL/glioma differentiation. |
| Chen et al., 2017 [29] | P | PCNSL, GBM | 96 (30 PCNSL, 66 GBM) | CE-T1WI | A, 3D | whole tumor | NR | 16,384 (SIFT features) | t-test, GA | SVM | LOOCV | yes | AUC = 0.991 | SIFT method produced more competitive PCNSL and GBM differentiation performance by using conventional MRI. |
| Wu et al., 2017 [30] | R | PCNSL, GBM | 102 (32 PCNSLs, 70 GBMs) | T2WI, CE-T1WI | A + S for small tumors, 3D | CE tumor and peritumoral edema | Matlab | NR (Deep learning features) | sparse representation-based feature selection method | sparse representation classification | LOOCV | yes | Acc = 98.51% | The SRR system had superior PCNSL/GBM differentiation performance compared to advanced imaging techniques. |
| Artzi et al., 2019 [31] | R | GBM, MET | 439 (212 GBM, 227 MET) | CE-T1WI | S, 3D | CE tumor | Matlab R2017a | 757 (Location, first-order, second-order, morphological, wavelet) | NCA, PCA | SVM, kNN, DT, ensemble classifiers, BoF | 5-fold CV | yes | AUC = 0.85 | GBM/MET differentiation showed a high success rate based on postcontrast T1W. GBM/MET subtypes classification may require additional MRI sequences. |
| Kang et al., 2018 [32] | R | PCNSL, GBM | 196 (119 GBM, 77 PCNSL) | CE-T1WI, ADC | S, 3D | CE tumor | Matlab R2014b | 1618 (first-order, shape, texture, wavelet) | 12 feature selection methods | KNN, NB, DT, LDA, RF, AB, boosting, linear SVM, radial basis function SVM | 10-fold CV | yes | AUC = 0.983 | The diffusion radiomics model yielded a better diagnostic performance than conventional radiomics or single advanced MRI in identifying atypical PCNSL mimicking GBM. |
| Kim et al., 2018 [21] | R | PCNSL, GBM | 143 () 86 (78 GBM, 65 PCNSL) | T2w, FLAIR, CE-T1WI, DWI | S, 3D | CE tumor and whole (enhancing or non-enhancing) tumor plus peritumoral edema | Matlab | 127 (16 shapebased, 57 histogram-based, and 54 texture-based) | mRMR, LASSO | 3 classifiers: logistic classifier, SVM, RF | 10-fold CV | yes | AUC = 0.979 in the discovery cohort and 0.956 in the validation cohort | Radiomics features derived from multi-parametric MRI can be used to differentiate PCNSL from glioblastoma effectively. |
| Kunimatsu et al., 2018 [33] | R | PCNSL, GBM | 60 (16 PCNSL, 44 GBM) | CE-T1WI | S, 2D | CE tumor | R | 67 (first-order, second-order features) | ICC, t-test | PCA | no | no | NR | Among MRI-based textures, first-order entropy, median, GLRLM-based run length non-uniformity, and run percentage are considered to enhance differences between GBM and PCNSL. |
| Nakagawa et al., 2018 [34] | R | PCNSL, GBM | 70 (45 GBM, 25 PCNSL) | T2, rCBV, CE-T1WIs, ADC | M, 2D | whole tumor | LIFEx | 48 (12 for each sequence) (histograms and texture parameters) | not performed | LR, multivariate XGBoost | 10-fold CV | no | AUC = 0.98 | mpMRI radiomics model outperformed conventional cut-off method and the board certified radiologists in distinguishing GBM from PCNSL. |
| Suh et al., 2018 [35] | R | PCNSL, GBM | 77 (54 PCNSL, 23 non-necrotic atypical GBM) | post-contrast T1- and T2-weighted, and FLAIR; ADC (10th percentile) | S, 3D | CE tumor, NE tumour tissue and edema | PyRadiomics | 6366 (shape, volume, first-order, texture, and wavelet) | t-test, recursive feature elimination | RF | nested CV | no | AUC = 0.921 | The radiomics model yields a better diagnostic performance than human radiologists and ADC values. |
| Xiao et al., 2018 [36] | R | PCNSL, GBM | 82 (22 PCNSL, 60 GBM) | T1WI, CE-T1WI | M, 3D | CE tumor and intratumoral cysts | PyRadiomics | 105 (92 texture features and 13 geometric features) | Weka CfsSubsetEval | ROC analysis; NB, SVM, LR, RF | 10-fold CV | no | AUC = 0.90 for NB; Acc = 0.92 for SVM | MRI-based 3D texture analysis has potential utility for preoperative GBM/PCNSL discrimination. |
| Bao et al., 2018 [37] | R | GBM, PCNSL | 20 (9 PCNSL, 11 GBM) | rCBV, ADC | S, 3D | CE tumor (no cysts and necrosis) | nordicICE | 11 (histogram) | no | Multivariate LR | no | no | AUC = 0.97 | Whole-tumor histogram analysis of nCBV and ADC was able to differentiate between GBM and PCNSL. |
| Chen et al., 2019 [38] | R | GBM, MET | 134 (77 gbm, 58 MET) | CE-T1WI | M, 3D | CE tumor | LIFEx | 43 (shape, first-order, texture) | five selection methods: RF, LASSO, XGBoost, GBDT | LDA, SVM, RF, KNN, Gaussian NB, LR. | 4-fold | yes | AUC = 0.80 | Radiomic-based machine learning has potential to be utilized in differentiating GBM from MET. |
| Dong et al., 2019 [39] | R | GBM, MET | 120 (60 GBM, 60 MET) | T1W, T2W, CE-T1WI | M, 3D | peri- enhancing oedema | PyRadiomics | 321 (shape, first-order, texture) | ICC, Boruta algorithm | DT, SVM, NN, NB, KNN | 10-fold CV | yes | AUC from 0.70 to 0.76, for the training dataset, and from 0.56 to 0.64 for the validation data set | Combined use of classifiers could confer extra benefits for GBM/MET differentiation. |
