Prognostic Biomarkers in Uveal Melanoma: The Status Quo, Recent Advances and Future Directions
Abstract
Simple Summary
Abstract
1. Introduction
2. Uveal Melanoma: Relevant Epidemiological, Clinical and Pathological Features
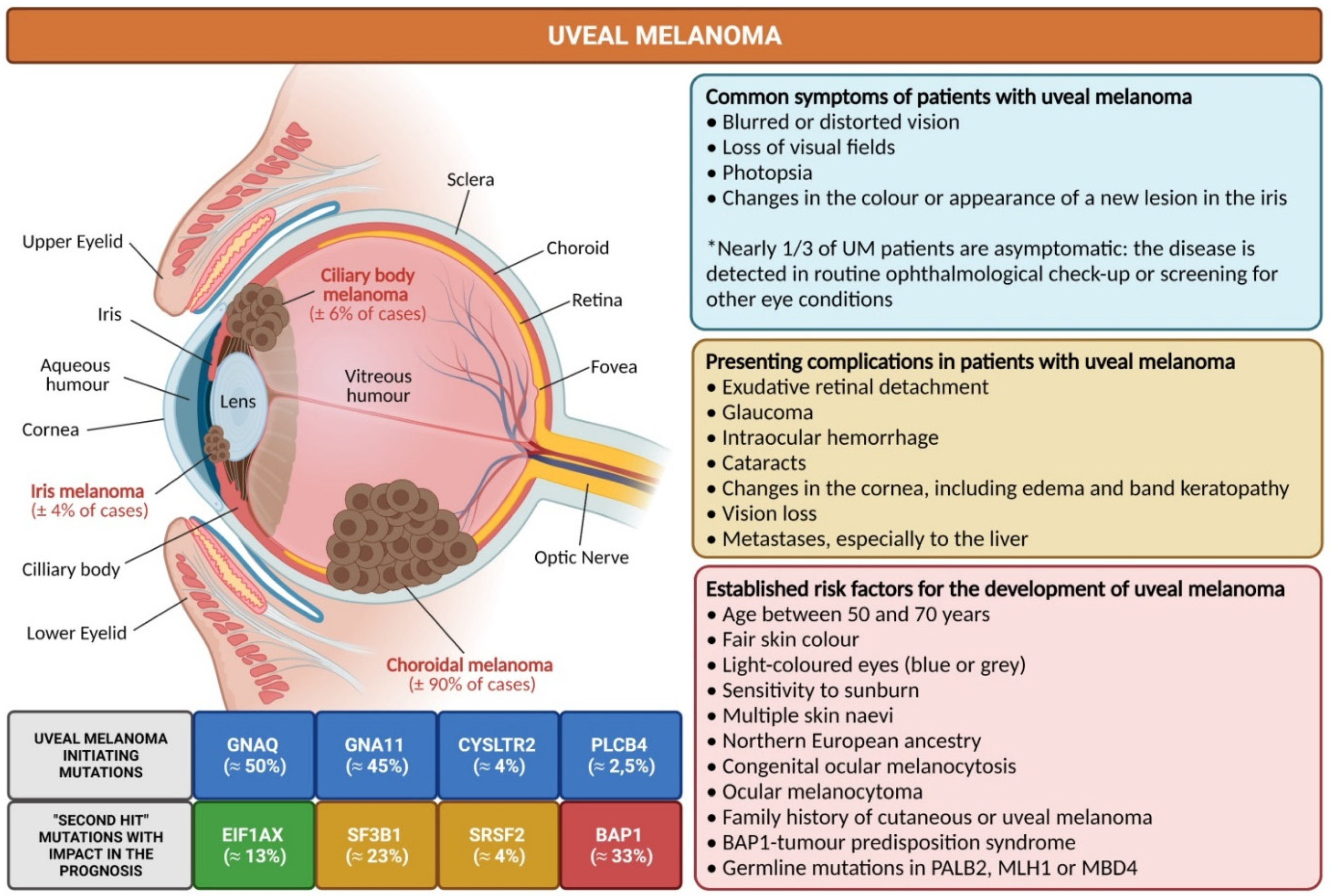
3. The Main Genetic Features of Uveal Melanoma
4. Current Well-Established Prognostic Biomarkers in Uveal Melanoma
| Factors Associated with a Worse Prognosis for Uveal Melanoma Patients |
|---|
| Higher age at diagnosis [77,78] |
| Male gender [17,79] |
| Ciliary body location and involvement [9,81,82] |
| Increased tumour size [Largest basal diameter (LBD) and tumour thickness (TT)] [73,82,85] |
| Epithelioid cell morphology [8,80,87] |
| Vascular invasion [83,88] |
| Extraocular spread [84,89] |
| Increased mitotic count [87,90,91] |
| Increased microvessel density [92,93] |
| Presence of tumour-infiltrating lymphocytes (TILs) [94,95,96] |
| Presence of tumour-infiltrating macrophages (TIMs) [94,95,96,97] |
| Presence of necrosis (in non-treated UM) [81] |
| Higher T stage (AJCC, TNM staging) [73,98] |
| Presence of uveal melanoma metastases [7,8,98] |
| Loss of nuclear BAP1 expression/BAP1 mutation [99,100,101,102] |
| PRAME expression [103,104,105,106] |
| Chromosomal abnormalities, especially M3, 8q gain, 6q loss and 1p loss [3,36,39,40,73,107,108,109] |
| Gene Expression Profiling (GEP) Class 2 [110,111,112,113] |
5. Gene Signatures as Novel Prognostic Biomarkers in Uveal Melanoma
6. Immunohistochemistry-Based Novel Prognostic Biomarkers in Uveal Melanoma
7. Additional Novel Promising Molecular Biomarkers in Uveal Melanoma with Prognostic Relevance
8. Current Challenges and Future Perspectives in Uveal Melanoma
8.1. Need for Accurate and Robust Models for Prognostication in Uveal Melanoma
8.2. The Promise of Liquid Biopsies for Uveal Melanoma
8.3. The Relevance of Non-Coding RNAs (ncRNAs) in Uveal Melanoma
8.4. Dissecting the Role of Tumour Infiltrating Immune Cells in Uveal Melanoma
8.5. Unravelling the Mystery of Preferential UM Metastization to the Liver
8.6. Novel In Vitro Cancer Models Will Likely Boost Research Efforts in Uveal Melanoma
8.7. The Need for Novel Effective Therapeutics for Metastatic Uveal Melanoma
9. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kaliki, S.; Shields, C.L. Uveal melanoma: Relatively rare but deadly cancer. Eye 2017, 31, 241–257. [Google Scholar] [CrossRef]
- Krantz, B.A.; Dave, N.; Komatsubara, K.M.; Marr, B.P.; Carvajal, R.D. Uveal melanoma: Epidemiology, etiology, and treatment of primary disease. Clin. Ophthalmol. 2017, 11, 279–289. [Google Scholar] [CrossRef]
- Shields, C.L.; Mayro, E.L.; Bas, Z.; Dockery, P.W.; Yaghy, A.; Lally, S.E.; Ganguly, A.; Shields, J.A. Ten-year outcomes of uveal melanoma based on The Cancer Genome Atlas (TCGA) classification in 1001 cases. Indian J. Ophthalmol. 2021, 69, 1839–1845. [Google Scholar] [CrossRef]
- Stalhammar, G. Forty-year prognosis after plaque brachytherapy of uveal melanoma. Sci. Rep. 2020, 10, 11297. [Google Scholar] [CrossRef] [PubMed]
- Rantala, E.S.; Hernberg, M.; Kivela, T.T. Overall survival after treatment for metastatic uveal melanoma: A systematic review and meta-analysis. Melanoma Res. 2019, 29, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Vidal, C.; Fernandez-Diaz, D.; Fernandez-Marta, B.; Lago-Baameiro, N.; Pardo, M.; Silva, P.; Paniagua, L.; Blanco-Teijeiro, M.J.; Pineiro, A.; Bande, M. Treatment of Metastatic Uveal Melanoma: Systematic Review. Cancers 2020, 12, 2557. [Google Scholar] [CrossRef] [PubMed]
- Lane, A.M.; Kim, I.K.; Gragoudas, E.S. Survival Rates in Patients After Treatment for Metastasis From Uveal Melanoma. JAMA Ophthalmol. 2018, 136, 981–986. [Google Scholar] [CrossRef] [PubMed]
- Kaliki, S.; Shields, C.L.; Shields, J.A. Uveal melanoma: Estimating prognosis. Indian J. Ophthalmol. 2015, 63, 93–102. [Google Scholar] [CrossRef]
- Jager, M.J.; Shields, C.L.; Cebulla, C.M.; Abdel-Rahman, M.H.; Grossniklaus, H.E.; Stern, M.H.; Carvajal, R.D.; Belfort, R.N.; Jia, R.; Shields, J.A.; et al. Uveal melanoma. Nat. Rev. Dis. Prim. 2020, 6, 24. [Google Scholar] [CrossRef]
- Carvajal, R.D.; Schwartz, G.K.; Tezel, T.; Marr, B.; Francis, J.H.; Nathan, P.D. Metastatic disease from uveal melanoma: Treatment options and future prospects. Br. J. Ophthalmol. 2017, 101, 38–44. [Google Scholar] [CrossRef]
- Dogrusoz, M.; Jager, M.J.; Damato, B. Uveal Melanoma Treatment and Prognostication. Asia-Pac. J. Ophthalmol. 2017, 6, 186–196. [Google Scholar] [CrossRef]
- Comito, F.; Marchese, P.V.; Ricci, A.D.; Tober, N.; Peterle, C.; Sperandi, F.; Melotti, B. Systemic and liver-directed therapies in metastatic uveal melanoma: State-of-the-art and novel perspectives. Future Oncol. 2021, 17, 4583–4606. [Google Scholar] [CrossRef]
- Bethlehem, M.S.; Katsarelias, D.; Olofsson Bagge, R. Meta-Analysis of Isolated Hepatic Perfusion and Percutaneous Hepatic Perfusion as a Treatment for Uveal Melanoma Liver Metastases. Cancers 2021, 13, 4726. [Google Scholar] [CrossRef] [PubMed]
- Rowcroft, A.; Loveday, B.P.T.; Thomson, B.N.J.; Banting, S.; Knowles, B. Systematic review of liver directed therapy for uveal melanoma hepatic metastases. HPB Off. J. Int. Hepato Pancreato Biliary Assoc. 2020, 22, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Kivela, T.T. The first description of the complete natural history of uveal melanoma by two Scottish surgeons, Allan Burns and James Wardrop. Acta Ophthalmol. 2018, 96, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Shields, C.L.; Furuta, M.; Thangappan, A.; Nagori, S.; Mashayekhi, A.; Lally, D.R.; Kelly, C.C.; Rudich, D.S.; Nagori, A.V.; Wakade, O.A.; et al. Metastasis of uveal melanoma millimeter-by-millimeter in 8033 consecutive eyes. Arch. Ophthalmol. 2009, 127, 989–998. [Google Scholar] [CrossRef] [PubMed]
- Alfaar, A.S.; Saad, A.; Wiedemann, P.; Rehak, M. The epidemiology of uveal melanoma in Germany: A nationwide report of incidence and survival between 2009 and 2015. Graefe’s Arch. Clin. Exp. Ophthalmol. 2021, 1–9. [Google Scholar] [CrossRef]
- Smidt-Nielsen, I.; Bagger, M.; Heegaard, S.; Andersen, K.K.; Kiilgaard, J.F. Posterior uveal melanoma incidence and survival by AJCC tumour size in a 70-year nationwide cohort. Acta Ophthalmol. 2021, 99, e1474–e1482. [Google Scholar] [CrossRef]
- Xu, Y.; Lou, L.; Wang, Y.; Miao, Q.; Jin, K.; Chen, M.; Ye, J. Epidemiological Study of Uveal Melanoma from US Surveillance, Epidemiology, and End Results Program (2010–2015). J. Ophthalmol. 2020, 2020, 3614039. [Google Scholar] [CrossRef]
- Mahendraraj, K.; Lau, C.S.; Lee, I.; Chamberlain, R.S. Trends in incidence, survival, and management of uveal melanoma: A population-based study of 7516 patients from the Surveillance, Epidemiology, and End Results database (1973–2012). Clin. Ophthalmol. 2016, 10, 2113–2119. [Google Scholar] [CrossRef]
- Shields, C.L.; Kaliki, S.; Furuta, M.; Fulco, E.; Alarcon, C.; Shields, J.A. American Joint Committee on Cancer classification of posterior uveal melanoma (tumor size category) predicts prognosis in 7731 patients. Ophthalmology 2013, 120, 2066–2071. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, C.; Kim, D.W.; Gombos, D.S.; Oba, J.; Qin, Y.; Williams, M.D.; Esmaeli, B.; Grimm, E.A.; Wargo, J.A.; Woodman, S.E.; et al. Uveal melanoma: From diagnosis to treatment and the science in between. Cancer 2016, 122, 2299–2312. [Google Scholar] [CrossRef] [PubMed]
- Jovanovic, P.; Mihajlovic, M.; Djordjevic-Jocic, J.; Vlajkovic, S.; Cekic, S.; Stefanovic, V. Ocular melanoma: An overview of the current status. Int. J. Clin. Exp. Pathol. 2013, 6, 1230–1244. [Google Scholar] [PubMed]
- Garg, G.; Finger, P.T.; Kivela, T.T.; Simpson, E.R.; Gallie, B.L.; Saakyan, S.; Amiryan, A.G.; Valskiy, V.; Chin, K.J.; Semenova, E.; et al. Patients presenting with metastases: Stage IV uveal melanoma, an international study. Br. J. Ophthalmol. 2021. [Google Scholar] [CrossRef]
- Derrien, A.C.; Rodrigues, M.; Eeckhoutte, A.; Dayot, S.; Houy, A.; Mobuchon, L.; Gardrat, S.; Lequin, D.; Ballet, S.; Pierron, G.; et al. Germline MBD4 Mutations and Predisposition to Uveal Melanoma. J. Natl. Cancer Inst. 2021, 113, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Rahman, M.H.; Sample, K.M.; Pilarski, R.; Walsh, T.; Grosel, T.; Kinnamon, D.; Boru, G.; Massengill, J.B.; Schoenfield, L.; Kelly, B.; et al. Whole Exome Sequencing Identifies Candidate Genes Associated with Hereditary Predisposition to Uveal Melanoma. Ophthalmology 2020, 127, 668–678. [Google Scholar] [CrossRef]
- Rodrigues, M.; Mobuchon, L.; Houy, A.; Fievet, A.; Gardrat, S.; Barnhill, R.L.; Popova, T.; Servois, V.; Rampanou, A.; Mouton, A.; et al. Outlier response to anti-PD1 in uveal melanoma reveals germline MBD4 mutations in hypermutated tumors. Nat. Commun. 2018, 9, 1866. [Google Scholar] [CrossRef] [PubMed]
- Johansson, P.A.; Brooks, K.; Newell, F.; Palmer, J.M.; Wilmott, J.S.; Pritchard, A.L.; Broit, N.; Wood, S.; Carlino, M.S.; Leonard, C.; et al. Whole genome landscapes of uveal melanoma show an ultraviolet radiation signature in iris tumours. Nat. Commun. 2020, 11, 2408. [Google Scholar] [CrossRef]
- Chalada, M.; Ramlogan-Steel, C.A.; Dhungel, B.P.; Layton, C.J.; Steel, J.C. The Impact of Ultraviolet Radiation on the Aetiology and Development of Uveal Melanoma. Cancers 2021, 13, 1700. [Google Scholar] [CrossRef]
- Karlsson, J.; Nilsson, L.M.; Mitra, S.; Alsen, S.; Shelke, G.V.; Sah, V.R.; Forsberg, E.M.V.; Stierner, U.; All-Eriksson, C.; Einarsdottir, B.; et al. Molecular profiling of driver events in metastatic uveal melanoma. Nat. Commun. 2020, 11, 1894. [Google Scholar] [CrossRef] [PubMed]
- Reichstein, D.A.; Brock, A.L. Radiation therapy for uveal melanoma: A review of treatment methods available in 2021. Curr. Opin. Ophthalmol. 2021, 32, 183–190. [Google Scholar] [CrossRef]
- Sikuade, M.J.; Salvi, S.; Rundle, P.A.; Errington, D.G.; Kacperek, A.; Rennie, I.G. Outcomes of treatment with stereotactic radiosurgery or proton beam therapy for choroidal melanoma. Eye 2015, 29, 1194–1198. [Google Scholar] [CrossRef]
- Papakostas, T.D.; Lane, A.M.; Morrison, M.; Gragoudas, E.S.; Kim, I.K. Long-term Outcomes After Proton Beam Irradiation in Patients With Large Choroidal Melanomas. JAMA Ophthalmol. 2017, 135, 1191–1196. [Google Scholar] [CrossRef]
- Van Beek, J.G.M.; Buitendijk, G.H.S.; Timman, R.; Muller, K.; Luyten, G.P.M.; Paridaens, D.; Naus, N.C.; Kilic, E. Quality of life: Fractionated stereotactic radiotherapy versus enucleation treatment in uveal melanoma patients. Acta Ophthalmol. 2018, 96, 841–848. [Google Scholar] [CrossRef]
- Marinkovic, M.; Horeweg, N.; Fiocco, M.; Peters, F.P.; Sommers, L.W.; Laman, M.S.; Bleeker, J.C.; Ketelaars, M.; Luyten, G.P.; Creutzberg, C.L. Ruthenium-106 brachytherapy for choroidal melanoma without transpupillary thermotherapy: Similar efficacy with improved visual outcome. Eur. J. Cancer 2016, 68, 106–113. [Google Scholar] [CrossRef]
- Bakhoum, M.F.; Esmaeli, B. Molecular Characteristics of Uveal Melanoma: Insights from the Cancer Genome Atlas (TCGA) Project. Cancers 2019, 11, 1061. [Google Scholar] [CrossRef] [PubMed]
- Carbone, M.; Ferris, L.K.; Baumann, F.; Napolitano, A.; Lum, C.A.; Flores, E.G.; Gaudino, G.; Powers, A.; Bryant-Greenwood, P.; Krausz, T.; et al. BAP1 cancer syndrome: Malignant mesothelioma, uveal and cutaneous melanoma, and MBAITs. J. Transl. Med. 2012, 10, 179. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Rahman, M.H.; Pilarski, R.; Cebulla, C.M.; Massengill, J.B.; Christopher, B.N.; Boru, G.; Hovland, P.; Davidorf, F.H. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J. Med. Genet. 2011, 48, 856–859. [Google Scholar] [CrossRef]
- Jager, M.J.; Brouwer, N.J.; Esmaeli, B. The Cancer Genome Atlas Project: An Integrated Molecular View of Uveal Melanoma. Ophthalmology 2018, 125, 1139–1142. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.G.; Shih, J.; Yau, C.; Gibb, E.A.; Oba, J.; Mungall, K.L.; Hess, J.M.; Uzunangelov, V.; Walter, V.; Danilova, L.; et al. Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma. Cancer Cell 2017, 32, 204–220.e15. [Google Scholar] [CrossRef] [PubMed]
- Diener-West, M.; Reynolds, S.M.; Agugliaro, D.J.; Caldwell, R.; Cumming, K.; Earle, J.D.; Hawkins, B.S.; Hayman, J.A.; Jaiyesimi, I.; Jampol, L.M.; et al. Development of metastatic disease after enrollment in the COMS trials for treatment of choroidal melanoma: Collaborative Ocular Melanoma Study Group Report No. 26. Arch. Ophthalmol. 2005, 123, 1639–1643. [Google Scholar] [CrossRef] [PubMed]
- Rajpal, S.; Moore, R.; Karakousis, C.P. Survival in metastatic ocular melanoma. Cancer 1983, 52, 334–336. [Google Scholar] [CrossRef]
- Kath, R.; Hayungs, J.; Bornfeld, N.; Sauerwein, W.; Hoffken, K.; Seeber, S. Prognosis and treatment of disseminated uveal melanoma. Cancer 1993, 72, 2219–2223. [Google Scholar] [CrossRef]
- Rietschel, P.; Panageas, K.S.; Hanlon, C.; Patel, A.; Abramson, D.H.; Chapman, P.B. Variates of survival in metastatic uveal melanoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2005, 23, 8076–8080. [Google Scholar] [CrossRef] [PubMed]
- Jochems, A.; van der Kooij, M.K.; Fiocco, M.; Schouwenburg, M.G.; Aarts, M.J.; van Akkooi, A.C.; van den Berkmortel, F.; Blank, C.U.; van den Eertwegh, A.J.M.; Franken, M.G.; et al. Metastatic Uveal Melanoma: Treatment Strategies and Survival-Results from the Dutch Melanoma Treatment Registry. Cancers 2019, 11, 1007. [Google Scholar] [CrossRef]