| Kong et al., 2019 [23] | R | PCNSL, GBM | 77 (24 lymphoma, 53 GBM) | SUV map, SUVncc map, SUVnbm map | M, 3D | whole tumor | PyRadiomics | 107 (shape, first-order, texture) | ICC | ROC analysis | 10-fold CV | no | AUC = 0.998 | 18F-FDG-PET-based radiomics is a reliable noninvasive method to distinguish PCNSL from GBM. |
| Kumimatsu et al., 2019 [40] | R | PCNSL, GBM | 76 (55 GBM, 21 PCNSL) | CE-T1WI | S, 2D | CE tumor | R | 67 (texture) | ICC, PCA | KNN, DT, LDA, SVM | 6-fold CV | yes | AUC = 0.99 on training set; Acc = 75% on test data | Radiomics MRI may provide complementary diagnostic information on routine brain MRI. |
| Petrujkic et al., 2019 [41] | R | GBM, MET | 55 (30 GBMs and 25 solitary MET) | T2W, SWI, CE-T1WI | M, 3D | CE tumor | ImageJ | 14 (Euclidian, fractal, texture (GLCM)) | no | ROC analysis | no | no | AUC = 0.908 | Texture features are more significant than fractal-based features in GBM/MET differentiation. |
| Qian et al., 2019 [42] | R | GBM, MET | 412 (242 GBM, 170 solitary brain MET) | T1W, T2W, CE-T1WI | M, 3D | CE tumor | PyRadiomics | 1303 (shape, first-order, texture, square, square root, logarithm, exponential, LoG, wavelet) | 12 methods (filter, wrapper, embedded methods). | 7 supervised machine-learning algorithms | 5-fold CV | yes | AUC ≥ 0.95 in the training set; AUC = 0.90 in the test set | Radiomic machine-learning technology could help in differentiating GBM from MET preoperatively. |
| Wang et al., 2019 [43] | R | PCNSL, GBM | 109 (28 PCNSL, 81 GBM) | T2W | M, 2D | CE tumor (no hemorrhage, necrosis, cysts, non- enhancement) | ImageJ | 5 (texture) | no | binary logistic regression | no | no | AUC = 0.917 | The texture features of T2WI and conventional imaging findings may be used to distinguish GBM from PCNSL. |
| Yun et al., 2019 [44] | R | PCNSL, GBM | 195 (119 GBM, 76 PCNSL) | CE-T1WI | S, 3D | CE tumor | Matlab | 936 (first- order, texture, wavelet) | Metric 1: mRMR, CFS, backward elimination; Metric 2: MLP network | Metric 1: SVM, the boosted generalized linear mixed model, regularized RF; Metric 2: MLP network | Metric 1: 10 fold CV Metric 2: 10 fold CV | yes | AUC > 0.82 | A combination of radiomic features and MLP network classifier serves a high-performing and generalizable model for PCNSL/GBM DDx. |
| Bae et al., 2020 [45] | R | GBM, MET | 248 (159 GBM, 89 MET) | CE mask on CE-T1WI, CE mask on T2WI, and PT mask on T2WI | S, 3D | CE tumors, non- enhancing T2 hyperintense tumors | PyRadiomics | 265 (first- order, texture) | five methods for feature selection | KNN, NB, RF, AdaBoost, L-SVM, SVM using radial basis function kernel, LDA; Multi input DNN | 10-fold CV | yes | AUC = 0.95 | The results demonstrated that deep learning using radiomic features can be useful for distinguishing GBM/MET. |
| Dastmalchian et al., 2020 [46] | P | GLIOMAS, MET | 31 (17 GBM, 6 LGG, 8 MET) | T1 and T2 maps | M, 2D | CE tumors and peritumoral white matter | Matlab | 39 (texture (GLCM, GLRLM)) | Spearman correlation filter, Wilcoxon | ROC analysis | no | no | AUC = 0.952 (LGG vs. MET); AUC = 0.877 (GBM vs. MET) | Texture analysis of MRF-derived maps can improve our ability to differentiate glioma from GBM. |
| Chen et al., 2020 [47] | R | GBM, PCNSL | 138 (76 GBM, 62 PCNSL) | CE-T1WI | A, 3D | whole tumor | lifeX | 43 (histogram, shape, texture) | distance correlation, RF, LASSO, XGBoost, GBDT | LDA, SVM, LR | validation set, 100 train-validation repetition times | yes | AUC = 0.98 | Radiomics-based machine-learning algorithms potentially have promising performances in differentiating GBM from PCNSL. |
| Dong et al., 2020 [48] | R | EP, MB | 51 (24 EPs, 27 MB) | CE-T1W, ADC | S, 3D | CE tumors | 3D Slicer | 188 (shape, first-order, texture) | t-test, multivariable LR, univariate analysis screening | kNN, AdaBoost, RF, SVM | 10-fold CV | no | AUC = 0.91 | The combination of radiomics and machine-learning approach on 3D multimodal MRI could well distinguish EP and MB. |
| Oritz-Ramon et al., 2020 [20] | R | GBM, MET | 100 (50 MET, 50 GBM) | T1w | M, 2D | CE tumors | Matlab | 88 (histogram, texture, and local binary patterns) | ICC, MWW, MIC, Relief-F | RF, SVM, KNN, NB, MLP | nested CV | no | AUC = 0.896 | The proposed radiomics MRI approach is able to discriminate between GBM and BM. |
| Xia et al., 2020 [49] | R | PCNSL, GBM | 240 (129 GBM, 111 PCNSL) | FLAIR, DWI, CE-T1WI, ADC | M, 3D | Tumor tissue and peritumoral edema | PyRadiomics | 851 (shape, first-order, texture, wavelet) | ICC, Spearman correlation filter, Mrmr, LASSO | LASSO | 10-fold CV | yes | AUC = 0.943 | The model combining MP-MRI and radiologists’ diagnoses had superior performance to the radiologists alone. |
| Zhou et al., 2020 [50] | R | MB, EP, PA | 288 (111 MB, 70 EP, 107 PA) | CE-T1WI, T2WI, DWI, ADC maps | S, 3D | CE and non-CE tumor and peritumoral edema | Matlab | 3087 (shape, first-order, texture) | multiclass classification: TPOT; binary classification: 13 different feature-selection methods | multiclass classification: TPOT; binary classification: 10 machine learning classifiers | 5-fold CV | yes | AUC = 0.84–0.94 | Automatic machine learning based on routine MRI classified pediatric posterior fossa tumors with high accuracy compared with manual expert pipeline optimization and qualitative expert MRI review. |
| Csutak et al., 2020 [51] | R | GLIOMAS, MET | 42 (16 HGGs, 26 MET) | T2WI | S, 3D | peritumoral region | MaZda | NS | Fisher, POE + ACC | univariate analysis (t-test), ROC analysis, multiple regression | no | no | 75–87.5% sen, 53.85–88.46% spec (univariate); 100% sens, 66.7% spec (multivariate) | Texture analysis can provide a quantitative description of the peritumoral zone encountered in solitary brain tumors. |
| Xia et al., 2021 [52] | R | GBM, PCNSL | 289 (136 PCNSL, 153 GBM) | CE-T1WI, FLAIR, ADC | M, 3D | whole tumor | PyRadiomics | 851 (NS) | mRMR, LASSO | Logistic regression | 5-fold CV | yes | AUC = 0.865 | A CNN model can differentiate PCNSL from GBM without tumor delineation, and comparable to the radiomics models and radiologists |
| Bathla et al., 2021 [53] | R | GBM, PCNSL | 94 (34 PCNSL, 60 GBM) | CE-T1WI, FLAIR, ADC | S, 3D | CE tumor and surrounding edema | PyRadiomics | 107 (shape, first-order, texture) | linear combination filter, high correlation filter, PCA | 12 classifiers (linear, non-linear, and ensemble) | 5-fold repeated CV | no | AUC = 0.98 | Radiomics-based diagnostic performance of various machine learning models for differentiating glioblastoma and PCNSL varies considerably. |
| Priya et al., 2021 [26] | R | GBM, PCNSL, MET | 253 (120 MET, 40 PCNSL, 93 GBM | T1W, T2W, FLAIR, ADC map, T1-CE | S, 3D | whole tumor, CE tumor, necrosis, peritumoral edema | PyRadiomics 3.0 | 107 (shape, first-order, texture) | linear combination filter, a high correlation filter, PCA | 12 classifiers (linear, non-linear, and ensemble) | 5-fold repeated CV | no | AUC = 0.91 for mpMRI, AUC = 0.90 for T1-CE | T1-CE is the single best sequence with comparable performance to that of MP-MRI. |
| De Causans et al., 2021 [54] | R | GBM, MET | 143 (71 GBM, 72 BM) | post-contrast T1-CE | S, 3D | CE tumor and necrotic region | PyRadiomics 2.1.2 | 100 (shape, first-order, texture) | 9 feature scaling methods | 16 classifiers | stratified 5-fold CV | yes | AUC = 0.92 in the training CV set, AUC = 0.85 in the test set | The proposed diagnostic support system helps in differentiating solitary BM from GBM with high diagnosis performance and generalizability. |
| Zhang et al., 2021 [24] | R | GBM, MET | 100 (50 GBM, 50 MET) | CE-T1WI, T2WI, ADC, 18F-FDG PET | S, 3D | CE tumor and perifocal edema | PyRadiomics | 4424 (shape, first-order, texture, LoG, wavelet) | t-test, PCA | partial least squares | 5-fold CV | yes | AUC = 0.98 in TS and 0.93 in VS | An integrated radiomics model incorporating DWI and F-FDG PET improved performances of GBM/MET differentiation. |
| Han et al., 2021 [55] | R | GBM, MET | 350 (182 GBM, 168 MET) | CE-T1WI | M, 3D | CE tumor | PyRadiomics v3.0 | 841 (shape, first-order, texture) | CMIM, MR R, DISR, Fisher, relief, MCFS, RFS | LR, SVM, DT, RF | 5-fold CV | yes | AUC = 0.764 | The combination models incorporating the radiomics signature and clinical-radiological characteristics were superior to the clinical-radiological models in differentiating between GBM and MET. |
| Han et al., 2021 [12] | R | GLIOMA, INFLAMMATION | 57 (39 grade II glioma, 18 inflammation) | T1W and T2W | M, 3D | whole tumor | MATLAB 2014b | 45 (shape, global, texture) | two-sample t-test, LASSO | linear regression | 10-fold CV | yes | AUC = 0.98–0.988 in primary cohort and 0.950, 0.925 in validation cohort | The radiomics signature helps to differentiate inflammation from grade II glioma and improved performance compared with experienced radiologists. |