- Aronow, M.E.; Topham, A.K.; Singh, A.D. Uveal Melanoma: 5-Year Update on Incidence, Treatment, and Survival (SEER 1973–2013). Ocul. Oncol. Pathol. 2018, 4, 145–151. [Google Scholar] [CrossRef]
- Singh, A.D.; Turell, M.E.; Topham, A.K. Uveal melanoma: Trends in incidence, treatment, and survival. Ophthalmology 2011, 118, 1881–1885. [Google Scholar] [CrossRef]
- Roelofsen, C.D.M.; Wierenga, A.P.A.; van Duinen, S.; Verdijk, R.M.; Bleeker, J.; Marinkovic, M.; Luyten, G.P.M.; Jager, M.J. Five Decades of Enucleations for Uveal Melanoma in One Center: More Tumors with High Risk Factors, No Improvement in Survival over Time. Ocul. Oncol. Pathol. 2021, 7, 133–141. [Google Scholar] [CrossRef]
- Eskelin, S.; Pyrhonen, S.; Summanen, P.; Hahka-Kemppinen, M.; Kivela, T. Tumor doubling times in metastatic malignant melanoma of the uvea: Tumor progression before and after treatment. Ophthalmology 2000, 107, 1443–1449. [Google Scholar] [CrossRef]
- Park, S.Y.; Nam, J.S. The force awakens: Metastatic dormant cancer cells. Exp. Mol. Med. 2020, 52, 569–581. [Google Scholar] [CrossRef]
- Neophytou, C.M.; Kyriakou, T.C.; Papageorgis, P. Mechanisms of Metastatic Tumor Dormancy and Implications for Cancer Therapy. Int. J. Mol. Sci. 2019, 20, 6158. [Google Scholar] [CrossRef] [PubMed]
- Collaborative Ocular Melanoma Study Group. Assessment of metastatic disease status at death in 435 patients with large choroidal melanoma in the Collaborative Ocular Melanoma Study (COMS): COMS report no. 15. Arch. Ophthalmol. 2001, 119, 670–676. [Google Scholar] [CrossRef] [PubMed]
- Smit, K.N.; Jager, M.J.; de Klein, A.; Kili, E. Uveal melanoma: Towards a molecular understanding. Prog. Retin. Eye Res. 2020, 75, 100800. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.; Xie, F.; Mao, J.; Bai, Y.; Wang, X. Significance of Tumor Mutation Burden in Immune Infiltration and Prognosis in Cutaneous Melanoma. Front. Oncol. 2020, 10, 573141. [Google Scholar] [CrossRef] [PubMed]
- Heeke, S.; Hofman, P. Tumor mutational burden assessment as a predictive biomarker for immunotherapy in lung cancer patients: Getting ready for prime-time or not? Transl. Lung Cancer Res. 2018, 7, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas, N. Genomic Classification of Cutaneous Melanoma. Cell 2015, 161, 1681–1696. [Google Scholar] [CrossRef]
- Nahon-Esteve, S.; Martel, A.; Maschi, C.; Caujolle, J.P.; Baillif, S.; Lassalle, S.; Hofman, P. The Molecular Pathology of Eye Tumors: A 2019 Update Main Interests for Routine Clinical Practice. Curr. Mol. Med. 2019, 19, 632–664. [Google Scholar] [CrossRef] [PubMed]
- Shoushtari, A.N.; Carvajal, R.D. GNAQ and GNA11 mutations in uveal melanoma. Melanoma Res. 2014, 24, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Van Raamsdonk, C.D.; Griewank, K.G.; Crosby, M.B.; Garrido, M.C.; Vemula, S.; Wiesner, T.; Obenauf, A.C.; Wackernagel, W.; Green, G.; Bouvier, N.; et al. Mutations in GNA11 in uveal melanoma. N. Engl. J. Med. 2010, 363, 2191–2199. [Google Scholar] [CrossRef]
- Van Raamsdonk, C.D.; Bezrookove, V.; Green, G.; Bauer, J.; Gaugler, L.; O’Brien, J.M.; Simpson, E.M.; Barsh, G.S.; Bastian, B.C. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 2009, 457, 599–602. [Google Scholar] [CrossRef]
- Moore, A.R.; Ceraudo, E.; Sher, J.J.; Guan, Y.; Shoushtari, A.N.; Chang, M.T.; Zhang, J.Q.; Walczak, E.G.; Kazmi, M.A.; Taylor, B.S.; et al. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nat. Genet. 2016, 48, 675–680. [Google Scholar] [CrossRef]
- Nell, R.J.; Menger, N.V.; Versluis, M.; Luyten, G.P.M.; Verdijk, R.M.; Madigan, M.C.; Jager, M.J.; van der Velden, P.A. Involvement of mutant and wild-type CYSLTR2 in the development and progression of uveal nevi and melanoma. BMC Cancer 2021, 21, 164. [Google Scholar] [CrossRef]
- Johansson, P.; Aoude, L.G.; Wadt, K.; Glasson, W.J.; Warrier, S.K.; Hewitt, A.W.; Kiilgaard, J.F.; Heegaard, S.; Isaacs, T.; Franchina, M.; et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget 2016, 7, 4624–4631. [Google Scholar] [CrossRef] [PubMed]
- Koopmans, A.E.; Vaarwater, J.; Paridaens, D.; Naus, N.C.; Kilic, E.; de Klein, A.; Rotterdam Ocular Melanoma Study Group. Patient survival in uveal melanoma is not affected by oncogenic mutations in GNAQ and GNA11. Br. J. Cancer 2013, 109, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Weng, L.; Bastian, B.C.; Chen, X. Functional characterization of uveal melanoma oncogenes. Oncogene 2021, 40, 806–820. [Google Scholar] [CrossRef]
- Vader, M.J.C.; Madigan, M.C.; Versluis, M.; Suleiman, H.M.; Gezgin, G.; Gruis, N.A.; Out-Luiting, J.J.; Bergman, W.; Verdijk, R.M.; Jager, M.J.; et al. GNAQ and GNA11 mutations and downstream YAP activation in choroidal nevi. Br. J. Cancer 2017, 117, 884–887. [Google Scholar] [CrossRef] [PubMed]
- Decatur, C.L.; Ong, E.; Garg, N.; Anbunathan, H.; Bowcock, A.M.; Field, M.G.; Harbour, J.W. Driver Mutations in Uveal Melanoma: Associations With Gene Expression Profile and Patient Outcomes. JAMA Ophthalmol. 2016, 134, 728–733. [Google Scholar] [CrossRef]
- Rodrigues, M.; Ait Rais, K.; Salviat, F.; Algret, N.; Simaga, F.; Barnhill, R.; Gardrat, S.; Servois, V.; Mariani, P.; Piperno-Neumann, S.; et al. Association of Partial Chromosome 3 Deletion in Uveal Melanomas With Metastasis-Free Survival. JAMA Ophthalmol. 2020, 138, 182–188. [Google Scholar] [CrossRef]
- Martin, M.; Masshofer, L.; Temming, P.; Rahmann, S.; Metz, C.; Bornfeld, N.; van de Nes, J.; Klein-Hitpass, L.; Hinnebusch, A.G.; Horsthemke, B.; et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat. Genet. 2013, 45, 933–936. [Google Scholar] [CrossRef]
- Nguyen, J.Q.N.; Drabarek, W.; Yavuzyigitoglu, S.; Medico Salsench, E.; Verdijk, R.M.; Naus, N.C.; de Klein, A.; Kilic, E.; Brosens, E. Spliceosome Mutations in Uveal Melanoma. Int. J. Mol. Sci. 2020, 21, 9546. [Google Scholar] [CrossRef]
- Yavuzyigitoglu, S.; Koopmans, A.E.; Verdijk, R.M.; Vaarwater, J.; Eussen, B.; van Bodegom, A.; Paridaens, D.; Kilic, E.; de Klein, A.; Rotterdam Ocular Melanoma Study, G. Uveal Melanomas with SF3B1 Mutations: A Distinct Subclass Associated with Late-Onset Metastases. Ophthalmology 2016, 123, 1118–1128. [Google Scholar] [CrossRef]
- Van Poppelen, N.M.; Drabarek, W.; Smit, K.N.; Vaarwater, J.; Brands, T.; Paridaens, D.; Kilic, E.; de Klein, A. SRSF2 Mutations in Uveal Melanoma: A Preference for In-Frame Deletions? Cancers 2019, 11, 1200. [Google Scholar] [CrossRef]