| Priya et al., 2021 [56] | R | GBM, MET | 120 (60 GBM, 60 MET) | T1W, T2W, FLAIR, ADC, CE-T1WI | S, 3D | CE tumor + necrosis, surrounding edema | PyRadiomics | 107 (shape, first-order, texture) | linear combinations filter, a high correlation filter, PCA | 20 different models grouped into: linear classifiers, non-linear classifiers, and ensemble classifiers | 5-fold CV | no | AUC = 0.951 | Radiomics based machine learning can classify GBM and IMD with excellent diagnostic performance. The performance of mpMRI and single FLAIR or combined T1-CE and FLAIR sequence is comparable. |
| Priya et al., 2021 [57] | R | PCNSL, GBM | 97 GBM and 46 PCNSL | T1W, T2W, FLAIR, ADC, CE-T1WI | M, 3D | CE tumor + necrosis | TexRAD | 72 (histogram first-order (LoG filtered)) | full-features, correlation, PCA | 12 models grouped into: linear classifiers, non-linear classifiers, and ensemble classifiers | 5-fold CV | no | LASSO model with correlation filter as selection method: AUC = 0.914 | T1-CE derived first-order texture analysis can differentiate between GBM and PCNSL with good diagnostic performance. |
| Sartoretti et al., 2021 [58] | R | GLIOMAS, MET | 48 (21 gliomas, 27 MET) | APTw | M; 3D | whole tumor | PyRadiomics | 110 (first-order features; shape features; texture features) | ICC, correlation-based (CfsSubsetEval by Weka) | Multilayer perceptron classifier, Random Forest | 10-fold CV | yes | AUC = 0.797 | The use of radiomics for APTw imaging is feasible and the differentiation of primary glial brain tumors from metastases is achievable with a high degree of accuracy. |
| Su et al., 2021 [59] | R | GBM, MET | 225 (157 GBM, 98 solitary brain MET) | CE-T1WI | M; 3D | CE tumor | AK software | 396 (first-order features; shape features; texture features) | ICC, Mmrmr, LASSO | logistic regression | 10-fold CV | yes | AUC of 0.82 and 0.81 in the training and validation cohort to distinguish between GBM and solitary brain MET | The radiomics model might be a useful supporting tool for the preoperative differentiation of GBM from solitary brain MET, which could aid pretreatment decision making. |
| Xiao et al., 2021 [60] | R | GBM, BRAIN ABSCESS | 118 (86 nGBM, 32 BRAIN AB) | CE-T1WI, T2 FLAIR | S, 3D | Peritumoral edema, tumor | PyRadiomics | 1004 (shape, first-order, texture, LoG, wavelet) | LASSO, PCA | logistic regression, RF | 5-fold CV with 1000 iterations | yes | AUC = 0.97 | The radiomic features combined with the peritumoral edema/tumor volume ratio provided the prediction model with the greatest diagnostic performance. |
| Bo et al., 2021 [61] | R | CYSTIC GLIOMA, BRAIN ABSCESS | 188 (102 BRAIN ABSCESS, 86 CYSTIC GLIOMA) | T1WI and T2WI | M, 3D | whole tumor | PyRadiomics | 1000 DTL + 105 radiomic (first-order features; shape features; texture features) | Spearman’s rank correlation, MI | LR, RFC, KNN, and SVM | nested 5-fold CV | yes | AUC = 0.86 in TS and 0.85 in VS | The combination of HCR and DTL features can lead to impressive performance for distinguishing brain abscess from GBM. |
| Marginean et al., 2022 [25] | R | HGGs, MET | 36 (HGGs, n = 17; MET, n = 19) | CT | S, 3D | Peritumoral zone | maZda | 275 (GLRLM, wavelet GLCM, histogram, absolute gradient, auto- regressive model) | POE + ACC and Fisher coefficients, Mann– Whitney | Univariate and multivariate regression analysis | no | no | AUC = 0.992 | The CT-based TA can be a useful tool for differentiating between HGG and MET. |
3.3. Radiomics for DDx of Glioma and PCNSL
3.4. Radiomics for DDx of Glioma and Metastases
3.5. Radiomics for DDx of Glioma and Other Brain Diseases
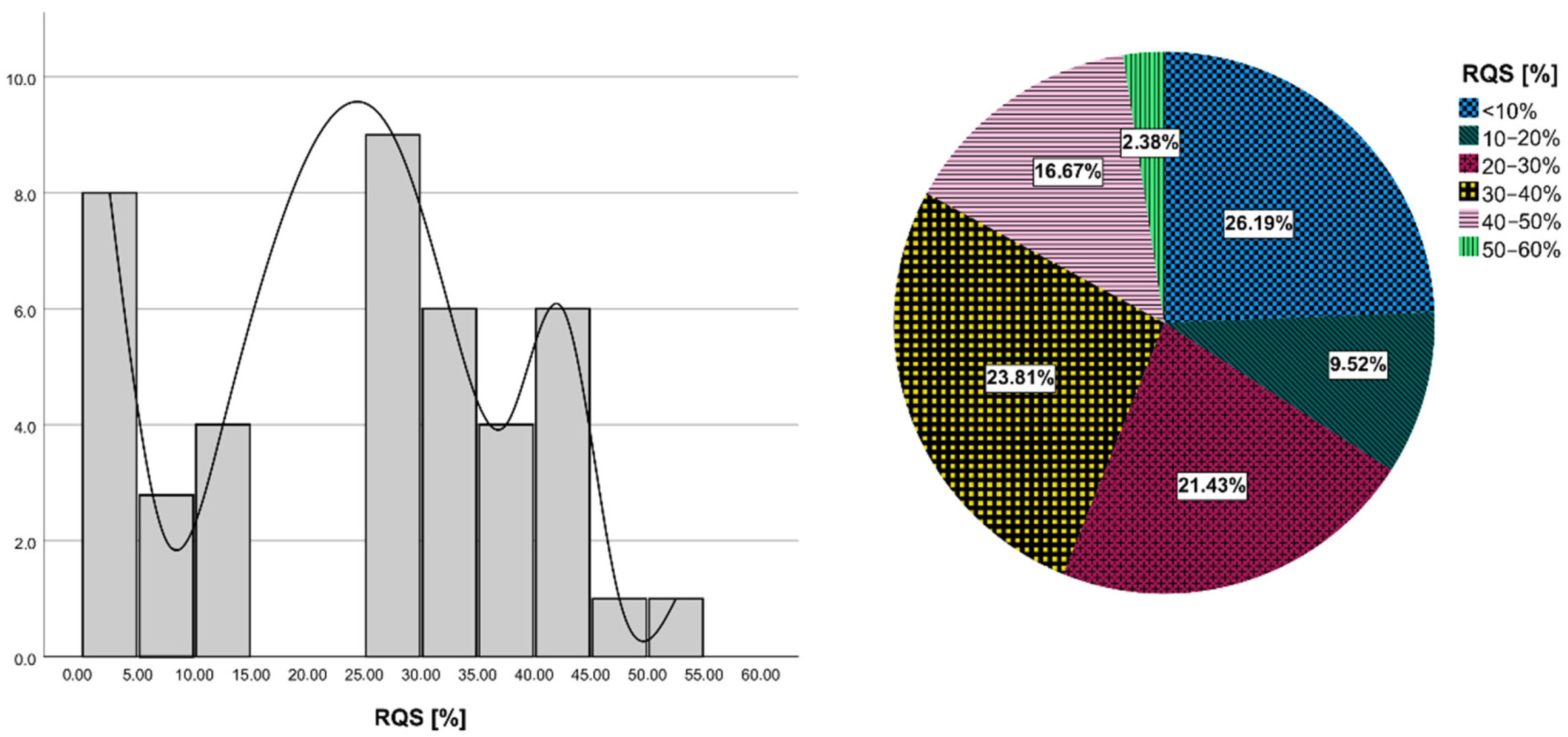
3.6. Quality Assessment with RQS
3.7. Statistical Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mesfin, F.B.; Al-Dhahir, M.A. Gliomas. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.-L.; Wei, X.-T. Advanced Diagnosis of Glioma by Using Emerging Magnetic Resonance Sequences. Front. Oncol. 2021, 11, 694498. [Google Scholar] [CrossRef]
- Gokden, M. If It Is Not a Glioblastoma, Then What Is It? A Differential Diagnostic Review. Adv. Anat. Pathol. 2017, 24, 379–391. [Google Scholar] [CrossRef]
- Jacobs, A.H.; Kracht, L.W.; Gossmann, A.; Rüger, M.A.; Thomas, A.V.; Thiel, A.; Herholz, K. Imaging in Neurooncology. Neurotherapeutics 2005, 2, 333–347. [Google Scholar] [CrossRef]
- Carrete, L.R.; Young, J.S.; Cha, S. Advanced Imaging Techniques for Newly Diagnosed and Recurrent Gliomas. Front. Neurosci. 2022, 16, 787755. [Google Scholar] [CrossRef]
- Brancato, V.; Nuzzo, S.; Tramontano, L.; Condorelli, G.; Salvatore, M.; Cavaliere, C. Predicting Survival in Glioblastoma Patients Using Diffusion MR Imaging Metrics—A Systematic Review. Cancers 2020, 12, 2858. [Google Scholar] [CrossRef]
- Overcast, W.B.; Davis, K.M.; Ho, C.Y.; Hutchins, G.D.; Green, M.A.; Graner, B.D.; Veronesi, M.C. Advanced Imaging Techniques for Neuro-Oncologic Tumor Diagnosis, with an Emphasis on PET-MRI Imaging of Malignant Brain Tumors. Curr. Oncol. Rep. 2021, 23, 34. [Google Scholar] [CrossRef]
- Deckert, M.; Brunn, A.; Montesinos-Rongen, M.; Terreni, M.R.; Ponzoni, M. Primary Lymphoma of the Central Nervous System—A Diagnostic Challenge. Hematol. Oncol. 2014, 32, 57–67. [Google Scholar] [CrossRef]
- Fordham, A.-J.; Hacherl, C.-C.; Patel, N.; Jones, K.; Myers, B.; Abraham, M.; Gendreau, J. Differentiating Glioblastomas from Solitary Brain Metastases: An Update on the Current Literature of Advanced Imaging Modalities. Cancers 2021, 13, 2960. [Google Scholar] [CrossRef]
- Han, Y.; Yang, Y.; Shi, Z.; Zhang, A.; Yan, L.; Hu, Y.; Feng, L.; Ma, J.; Wang, W.; Cui, G. Distinguishing Brain Inflammation from Grade II Glioma in Population without Contrast Enhancement: A Radiomics Analysis Based on Conventional MRI. Eur. J. Radiol. 2021, 134, 109467. [Google Scholar] [CrossRef] [PubMed]
- Jekel, L.; Brim, W.R.; von Reppert, M.; Staib, L.; Cassinelli Petersen, G.; Merkaj, S.; Subramanian, H.; Zeevi, T.; Payabvash, S.; Bousabarah, K.; et al. Machine Learning Applications for Differentiation of Glioma from Brain Metastasis—A Systematic Review. Cancers 2022, 14, 1369. [Google Scholar] [CrossRef] [PubMed]
- Bapuraj, J.R.; Wang, N.; Srinivasan, A.; Rao, A. Advanced Imaging and Computational Techniques for the Diagnostic and Prognostic Assessment of Malignant Gliomas. Cancer J. 2021, 27, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The Bridge between Medical Imaging and Personalized Medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting More Information from Medical Images Using Advanced Feature Analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rizzo, S.; Botta, F.; Raimondi, S.; Origgi, D.; Fanciullo, C.; Morganti, A.G.; Bellomi, M. Radiomics: The Facts and the Challenges of Image Analysis. Eur. Radiol. Exp. 2018, 2, 36. [Google Scholar] [CrossRef]
- van Timmeren, J.E.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baessler, B. Radiomics in Medical Imaging—“How-to” Guide and Critical Reflection. Insights Imaging 2020, 11, 91. [Google Scholar] [CrossRef]
- Lohmann, P.; Galldiks, N.; Kocher, M.; Heinzel, A.; Filss, C.P.; Stegmayr, C.; Mottaghy, F.M.; Fink, G.R.; Jon Shah, N.; Langen, K.-J. Radiomics in Neuro-Oncology: Basics, Workflow, and Applications. Methods 2021, 188, 112–121. [Google Scholar] [CrossRef]
- Ortiz-Ramón, R.; Ruiz-España, S.; Mollá-Olmos, E.; Moratal, D. Glioblastomas and Brain Metastases Differentiation Following an MRI Texture Analysis-Based Radiomics Approach. Phys. Med. 2020, 76, 44–54. [Google Scholar] [CrossRef]
- Kim, Y.; Cho, H.; Kim, S.T.; Park, H.; Nam, D.; Kong, D.-S. Radiomics Features to Distinguish Glioblastoma from Primary Central Nervous System Lymphoma on Multi-Parametric MRI. Neuroradiology 2018, 60, 1297–1305. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Kong, Z.; Jiang, C.; Zhu, R.; Feng, S.; Wang, Y.; Li, J.; Chen, W.; Liu, P.; Zhao, D.; Ma, W.; et al. 18F-FDG-PET-Based Radiomics Features to Distinguish Primary Central Nervous System Lymphoma from Glioblastoma. NeuroImage Clin. 2019, 23, 101912. [Google Scholar] [CrossRef]
- Zhang, L.; Yao, R.; Gao, J.; Tan, D.; Yang, X.; Wen, M.; Wang, J.; Xie, X.; Liao, R.; Tang, Y.; et al. An Integrated Radiomics Model Incorporating Diffusion-Weighted Imaging and 18F-FDG PET Imaging Improves the Performance of Differentiating Glioblastoma from Solitary Brain Metastases. Front. Oncol. 2021, 11, 732704. [Google Scholar] [CrossRef] [PubMed]
- Mărginean, L.; Ștefan, P.A.; Lebovici, A.; Opincariu, I.; Csutak, C.; Lupean, R.A.; Coroian, P.A.; Suciu, B.A. CT in the Differentiation of Gliomas from Brain Metastases: The Radiomics Analysis of the Peritumoral Zone. Brain Sci. 2022, 12, 109. [Google Scholar] [CrossRef] [PubMed]
- Priya, S.; Liu, Y.; Ward, C.; Le, N.H.; Soni, N.; Pillenahalli Maheshwarappa, R.; Monga, V.; Zhang, H.; Sonka, M.; Bathla, G. Radiomic Based Machine Learning Performance for a Three Class Problem in Neuro-Oncology: Time to Test the Waters? Cancers 2021, 13, 2568. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.S.; Lee, H.-J.; Ahn, S.S.; Chang, J.H.; Kang, S.-G.; Kim, E.H.; Kim, S.H.; Lee, S.-K. Primary Central Nervous System Lymphoma and Atypical Glioblastoma: Differentiation Using the Initial Area under the Curve Derived from Dynamic Contrast-Enhanced MR and the Apparent Diffusion Coefficient. Eur. Radiol. 2017, 27, 1344–1351. [Google Scholar] [CrossRef]