- Shields, C.L.; Dalvin, L.A.; Vichitvejpaisal, P.; Mazloumi, M.; Ganguly, A.; Shields, J.A. Prognostication of uveal melanoma is simple and highly predictive using The Cancer Genome Atlas (TCGA) classification: A review. Indian J. Ophthalmol. 2019, 67, 1959–1963. [Google Scholar] [CrossRef] [PubMed]
- Correa, Z.M. Assessing Prognosis in Uveal Melanoma. Cancer Control. J. Moffitt Cancer Cent. 2016, 23, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Sussman, T.A.; Funchain, P.; Singh, A. Clinical Trials in Metastatic Uveal Melanoma: Current Status. Ocul. Oncol. Pathol. 2020, 6, 381–387. [Google Scholar] [CrossRef]
- Beran, T.M.; McCannel, T.A.; Stanton, A.L.; Straatsma, B.R.; Burgess, B.L. Reactions to and desire for prognostic testing in choroidal melanoma patients. J. Genet. Couns. 2009, 18, 265–274. [Google Scholar] [CrossRef]
- Kaliki, S.; Shields, C.L.; Mashayekhi, A.; Ganesh, A.; Furuta, M.; Shields, J.A. Influence of age on prognosis of young patients with uveal melanoma: A matched retrospective cohort study. Eur. J. Ophthalmol. 2013, 23, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Shields, C.L.; Kaliki, S.; Furuta, M.; Mashayekhi, A.; Shields, J.A. Clinical spectrum and prognosis of uveal melanoma based on age at presentation in 8033 cases. Retina 2012, 32, 1363–1372. [Google Scholar] [CrossRef] [PubMed]
- Zloto, O.; Pe’er, J.; Frenkel, S. Gender differences in clinical presentation and prognosis of uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2013, 54, 652–656. [Google Scholar] [CrossRef] [PubMed]
- McLean, I.W.; Foster, W.D.; Zimmerman, L.E. Uveal melanoma: Location, size, cell type, and enucleation as risk factors in metastasis. Hum. Pathol. 1982, 13, 123–132. [Google Scholar] [CrossRef]
- Albert, D.M.; Diener-West, M.; Robinson, N.L.; Grossniklaus, H.E.; Green, W.R.; Vine, A.K.; Willis, J.; Frueh, B.; Kurtz, R.M.; Elner, S.; et al. Histopathologic characteristics of uveal melanomas in eyes enucleated from the Collaborative Ocular Melanoma Study COMS report no. 6. Am. J. Ophthalmol. 1998, 125, 745–766. [Google Scholar] [CrossRef]
- Singh, M.; Durairaj, P.; Yeung, J. Uveal Melanoma: A Review of the Literature. Oncol. Ther. 2018, 6, 87–104. [Google Scholar] [CrossRef] [PubMed]
- Barnhill, R.L.; Ye, M.; Batistella, A.; Stern, M.H.; Roman-Roman, S.; Dendale, R.; Lantz, O.; Piperno-Neumann, S.; Desjardins, L.; Cassoux, N.; et al. The biological and prognostic significance of angiotropism in uveal melanoma. Lab. Investig. 2017, 97, 746–759. [Google Scholar] [CrossRef] [PubMed]
- Coupland, S.E.; Campbell, I.; Damato, B. Routes of extraocular extension of uveal melanoma: Risk factors and influence on survival probability. Ophthalmology 2008, 115, 1778–1785. [Google Scholar] [CrossRef]
- Force, A.O.O.T. International Validation of the American Joint Committee on Cancer’s 7th Edition Classification of Uveal Melanoma. JAMA Ophthalmol. 2015, 133, 376–383. [Google Scholar] [CrossRef]
- Coleman, K.; Baak, J.P.; Van Diest, P.; Mullaney, J.; Farrell, M.; Fenton, M. Prognostic factors following enucleation of 111 uveal melanomas. Br. J. Ophthalmol. 1993, 77, 688–692. [Google Scholar] [CrossRef][Green Version]
- Damato, B.; Dopierala, J.A.; Coupland, S.E. Genotypic profiling of 452 choroidal melanomas with multiplex ligation-dependent probe amplification. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2010, 16, 6083–6092. [Google Scholar] [CrossRef]
- Ly, L.V.; Odish, O.F.; Wolff-Rouendaal, D.; Missotten, G.S.; Luyten, G.P.; Jager, M.J. Intravascular presence of tumor cells as prognostic parameter in uveal melanoma: A 35-year survey. Investig. Ophthalmol. Vis. Sci. 2010, 51, 658–665. [Google Scholar] [CrossRef]
- Van Beek, J.G.; Koopmans, A.E.; Vaarwater, J.; de Rooi, J.J.; Paridaens, D.; Naus, N.C.; de Klein, A.; Verdijk, R.M.; Kilic, E. The prognostic value of extraocular extension in relation to monosomy 3 and gain of chromosome 8q in uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2014, 55, 1284–1291. [Google Scholar] [CrossRef]
- Lattman, J.; Kroll, S.; Char, D.H.; Ghazvini, S.; Frigillana, H.; O’Brien, J.M.; Elbakri, H.R. Cell cycling and prognosis in uveal melanoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 1995, 1, 41–47. [Google Scholar]
- Angi, M.; Damato, B.; Kalirai, H.; Dodson, A.; Taktak, A.; Coupland, S.E. Immunohistochemical assessment of mitotic count in uveal melanoma. Acta Ophthalmol. 2011, 89, e155–e160. [Google Scholar] [CrossRef]
- Foss, A.J.; Alexander, R.A.; Jefferies, L.W.; Hungerford, J.L.; Harris, A.L.; Lightman, S. Microvessel count predicts survival in uveal melanoma. Cancer Res. 1996, 56, 2900–2903. [Google Scholar] [PubMed]
- Makitie, T.; Summanen, P.; Tarkkanen, A.; Kivela, T. Microvascular density in predicting survival of patients with choroidal and ciliary body melanoma. Investig. Ophthalmol. Vis. Sci. 1999, 40, 2471–2480. [Google Scholar]
- De la Cruz Jr, P.O.; Specht, C.S.; McLean, I.W. Lymphocytic infiltration in uveal malignant melanoma. Cancer 1990, 65, 112–115. [Google Scholar] [CrossRef]
- Whelchel, J.C.; Farah, S.E.; McLean, I.W.; Burnier, M.N. Immunohistochemistry of infiltrating lymphocytes in uveal malignant melanoma. Investig. Ophthalmol. Vis. Sci. 1993, 34, 2603–2606. [Google Scholar]
- Makitie, T.; Summanen, P.; Tarkkanen, A.; Kivela, T. Tumor-infiltrating macrophages (CD68(+) cells) and prognosis in malignant uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2001, 42, 1414–1421. [Google Scholar]
- Bronkhorst, I.H.; Ly, L.V.; Jordanova, E.S.; Vrolijk, J.; Versluis, M.; Luyten, G.P.; Jager, M.J. Detection of M2-macrophages in uveal melanoma and relation with survival. Investig. Ophthalmol. Vis. Sci. 2011, 52, 643–650. [Google Scholar] [CrossRef]
- Keung, E.Z.; Gershenwald, J.E. The eighth edition American Joint Committee on Cancer (AJCC) melanoma staging system: Implications for melanoma treatment and care. Expert Rev. Anticancer. Ther. 2018, 18, 775–784. [Google Scholar] [CrossRef]
- Shah, A.A.; Bourne, T.D.; Murali, R. BAP1 protein loss by immunohistochemistry: A potentially useful tool for prognostic prediction in patients with uveal melanoma. Pathology 2013, 45, 651–656. [Google Scholar] [CrossRef]
- Koopmans, A.E.; Verdijk, R.M.; Brouwer, R.W.; van den Bosch, T.P.; van den Berg, M.M.; Vaarwater, J.; Kockx, C.E.; Paridaens, D.; Naus, N.C.; Nellist, M.; et al. Clinical significance of immunohistochemistry for detection of BAP1 mutations in uveal melanoma. Mod. Pathol. Off. J. United States Can. Acad. Pathol. Inc 2014, 27, 1321–1330. [Google Scholar] [CrossRef]
- Kalirai, H.; Dodson, A.; Faqir, S.; Damato, B.E.; Coupland, S.E. Lack of BAP1 protein expression in uveal melanoma is associated with increased metastatic risk and has utility in routine prognostic testing. Br. J. Cancer 2014, 111, 1373–1380. [Google Scholar] [CrossRef]
- Smit, K.N.; van Poppelen, N.M.; Vaarwater, J.; Verdijk, R.; van Marion, R.; Kalirai, H.; Coupland, S.E.; Thornton, S.; Farquhar, N.; Dubbink, H.J.; et al. Combined mutation and copy-number variation detection by targeted next-generation sequencing in uveal melanoma. Mod. Pathol. Off. J. United States Can. Acad. Pathol. Inc 2018, 31, 763–771. [Google Scholar] [CrossRef]
- Field, M.G.; Decatur, C.L.; Kurtenbach, S.; Gezgin, G.; van der Velden, P.A.; Jager, M.J.; Kozak, K.N.; Harbour, J.W. PRAME as an Independent Biomarker for Metastasis in Uveal Melanoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 1234–1242. [Google Scholar] [CrossRef]
- Field, M.G.; Durante, M.A.; Decatur, C.L.; Tarlan, B.; Oelschlager, K.M.; Stone, J.F.; Kuznetsov, J.; Bowcock, A.M.; Kurtenbach, S.; Harbour, J.W. Epigenetic reprogramming and aberrant expression of PRAME are associated with increased metastatic risk in Class 1 and Class 2 uveal melanomas. Oncotarget 2016, 7, 59209–59219. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Paez-Escamilla, M.; Walter, S.D.; Tarlan, B.; Decatur, C.L.; Perez, B.M.; Harbour, J.W. Gene Expression Profiling and PRAME Status Versus Tumor-Node-Metastasis Staging for Prognostication in Uveal Melanoma. Am. J. Ophthalmol. 2018, 195, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Schefler, A.C.; Koca, E.; Bernicker, E.H.; Correa, Z.M. Relationship between clinical features, GEP class, and PRAME expression in uveal melanoma. Graefe’s Arch. Clin. Exp. Ophthalmol. 2019, 257, 1541–1545. [Google Scholar] [CrossRef]
- Prescher, G.; Bornfeld, N.; Horsthemke, B.; Becher, R. Chromosomal aberrations defining uveal melanoma of poor prognosis. Lancet 1992, 339, 691–692. [Google Scholar] [CrossRef]
- Horsman, D.E.; Sroka, H.; Rootman, J.; White, V.A. Monosomy 3 and isochromosome 8q in a uveal melanoma. Cancer Genet. Cytogenet. 1990, 45, 249–253. [Google Scholar] [CrossRef]
- Prescher, G.; Bornfeld, N.; Becher, R. Nonrandom chromosomal abnormalities in primary uveal melanoma. J. Natl. Cancer Inst. 1990, 82, 1765–1769. [Google Scholar] [CrossRef]
- Onken, M.D.; Worley, L.A.; Ehlers, J.P.; Harbour, J.W. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004, 64, 7205–7209. [Google Scholar] [CrossRef]
- Onken, M.D.; Worley, L.A.; Tuscan, M.D.; Harbour, J.W. An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J. Mol. Diagn. JMD 2010, 12, 461–468. [Google Scholar] [CrossRef]
- Onken, M.D.; Worley, L.A.; Char, D.H.; Augsburger, J.J.; Correa, Z.M.; Nudleman, E.; Aaberg, T.M., Jr.; Altaweel, M.M.; Bardenstein, D.S.; Finger, P.T.; et al. Collaborative Ocular Oncology Group report number 1: Prospective validation of a multi-gene prognostic assay in uveal melanoma. Ophthalmology 2012, 119, 1596–1603. [Google Scholar] [CrossRef] [PubMed]
- Aaberg, T.M.; Covington, K.R.; Tsai, T.; Shildkrot, Y.; Plasseraud, K.M.; Alsina, K.M.; Oelschlager, K.M.; Monzon, F.A. Gene Expression Profiling in Uveal Melanoma: Five-Year Prospective Outcomes and Meta-Analysis. Ocul. Oncol. Pathol. 2020, 6, 360–367. [Google Scholar] [CrossRef]
- Kashyap, S.; Meel, R.; Singh, L.; Singh, M. Uveal melanoma. Semin. Diagn. Pathol. 2016, 33, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Onken, M.D.; Blumer, K.J.; Cooper, J.A. Uveal melanoma cells use ameboid and mesenchymal mechanisms of cell motility crossing the endothelium. Mol. Biol. Cell 2021, 32, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Dithmar, S.; Diaz, C.E.; Grossniklaus, H.E. Intraocular melanoma spread to regional lymph nodes: Report of two cases. Retina 2000, 20, 76–79. [Google Scholar] [CrossRef]
- Van Beek, J.G.M.; van den Bosch, Q.C.C.; Naus, N.; Paridaens, D.; de Klein, A.; Kilic, E.; Verdijk, R.M. Absence of Intraocular Lymphatic Vessels in Uveal Melanomas with Extrascleral Growth. Cancers 2019, 11, 228. [Google Scholar] [CrossRef]
- Brouwer, N.J.; Gezgin, G.; Wierenga, A.P.A.; Bronkhorst, I.H.G.; Marinkovic, M.; Luyten, G.P.M.; Versluis, M.; Kroes, W.G.M.; van der Velden, P.A.; Verdijk, R.M.; et al. Tumour Angiogenesis in Uveal Melanoma Is Related to Genetic Evolution. Cancers 2019, 11, 979. [Google Scholar] [CrossRef]
- Taylor, A.W. Ocular immune privilege. Eye 2009, 23, 1885–1889. [Google Scholar] [CrossRef]
- Taylor, A.W. Ocular Immune Privilege and Transplantation. Front. Immunol. 2016, 7, 37. [Google Scholar] [CrossRef]
- Kuchle, M.; Nguyen, N.X.; Naumann, G.O. Quantitative assessment of the blood-aqueous barrier in human eyes with malignant or benign uveal tumors. Am. J. Ophthalmol. 1994, 117, 521–528. [Google Scholar] [CrossRef]
- Bronkhorst, I.H.; Jager, M.J. Inflammation in uveal melanoma. Eye 2013, 27, 217–223. [Google Scholar] [CrossRef]
- Wessely, A.; Steeb, T.; Erdmann, M.; Heinzerling, L.; Vera, J.; Schlaak, M.; Berking, C.; Heppt, M.V. The Role of Immune Checkpoint Blockade in Uveal Melanoma. Int. J. Mol. Sci. 2020, 21, 879. [Google Scholar] [CrossRef] [PubMed]
- Zane, K.E.; Cloyd, J.M.; Mumtaz, K.S.; Wadhwa, V.; Makary, M.S. Metastatic disease to the liver: Locoregional therapy strategies and outcomes. World J. Clin. Oncol. 2021, 12, 725–745. [Google Scholar] [CrossRef] [PubMed]
- Hoppener, D.J.; Grunhagen, D.J.; Eggermont, A.M.M.; van der Veldt, A.A.M.; Verhoef, C. An Overview of Liver Directed Locoregional Therapies. Surg. Oncol. Clin. 2021, 30, 103–123. [Google Scholar] [CrossRef] [PubMed]
- Bakhoum, M.F.; Curtis, E.J.; Goldbaum, M.H.; Mischel, P.S. BAP1 methylation: A prognostic marker of uveal melanoma metastasis. NPJ Precis. Oncol. 2021, 5, 89. [Google Scholar] [CrossRef] [PubMed]
- Uner, O.E.; See, T.R.O.; Szalai, E.; Grossniklaus, H.E.; Stalhammar, G. Estimation of the timing of BAP1 mutation in uveal melanoma progression. Sci. Rep. 2021, 11, 8923. [Google Scholar] [CrossRef]
- Ikeda, H.; Lethe, B.; Lehmann, F.; van Baren, N.; Baurain, J.F.; de Smet, C.; Chambost, H.; Vitale, M.; Moretta, A.; Boon, T.; et al. Characterization of an antigen that is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity 1997, 6, 199–208. [Google Scholar] [CrossRef]
- Lezcano, C.; Jungbluth, A.A.; Nehal, K.S.; Hollmann, T.J.; Busam, K.J. PRAME Expression in Melanocytic Tumors. Am. J. Surg. Pathol. 2018, 42, 1456–1465. [Google Scholar] [CrossRef]
- Sekoranja, D.; Hawlina, G.; Pizem, J. PRAME expression in melanocytic lesions of the conjunctiva. Histopathology 2021, 79, 989–996. [Google Scholar] [CrossRef]
- Epping, M.T.; Hart, A.A.; Glas, A.M.; Krijgsman, O.; Bernards, R. PRAME expression and clinical outcome of breast cancer. Br. J. Cancer 2008, 99, 398–403. [Google Scholar] [CrossRef]