- Alcaide-Leon, P.; Dufort, P.; Geraldo, A.F.; Alshafai, L.; Maralani, P.J.; Spears, J.; Bharatha, A. Differentiation of Enhancing Glioma and Primary Central Nervous System Lymphoma by Texture-Based Machine Learning. AJNR Am. J. Neuroradiol. 2017, 38, 1145–1150. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Li, Z.; Wu, G.; Yu, J.; Wang, Y.; Lv, X.; Ju, X.; Chen, Z. Primary Central Nervous System Lymphoma and Glioblastoma Differentiation Based on Conventional Magnetic Resonance Imaging by High-Throughput SIFT Features. Int. J. Neurosci. 2018, 128, 608–618. [Google Scholar] [CrossRef]
- Wu, G.; Chen, Y.; Wang, Y.; Yu, J.; Lv, X.; Ju, X.; Shi, Z.; Chen, L.; Chen, Z. Sparse Representation-Based Radiomics for the Diagnosis of Brain Tumors. IEEE Trans. Med. Imaging 2018, 37, 893–905. [Google Scholar] [CrossRef]
- Artzi, M.; Bressler, I.; Ben Bashat, D. Differentiation between Glioblastoma, Brain Metastasis and Subtypes Using Radiomics Analysis: Radiomics Classification of Brain Tumors. J. Magn. Reson. Imaging 2019, 50, 519–528. [Google Scholar] [CrossRef]
- Kang, D.; Park, J.E.; Kim, Y.-H.; Kim, J.H.; Oh, J.Y.; Kim, J.; Kim, Y.; Kim, S.T.; Kim, H.S. Diffusion Radiomics as a Diagnostic Model for Atypical Manifestation of Primary Central Nervous System Lymphoma: Development and Multicenter External Validation. Neuro-Oncology 2018, 20, 1251–1261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunimatsu, A.; Kunimatsu, N.; Kamiya, K.; Watadani, T.; Mori, H.; Abe, O. Comparison between Glioblastoma and Primary Central Nervous System Lymphoma Using MR Image-Based Texture Analysis. MRMS 2018, 17, 50–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakagawa, M.; Nakaura, T.; Namimoto, T.; Kitajima, M.; Uetani, H.; Tateishi, M.; Oda, S.; Utsunomiya, D.; Makino, K.; Nakamura, H.; et al. Machine Learning Based on Multi-Parametric Magnetic Resonance Imaging to Differentiate Glioblastoma Multiforme from Primary Cerebral Nervous System Lymphoma. Eur. J. Radiol. 2018, 108, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Suh, H.B.; Choi, Y.S.; Bae, S.; Ahn, S.S.; Chang, J.H.; Kang, S.-G.; Kim, E.H.; Kim, S.H.; Lee, S.-K. Primary Central Nervous System Lymphoma and Atypical Glioblastoma: Differentiation Using Radiomics Approach. Eur. Radiol. 2018, 28, 3832–3839. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.-D.; Yan, P.-F.; Wang, Y.-X.; Osman, M.S.; Zhao, H.-Y. Glioblastoma and Primary Central Nervous System Lymphoma: Preoperative Differentiation by Using MRI-Based 3D Texture Analysis. Clin. Neurol. Neurosurg. 2018, 173, 84–90. [Google Scholar] [CrossRef]
- Bao, S.; Watanabe, Y.; Takahashi, H.; Tanaka, H.; Arisawa, A.; Matsuo, C.; Wu, R.; Fujimoto, Y.; Tomiyama, N. Differentiating between Glioblastoma and Primary CNS Lymphoma Using Combined Whole-Tumor Histogram Analysis of the Normalized Cerebral Blood Volume and the Apparent Diffusion Coefficient. MRMS 2019, 18, 53–61. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Ou, X.; Wang, J.; Guo, W.; Ma, X. Radiomics-Based Machine Learning in Differentiation Between Glioblastoma and Metastatic Brain Tumors. Front. Oncol. 2019, 9, 806. [Google Scholar] [CrossRef] [Green Version]
- Dong, F.; Li, Q.; Jiang, B.; Zhu, X.; Zeng, Q.; Huang, P.; Chen, S.; Zhang, M. Differentiation of Supratentorial Single Brain Metastasis and Glioblastoma by Using Peri-Enhancing Oedema Region–Derived Radiomic Features and Multiple Classifiers. Eur. Radiol. 2020, 30, 3015–3022. [Google Scholar] [CrossRef]
- Kunimatsu, A.; Kunimatsu, N.; Yasaka, K.; Akai, H.; Kamiya, K.; Watadani, T.; Mori, H.; Abe, O. Machine Learning-Based Texture Analysis of Contrast-Enhanced MR Imaging to Differentiate between Glioblastoma and Primary Central Nervous System Lymphoma. MRMS 2019, 18, 44–52. [Google Scholar] [CrossRef] [Green Version]
- Petrujkić, K.; Milošević, N.; Rajković, N.; Stanisavljević, D.; Gavrilović, S.; Dželebdžić, D.; Ilić, R.; Di Ieva, A.; Maksimović, R. Computational Quantitative MR Image Features—A Potential Useful Tool in Differentiating Glioblastoma from Solitary Brain Metastasis. Eur. J. Radiol. 2019, 119, 108634. [Google Scholar] [CrossRef]
- Qian, Z.; Li, Y.; Wang, Y.; Li, L.; Li, R.; Wang, K.; Li, S.; Tang, K.; Zhang, C.; Fan, X.; et al. Differentiation of Glioblastoma from Solitary Brain Metastases Using Radiomic Machine-Learning Classifiers. Cancer Lett. 2019, 451, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.-T.; Liu, M.-X.; Chen, Z.-Y. Differential Diagnostic Value of Texture Feature Analysis of Magnetic Resonance T2 Weighted Imaging between Glioblastoma and Primary Central Neural System Lymphoma. Chin. Med. Sci. J. 2019, 34, 10–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yun, J.; Park, J.E.; Lee, H.; Ham, S.; Kim, N.; Kim, H.S. Radiomic Features and Multilayer Perceptron Network Classifier: A Robust MRI Classification Strategy for Distinguishing Glioblastoma from Primary Central Nervous System Lymphoma. Sci. Rep. 2019, 9, 5746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bae, S.; An, C.; Ahn, S.S.; Kim, H.; Han, K.; Kim, S.W.; Park, J.E.; Kim, H.S.; Lee, S.-K. Robust Performance of Deep Learning for Distinguishing Glioblastoma from Single Brain Metastasis Using Radiomic Features: Model Development and Validation. Sci. Rep. 2020, 10, 12110. [Google Scholar] [CrossRef]