- Szczepanski, M.J.; DeLeo, A.B.; Luczak, M.; Molinska-Glura, M.; Misiak, J.; Szarzynska, B.; Dworacki, G.; Zagor, M.; Rozwadowska, N.; Kurpisz, M.; et al. PRAME expression in head and neck cancer correlates with markers of poor prognosis and might help in selecting candidates for retinoid chemoprevention in pre-malignant lesions. Oral Oncol. 2013, 49, 144–151. [Google Scholar] [CrossRef]
- Oberthuer, A.; Hero, B.; Spitz, R.; Berthold, F.; Fischer, M. The tumor-associated antigen PRAME is universally expressed in high-stage neuroblastoma and associated with poor outcome. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2004, 10, 4307–4313. [Google Scholar] [CrossRef] [PubMed]
- Tan, P.; Zou, C.; Yong, B.; Han, J.; Zhang, L.; Su, Q.; Yin, J.; Wang, J.; Huang, G.; Peng, T.; et al. Expression and prognostic relevance of PRAME in primary osteosarcoma. Biochem. Biophys. Res. Commun. 2012, 419, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Prescher, G.; Bornfeld, N.; Hirche, H.; Horsthemke, B.; Jockel, K.H.; Becher, R. Prognostic implications of monosomy 3 in uveal melanoma. Lancet 1996, 347, 1222–1225. [Google Scholar] [CrossRef] [PubMed]
- Kilic, E.; Naus, N.C.; van Gils, W.; Klaver, C.C.; van Til, M.E.; Verbiest, M.M.; Stijnen, T.; Mooy, C.M.; Paridaens, D.; Beverloo, H.B.; et al. Concurrent loss of chromosome arm 1p and chromosome 3 predicts a decreased disease-free survival in uveal melanoma patients. Investig. Ophthalmol. Vis. Sci. 2005, 46, 2253–2257. [Google Scholar] [CrossRef] [PubMed]
- Hausler, T.; Stang, A.; Anastassiou, G.; Jockel, K.H.; Mrzyk, S.; Horsthemke, B.; Lohmann, D.R.; Zeschnigk, M. Loss of heterozygosity of 1p in uveal melanomas with monosomy 3. Int. J. Cancer 2005, 116, 909–913. [Google Scholar] [CrossRef]
- Dogrusoz, M.; Jager, M.J. Genetic prognostication in uveal melanoma. Acta Ophthalmol. 2018, 96, 331–347. [Google Scholar] [CrossRef]
- Honavar, S.G. Genetics of uveal melanoma—Breed is stronger than pasture. Indian J. Ophthalmol. 2021, 69, 1643–1644. [Google Scholar] [CrossRef]
- See, T.R.; Stalhammar, G.; Phillips, S.; Grossniklaus, H.E. BAP1 Immunoreactivity Correlates with Gene Expression Class in Uveal Melanoma. Ocul. Oncol. Pathol. 2020, 6, 129–137. [Google Scholar] [CrossRef]
- Petrausch, U.; Martus, P.; Tonnies, H.; Bechrakis, N.E.; Lenze, D.; Wansel, S.; Hummel, M.; Bornfeld, N.; Thiel, E.; Foerster, M.H.; et al. Significance of gene expression analysis in uveal melanoma in comparison to standard risk factors for risk assessment of subsequent metastases. Eye 2008, 22, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Correa, Z.M.; Augsburger, J.J. Independent Prognostic Significance of Gene Expression Profile Class and Largest Basal Diameter of Posterior Uveal Melanomas. Am. J. Ophthalmol. 2016, 162, 20–27.e1. [Google Scholar] [CrossRef] [PubMed]
- DeParis, S.W.; Taktak, A.; Eleuteri, A.; Enanoria, W.; Heimann, H.; Coupland, S.E.; Damato, B. External Validation of the Liverpool Uveal Melanoma Prognosticator Online. Investig. Ophthalmol. Vis. Sci. 2016, 57, 6116–6122. [Google Scholar] [CrossRef] [PubMed]
- Laurent, C.; Valet, F.; Planque, N.; Silveri, L.; Maacha, S.; Anezo, O.; Hupe, P.; Plancher, C.; Reyes, C.; Albaud, B.; et al. High PTP4A3 phosphatase expression correlates with metastatic risk in uveal melanoma patients. Cancer Res. 2011, 71, 666–674. [Google Scholar] [CrossRef] [PubMed]
- Gangemi, R.; Mirisola, V.; Barisione, G.; Fabbi, M.; Brizzolara, A.; Lanza, F.; Mosci, C.; Salvi, S.; Gualco, M.; Truini, M.; et al. Mda-9/syntenin is expressed in uveal melanoma and correlates with metastatic progression. PLoS ONE 2012, 7, e29989. [Google Scholar] [CrossRef] [PubMed]
- Worley, L.A.; Onken, M.D.; Person, E.; Robirds, D.; Branson, J.; Char, D.H.; Perry, A.; Harbour, J.W. Transcriptomic versus chromosomal prognostic markers and clinical outcome in uveal melanoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2007, 13, 1466–1471. [Google Scholar] [CrossRef]
- Van Essen, T.H.; van Pelt, S.I.; Bronkhorst, I.H.; Versluis, M.; Nemati, F.; Laurent, C.; Luyten, G.P.; van Hall, T.; van den Elsen, P.J.; van der Velden, P.A.; et al. Upregulation of HLA Expression in Primary Uveal Melanoma by Infiltrating Leukocytes. PLoS ONE 2016, 11, e0164292. [Google Scholar] [CrossRef]
- Wan, Q.; Tang, J.; Han, Y.; Wang, D. Co-expression modules construction by WGCNA and identify potential prognostic markers of uveal melanoma. Exp. Eye Res. 2018, 166, 13–20. [Google Scholar] [CrossRef]
- Xue, M.; Shang, J.; Chen, B.; Yang, Z.; Song, Q.; Sun, X.; Chen, J.; Yang, J. Identification of Prognostic Signatures for Predicting the Overall Survival of Uveal Melanoma Patients. J. Cancer 2019, 10, 4921–4931. [Google Scholar] [CrossRef]
- Ni, Y.; Zhang, Z.; Chen, G.; Long, W.; Tong, L.; Zeng, J. Integrated analyses identify potential prognostic markers for uveal melanoma. Exp. Eye Res. 2019, 187, 107780. [Google Scholar] [CrossRef]
- Choi, S.; Ha, M.; Lee, J.S.; Heo, H.J.; Kim, G.H.; Oh, S.O.; Lee, J.J.; Goh, T.S.; Kim, Y.H. Novel Prognostic Factor for Uveal Melanoma: Bioinformatics Analysis of Three Independent Cohorts. Anticancer. Res. 2020, 40, 3839–3846. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Ma, C.; Shao, J.; Cao, J. Prognostic Implications of Novel Ten-Gene Signature in Uveal Melanoma. Front. Oncol. 2020, 10, 567512. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.; Tang, J.; Lu, J.; Jin, L.; Su, Y.; Wang, S.; Cheng, Y.; Liu, Y.; Li, C.; Wang, Z. Six-gene-based prognostic model predicts overall survival in patients with uveal melanoma. Cancer Biomark. Sect. A Dis. Markers 2020, 27, 343–356. [Google Scholar] [CrossRef]
- Tang, Z.; Cai, K. A Novel 8-Gene Prognostic Signature for Survival Prediction of Uveal Melanoma. Anal. Cell. Pathol. 2021, 2021, 6693219. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lu, J.; Li, W. A Comprehensive Prognostic and Immunological Analysis of a Six-Gene Signature Associated With Glycolysis and Immune Response in Uveal Melanoma. Front. Immunol. 2021, 12, 738068. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst. Biol. 2007, 1, 54. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Sato, K.; Mano, T.; Matsuda, H.; Senda, M.; Ihara, R.; Suzuki, K.; Arai, H.; Ishii, K.; Ito, K.; Ikeuchi, T.; et al. Visualizing modules of coordinated structural brain atrophy during the course of conversion to Alzheimer’s disease by applying methodology from gene co-expression analysis. NeuroImage. Clin. 2019, 24, 101957. [Google Scholar] [CrossRef]
- Saris, C.G.; Horvath, S.; van Vught, P.W.; van Es, M.A.; Blauw, H.M.; Fuller, T.F.; Langfelder, P.; DeYoung, J.; Wokke, J.H.; Veldink, J.H.; et al. Weighted gene co-expression network analysis of the peripheral blood from Amyotrophic Lateral Sclerosis patients. BMC Genom. 2009, 10, 405. [Google Scholar] [CrossRef]
- Xie, Y.; Luo, X.; He, H.; Tang, M. Novel Insight Into the Role of Immune Dysregulation in Amyotrophic Lateral Sclerosis Based on Bioinformatic Analysis. Front. Neurosci. 2021, 15, 657465. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Yang, Y.; Li, C.; Xie, L.; Hu, R.; Qin, X.; Zhang, M. A Novel Prognostic Model of Early-Stage Lung Adenocarcinoma Integrating Methylation and Immune Biomarkers. Front. Genet. 2020, 11, 634634. [Google Scholar] [CrossRef]
- Niemira, M.; Collin, F.; Szalkowska, A.; Bielska, A.; Chwialkowska, K.; Reszec, J.; Niklinski, J.; Kwasniewski, M.; Kretowski, A. Molecular Signature of Subtypes of Non-Small-Cell Lung Cancer by Large-Scale Transcriptional Profiling: Identification of Key Modules and Genes by Weighted Gene Co-Expression Network Analysis (WGCNA). Cancers 2019, 12, 37. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Liu, C.; Liu, Y.; Su, Y.; Wang, S.; Jin, L.; Wan, Q.; Liu, Y.; Li, C.; Sang, X.; et al. Immune Microenvironment Related Competitive Endogenous RNA Network as Powerful Predictors for Melanoma Prognosis Based on WGCNA Analysis. Front. Oncol. 2020, 10, 577072. [Google Scholar] [CrossRef]
- Shi, Z.; Derow, C.K.; Zhang, B. Co-expression module analysis reveals biological processes, genomic gain, and regulatory mechanisms associated with breast cancer progression. BMC Syst. Biol. 2010, 4, 74. [Google Scholar] [CrossRef] [PubMed]
- Karami, H.; Derakhshani, A.; Ghasemigol, M.; Fereidouni, M.; Miri-Moghaddam, E.; Baradaran, B.; Tabrizi, N.J.; Najafi, S.; Solimando, A.G.; Marsh, L.M.; et al. Weighted Gene Co-Expression Network Analysis Combined with Machine Learning Validation to Identify Key Modules and Hub Genes Associated with SARS-CoV-2 Infection. J. Clin. Med. 2021, 10, 3567. [Google Scholar] [CrossRef]
- Zhao, W.; Langfelder, P.; Fuller, T.; Dong, J.; Li, A.; Hovarth, S. Weighted gene coexpression network analysis: State of the art. J. Biopharm. Stat. 2010, 20, 281–300. [Google Scholar] [CrossRef]
- Xu, J.; Bankov, G.; Kim, M.; Wretlind, A.; Lord, J.; Green, R.; Hodges, A.; Hye, A.; Aarsland, D.; Velayudhan, L.; et al. Integrated lipidomics and proteomics network analysis highlights lipid and immunity pathways associated with Alzheimer’s disease. Transl. Neurodegener. 2020, 9, 36. [Google Scholar] [CrossRef]
- Pei, G.; Chen, L.; Zhang, W. WGCNA Application to Proteomic and Metabolomic Data Analysis. Methods Enzymol. 2017, 585, 135–158. [Google Scholar] [CrossRef]
- Coupland, S.E.; Anastassiou, G.; Stang, A.; Schilling, H.; Anagnostopoulos, I.; Bornfeld, N.; Stein, H. The prognostic value of cyclin D1, p53, and MDM2 protein expression in uveal melanoma. J. Pathol. 2000, 191, 120–126. [Google Scholar] [CrossRef]
- Liu, Y.; Li, M.; Shi, D.; Zhu, Y. Higher expression of cation transport regulator-like protein 1 (CHAC1) predicts of poor outcomes in uveal melanoma (UM) patients. Int. Ophthalmol. 2019, 39, 2825–2832. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.C.; Lo, E.S.; Lee, K.C.; Chan, J.K.; Hsiao, W.L. Prognostic and diagnostic significance of beta-catenin nuclear immunostaining in colorectal cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2004, 10, 1401–1408. [Google Scholar] [CrossRef] [PubMed]
- Bian, J.; Dannappel, M.; Wan, C.; Firestein, R. Transcriptional Regulation of Wnt/beta-Catenin Pathway in Colorectal Cancer. Cells 2020, 9, 2125. [Google Scholar] [CrossRef] [PubMed]
- Tetsu, O.; McCormick, F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 1999, 398, 422–426. [Google Scholar] [CrossRef]
- De La Coste, A.; Romagnolo, B.; Billuart, P.; Renard, C.A.; Buendia, M.A.; Soubrane, O.; Fabre, M.; Chelly, J.; Beldjord, C.; Kahn, A.; et al. Somatic mutations of the beta-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc. Natl. Acad. Sci. USA 1998, 95, 8847–8851. [Google Scholar] [CrossRef]
- Miyoshi, Y.; Iwao, K.; Nagasawa, Y.; Aihara, T.; Sasaki, Y.; Imaoka, S.; Murata, M.; Shimano, T.; Nakamura, Y. Activation of the beta-catenin gene in primary hepatocellular carcinomas by somatic alterations involving exon 3. Cancer Res. 1998, 58, 2524–2527. [Google Scholar]
- Khalaf, A.M.; Fuentes, D.; Morshid, A.I.; Burke, M.R.; Kaseb, A.O.; Hassan, M.; Hazle, J.D.; Elsayes, K.M. Role of Wnt/beta-catenin signaling in hepatocellular carcinoma, pathogenesis, and clinical significance. J. Hepatocell. Carcinoma 2018, 5, 61–73. [Google Scholar] [CrossRef]
- Kageshita, T.; Hamby, C.V.; Ishihara, T.; Matsumoto, K.; Saida, T.; Ono, T. Loss of beta-catenin expression associated with disease progression in malignant melanoma. Br. J. Dermatol. 2001, 145, 210–216. [Google Scholar] [CrossRef]
- Gajos-Michniewicz, A.; Czyz, M. WNT Signaling in Melanoma. Int. J. Mol. Sci. 2020, 21, 4852. [Google Scholar] [CrossRef]
- Kovacs, D.; Migliano, E.; Muscardin, L.; Silipo, V.; Catricala, C.; Picardo, M.; Bellei, B. The role of Wnt/beta-catenin signaling pathway in melanoma epithelial-to-mesenchymal-like switching: Evidences from patients-derived cell lines. Oncotarget 2016, 7, 43295–43314. [Google Scholar] [CrossRef]
- McKenney, J.K.; Hornick, J.L. Immunohistochemistry in Surgical Pathology. Adv. Anat. Pathol. 2018, 25, 373. [Google Scholar] [CrossRef]
- Taylor, C.R. Immunohistochemistry in surgical pathology: Principles and practice. Methods Mol. Biol. 2014, 1180, 81–109. [Google Scholar] [CrossRef] [PubMed]
- Farquhar, N.; Thornton, S.; Coupland, S.E.; Coulson, J.M.; Sacco, J.J.; Krishna, Y.; Heimann, H.; Taktak, A.; Cebulla, C.M.; Abdel-Rahman, M.H.; et al. Patterns of BAP1 protein expression provide insights into prognostic significance and the biology of uveal melanoma. J. Pathology. Clin. Res. 2018, 4, 26–38. [Google Scholar] [CrossRef]
- Broggi, G.; Musumeci, G.; Puzzo, L.; Russo, A.; Reibaldi, M.; Ragusa, M.; Longo, A.; Caltabiano, R. Immunohistochemical Expression of ABCB5 as a Potential Prognostic Factor in Uveal Melanoma. Appl. Sci. 2019, 9, 1316. [Google Scholar] [CrossRef]
- Tura, A.; Thieme, C.; Brosig, A.; Merz, H.; Ranjbar, M.; Vardanyan, S.; Zuo, H.; Maassen, T.; Kakkassery, V.; Grisanti, S. Lower Levels of Adiponectin and Its Receptor Adipor1 in the Uveal Melanomas With Monosomy-3. Investig. Ophthalmol. Vis. Sci. 2020, 61, 12. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, S.; Jha, J.; Singh, M.K.; Singh, L.; Sen, S.; Kaur, J.; Bajaj, M.S.; Pushker, N. DNA damage response proteins and its role in tumor progression of uveal melanoma with patient outcome. Clin. Transl. Oncol. Off. Publ. Fed. Span. Oncol. Soc. Natl. Cancer Inst. Mex. 2020, 22, 1472–1480. [Google Scholar] [CrossRef]