- Dastmalchian, S.; Kilinc, O.; Onyewadume, L.; Tippareddy, C.; McGivney, D.; Ma, D.; Griswold, M.; Sunshine, J.; Gulani, V.; Barnholtz-Sloan, J.S.; et al. Radiomic Analysis of Magnetic Resonance Fingerprinting in Adult Brain Tumors. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 683–693. [Google Scholar] [CrossRef]
- Chen, C.; Zheng, A.; Ou, X.; Wang, J.; Ma, X. Comparison of Radiomics-Based Machine-Learning Classifiers in Diagnosis of Glioblastoma from Primary Central Nervous System Lymphoma. Front. Oncol. 2020, 10, 1151. [Google Scholar] [CrossRef]
- Dong, J.; Li, L.; Liang, S.; Zhao, S.; Zhang, B.; Meng, Y.; Zhang, Y.; Li, S. Differentiation Between Ependymoma and Medulloblastoma in Children with Radiomics Approach. Acad. Radiol. 2021, 28, 318–327. [Google Scholar] [CrossRef]
- Xia, W.; Hu, B.; Li, H.; Geng, C.; Wu, Q.; Yang, L.; Yin, B.; Gao, X.; Li, Y.; Geng, D. Multiparametric-MRI-Based Radiomics Model for Differentiating Primary Central Nervous System Lymphoma from Glioblastoma: Development and Cross-Vendor Validation. J. Magn. Reson. Imaging 2021, 53, 242–250. [Google Scholar] [CrossRef]
- Zhou, H.; Hu, R.; Tang, O.; Hu, C.; Tang, L.; Chang, K.; Shen, Q.; Wu, J.; Zou, B.; Xiao, B.; et al. Automatic Machine Learning to Differentiate Pediatric Posterior Fossa Tumors on Routine MR Imaging. AJNR Am. J. Neuroradiol. 2020, 41, 1279–1285. [Google Scholar] [CrossRef]
- Csutak, C.; Ștefan, P.-A.; Lenghel, L.M.; Moroșanu, C.O.; Lupean, R.-A.; Șimonca, L.; Mihu, C.M.; Lebovici, A. Differentiating High-Grade Gliomas from Brain Metastases at Magnetic Resonance: The Role of Texture Analysis of the Peritumoral Zone. Brain Sci. 2020, 10, 638. [Google Scholar] [CrossRef]
- Xia, W.; Hu, B.; Li, H.; Shi, W.; Tang, Y.; Yu, Y.; Geng, C.; Wu, Q.; Yang, L.; Yu, Z.; et al. Deep Learning for Automatic Differential Diagnosis of Primary Central Nervous System Lymphoma and Glioblastoma: Multi-Parametric Magnetic Resonance Imaging Based Convolutional Neural Network Model. Magn. Reson. Imaging 2021, 54, 880–887. [Google Scholar] [CrossRef] [PubMed]
- Bathla, G.; Priya, S.; Liu, Y.; Ward, C.; Le, N.H.; Soni, N.; Maheshwarappa, R.P.; Monga, V.; Zhang, H.; Sonka, M. Radiomics-Based Differentiation between Glioblastoma and Primary Central Nervous System Lymphoma: A Comparison of Diagnostic Performance across Different MRI Sequences and Machine Learning Techniques. Eur. Radiol. 2021, 31, 8703–8713. [Google Scholar] [CrossRef] [PubMed]
- De Causans, A.; Carré, A.; Roux, A.; Tauziède-Espariat, A.; Ammari, S.; Dezamis, E.; Dhermain, F.; Reuzé, S.; Deutsch, E.; Oppenheim, C.; et al. Development of a Machine Learning Classifier Based on Radiomic Features Extracted from Post-Contrast 3D T1-Weighted MR Images to Distinguish Glioblastoma From Solitary Brain Metastasis. Front. Oncol. 2021, 11, 638262. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhang, L.; Niu, S.; Chen, S.; Yang, B.; Chen, H.; Zheng, F.; Zang, Y.; Zhang, H.; Xin, Y.; et al. Differentiation Between Glioblastoma Multiforme and Metastasis from the Lungs and Other Sites Using Combined Clinical/Routine MRI Radiomics. Front. Cell Dev. Biol. 2021, 9, 710461. [Google Scholar] [CrossRef]
- Priya, S.; Liu, Y.; Ward, C.; Le, N.H.; Soni, N.; Pillenahalli Maheshwarappa, R.; Monga, V.; Zhang, H.; Sonka, M.; Bathla, G. Machine Learning Based Differentiation of Glioblastoma from Brain Metastasis Using MRI Derived Radiomics. Sci. Rep. 2021, 11, 10478. [Google Scholar] [CrossRef]
- Priya, S.; Ward, C.; Locke, T.; Soni, N.; Maheshwarappa, R.P.; Monga, V.; Agarwal, A.; Bathla, G. Glioblastoma and Primary Central Nervous System Lymphoma: Differentiation Using MRI Derived First-Order Texture Analysis—A Machine Learning Study. Neuroradiol. J. 2021, 34, 320–328. [Google Scholar] [CrossRef]
- Sartoretti, E.; Sartoretti, T.; Wyss, M.; Reischauer, C.; van Smoorenburg, L.; Binkert, C.A.; Sartoretti-Schefer, S.; Mannil, M. Amide Proton Transfer Weighted (APTw) Imaging Based Radiomics Allows for the Differentiation of Gliomas from Metastases. Sci. Rep. 2021, 11, 5506. [Google Scholar] [CrossRef]
- Su, C.-Q.; Chen, X.-T.; Duan, S.-F.; Zhang, J.-X.; You, Y.-P.; Lu, S.-S.; Hong, X.-N. A Radiomics-Based Model to Differentiate Glioblastoma from Solitary Brain Metastases. Clin. Radiol. 2021, 76, 629.e11–629.e18. [Google Scholar] [CrossRef]
- Xiao, D.; Wang, J.; Wang, X.; Fu, P.; Zhao, H.; Yan, P.; Jiang, X. Distinguishing Brain Abscess from Necrotic Glioblastoma Using MRI-Based Intranodular Radiomic Features and Peritumoral Edema/Tumor Volume Ratio. J. Integr. Neurosci. 2021, 20, 623–634. [Google Scholar] [CrossRef]
- Bo, L.; Zhang, Z.; Jiang, Z.; Yang, C.; Huang, P.; Chen, T.; Wang, Y.; Yu, G.; Tan, X.; Cheng, Q.; et al. Differentiation of Brain Abscess from Cystic Glioma Using Conventional MRI Based on Deep Transfer Learning Features and Hand-Crafted Radiomics Features. Front. Med. 2021, 8, 748144. [Google Scholar] [CrossRef]
- Stanzione, A.; Gambardella, M.; Cuocolo, R.; Ponsiglione, A.; Romeo, V.; Imbriaco, M. Prostate MRI Radiomics: A Systematic Review and Radiomic Quality Score Assessment. Eur. J. Radiol. 2020, 129, 109095. [Google Scholar] [CrossRef] [PubMed]
- Granzier, R.W.Y.; van Nijnatten, T.J.A.; Woodruff, H.C.; Smidt, M.L.; Lobbes, M.B.I. Exploring Breast Cancer Response Prediction to Neoadjuvant Systemic Therapy Using MRI-Based Radiomics: A Systematic Review. Eur. J. Radiol. 2019, 121, 108736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ursprung, S.; Beer, L.; Bruining, A.; Woitek, R.; Stewart, G.D.; Gallagher, F.A.; Sala, E. Radiomics of Computed Tomography and Magnetic Resonance Imaging in Renal Cell Carcinoma—A Systematic Review and Meta-Analysis. Eur. Radiol. 2020, 30, 3558–3566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.E.; Kim, H.S.; Kim, D.; Park, S.Y.; Kim, J.Y.; Cho, S.J.; Kim, J.H. A Systematic Review Reporting Quality of Radiomics Research in Neuro-Oncology: Toward Clinical Utility and Quality Improvement Using High-Dimensional Imaging Features. BMC Cancer 2020, 20, 29. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Park, S.Y.; Kim, H.J.; Kim, H.S. Reproducibility and Generalizability in Radiomics Modeling: Possible Strategies in Radiologic and Statistical Perspectives. Korean J. Radiol. 2019, 20, 1124. [Google Scholar] [CrossRef] [PubMed]
- Vesteghem, C.; Brøndum, R.F.; Sønderkær, M.; Sommer, M.; Schmitz, A.; Bødker, J.S.; Dybkær, K.; El-Galaly, T.C.; Bøgsted, M. Implementing the FAIR Data Principles in Precision Oncology: Review of Supporting Initiatives. Brief. Bioinform. 2020, 21, 936–945. [Google Scholar] [CrossRef]
- Hasselbring, W.; Carr, L.; Hettrick, S.; Packer, H.; Tiropanis, T. From FAIR Research Data toward FAIR and Open Research Software. IT Inform. Technol. 2020, 62, 39–47. [Google Scholar] [CrossRef] [Green Version]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-Based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Zhou, Y.; Li, L.; Hou, W.; Ma, X.; Tian, R. Current Status and Quality of Radiomics Studies in Lymphoma: A Systematic Review. Eur. Radiol. 2020, 30, 6228–6240. [Google Scholar] [CrossRef]
- Sanduleanu, S.; Woodruff, H.C.; de Jong, E.E.C.; van Timmeren, J.E.; Jochems, A.; Dubois, L.; Lambin, P. Tracking Tumor Biology with Radiomics: A Systematic Review Utilizing a Radiomics Quality Score. Radiother. Oncol. 2018, 127, 349–360. [Google Scholar] [CrossRef]
- Norvell, D. Study Types and Bias—Don’t Judge a Study by the Abstract’s Conclusion Alone. Evid. Based Spine-Care J. 2010, 1, 7–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripepi, G.; Jager, K.J.; Dekker, F.W.; Zoccali, C. Selection Bias and Information Bias in Clinical Research. Nephron Clin. Pract. 2010, 115, c94–c99. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brancato, V.; Cerrone, M.; Lavitrano, M.; Salvatore, M.; Cavaliere, C. A Systematic Review of the Current Status and Quality of Radiomics for Glioma Differential Diagnosis. Cancers 2022, 14, 2731. https://doi.org/10.3390/cancers14112731
Brancato V, Cerrone M, Lavitrano M, Salvatore M, Cavaliere C. A Systematic Review of the Current Status and Quality of Radiomics for Glioma Differential Diagnosis. Cancers. 2022; 14(11):2731. https://doi.org/10.3390/cancers14112731
Chicago/Turabian StyleBrancato, Valentina, Marco Cerrone, Marialuisa Lavitrano, Marco Salvatore, and Carlo Cavaliere. 2022. "A Systematic Review of the Current Status and Quality of Radiomics for Glioma Differential Diagnosis" Cancers 14, no. 11: 2731. https://doi.org/10.3390/cancers14112731
APA StyleBrancato, V., Cerrone, M., Lavitrano, M., Salvatore, M., & Cavaliere, C. (2022). A Systematic Review of the Current Status and Quality of Radiomics for Glioma Differential Diagnosis. Cancers, 14(11), 2731. https://doi.org/10.3390/cancers14112731