- Jha, J.; Singh, M.K.; Singh, L.; Pushker, N.; Bajaj, M.S.; Sen, S.; Kashyap, S. Prognostic relevance of ATM protein in uveal melanoma and its association with clinicopathological factors. Int. J. Clin. Oncol. 2019, 24, 1526–1535. [Google Scholar] [CrossRef] [PubMed]
- Broggi, G.; Ieni, A.; Russo, D.; Varricchio, S.; Puzzo, L.; Russo, A.; Reibaldi, M.; Longo, A.; Tuccari, G.; Staibano, S.; et al. The Macro-Autophagy-Related Protein Beclin-1 Immunohistochemical Expression Correlates With Tumor Cell Type and Clinical Behavior of Uveal Melanoma. Front. Oncol. 2020, 10, 589849. [Google Scholar] [CrossRef]
- Jiang, Z.; Yu, F.; Li, M. Upregulation of BCL2 19 kD Protein-Interacting Protein 3 (BNIP3) is Predictive of Unfavorable Prognosis in Uveal Melanoma. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2018, 24, 4711–4717. [Google Scholar] [CrossRef]
- Jiang, Z.; Liu, F. Butyrophilin-Like 9 (BTNL9) Suppresses Invasion and Correlates with Favorable Prognosis of Uveal Melanoma. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2019, 25, 3190–3198. [Google Scholar] [CrossRef]
- Singh, M.K.; Singh, L.; Pushker, N.; Chosdol, K.; Bakhshi, S.; Meel, R.; Sen, S.; Kashyap, S. Constitutive expression of c-REL in uveal melanoma patients: Correlation with clinicopathological parameters and patient outcome. Clin. Transl. Oncol. Off. Publ. Fed. Span. Oncol. Soc. Natl. Cancer Inst. Mex. 2020, 22, 1193–1204. [Google Scholar] [CrossRef] [PubMed]
- Gajdzis, M.; Theocharis, S.; Gajdzis, P.; Cassoux, N.; Gardrat, S.; Donizy, P.; Klijanienko, J.; Kaczmarek, R. Ephrin Receptors (Eph): EphA1, EphA5, and EphA7 Expression in Uveal Melanoma-Associations with Clinical Parameters and Patient Survival. Life 2020, 10, 225. [Google Scholar] [CrossRef] [PubMed]
- Levidou, G.; Gajdzis, P.; Cassoux, N.; Donizy, P.; Masaoutis, C.; Gajdzis, M.; Gardrat, S.; Pergaris, A.; Danas, E.; Klijanienko, J.; et al. Histone Deacetylase (HDAC)-1, -2, -4, and -6 in Uveal Melanomas: Associations with Clinicopathological Parameters and Patients’ Survival. Cancers 2021, 13, 4763. [Google Scholar] [CrossRef]
- Djirackor, L.; Shakir, D.; Kalirai, H.; Petrovski, G.; Coupland, S.E. Nestin expression in primary and metastatic uveal melanoma—Possible biomarker for high-risk uveal melanoma. Acta Ophthalmol. 2018, 96, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.K.; Singh, L.; Pushker, N.; Saini, N.; Meel, R.; Chosdol, K.; Bakhshi, S.; Sen, S.; Venkatesh, P.; Chawla, B.; et al. Identification of canonical NFkappaB (C-NFkappaB) pathway in uveal melanoma and their relation with patient outcome. Clin. Exp. Metastasis 2019, 36, 271–290. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.K.; Singh, L.; Chosdol, K.; Pushker, N.; Saini, N.; Meel, R.; Bakhshi, S.; Sen, S.; Kashyap, S. Differential expression of p52 and RelB proteins in the metastatic and non-metastatic groups of uveal melanoma with patient outcome. J. Cancer Res. Clin. Oncol. 2019, 145, 2969–2982. [Google Scholar] [CrossRef] [PubMed]
- Gajdzis, M.; Theocharis, S.; Klijanienko, J.; Cassoux, N.; Gardrat, S.; Donizy, P.; Kaczmarek, R.; Gajdzis, P. The Prognostic Values of PARP-1 Expression in Uveal Melanoma. Cells 2021, 10, 285. [Google Scholar] [CrossRef]
- Jiang, Z.; Yan, Y.; Dong, J.; Duan, L. PD-1 expression on uveal melanoma induces tumor proliferation and predicts poor patient survival. Int. J. Biol. Markers 2020, 35, 50–58. [Google Scholar] [CrossRef]
- Singh, L.; Singh, M.K.; Kenney, M.C.; Jager, M.J.; Rizvi, M.A.; Meel, R.; Lomi, N.; Bakhshi, S.; Sen, S.; Kashyap, S. Prognostic significance of PD-1/PD-L1 expression in uveal melanoma: Correlation with tumor-infiltrating lymphocytes and clinicopathological parameters. Cancer Immunol. Immunother. CII 2021, 70, 1291–1303. [Google Scholar] [CrossRef]
- Berus, T.; Markiewicz, A.; Kobylinska, K.; Biecek, P.; Orlowska-Heitzman, J.; Romanowska-Dixon, B.; Donizy, P. Downregulation of Polo-like kinase-1 (PLK-1) expression is associated with poor clinical outcome in uveal melanoma patients. Folia Histochem. Cytobiol. 2020, 58, 108–116. [Google Scholar] [CrossRef]
- Ramasamy, P.; Larkin, A.M.; Linge, A.; Tiernan, D.; McAree, F.; Horgan, N.; Moriarty, P.; Beatty, S.; Murphy, C.C.; Clynes, M.; et al. PRDX3 is associated with metastasis and poor survival in uveal melanoma. J. Clin. Pathol. 2020, 73, 408–412. [Google Scholar] [CrossRef]
- Salvatorelli, L.; Puzzo, L.; Russo, A.; Reibaldi, M.; Longo, A.; Ragusa, M.; Aldo, C.; Rappazzo, G.; Caltabiano, R.; Salemi, M. Immunoexpression of SPANX-C in metastatic uveal melanoma. Pathol. Res. Pract. 2019, 215, 152431. [Google Scholar] [CrossRef] [PubMed]
- Kadioglu, O.; Saeed, M.E.M.; Munder, M.; Spuller, A.; Greten, H.J.; Efferth, T. Effect of ABC transporter expression and mutational status on survival rates of cancer patients. Biomed. Pharmacother. 2020, 131, 110718. [Google Scholar] [CrossRef] [PubMed]
- Tangella, L.P.; Arooj, M.; Deplazes, E.; Gray, E.S.; Mancera, R.L. Identification and characterisation of putative drug binding sites in human ATP-binding cassette B5 (ABCB5) transporter. Comput. Struct. Biotechnol. J. 2021, 19, 691–704. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Grimmig, T.; Gonzalez, G.; Giobbie-Hurder, A.; Berg, G.; Carr, N.; Wilson, B.J.; Banerjee, P.; Ma, J.; Gold, J.S.; et al. ATP-binding cassette member B5 (ABCB5) promotes tumor cell invasiveness in human colorectal cancer. J. Biol. Chem. 2018, 293, 11166–11178. [Google Scholar] [CrossRef] [PubMed]
- Gambichler, T.; Petig, A.L.; Stockfleth, E.; Stucker, M. Expression of SOX10, ABCB5 and CD271 in melanocytic lesions and correlation with survival data of patients with melanoma. Clin. Exp. Dermatol. 2016, 41, 709–716. [Google Scholar] [CrossRef]
- Wang, S.; Tang, L.; Lin, J.; Shen, Z.; Yao, Y.; Wang, W.; Tao, S.; Gu, C.; Ma, J.; Xie, Y.; et al. ABCB5 promotes melanoma metastasis through enhancing NF-kappaB p65 protein stability. Biochem. Biophys. Res. Commun. 2017, 492, 18–26. [Google Scholar] [CrossRef]
- Cheung, S.T.; Cheung, P.F.; Cheng, C.K.; Wong, N.C.; Fan, S.T. Granulin-epithelin precursor and ATP-dependent binding cassette (ABC)B5 regulate liver cancer cell chemoresistance. Gastroenterology 2011, 140, 344–355. [Google Scholar] [CrossRef]
- Kleffel, S.; Lee, N.; Lezcano, C.; Wilson, B.J.; Sobolewski, K.; Saab, K.R.; Mueller, H.; Zhan, Q.; Posch, C.; Elco, C.P.; et al. ABCB5-Targeted Chemoresistance Reversal Inhibits Merkel Cell Carcinoma Growth. J. Investig. Dermatol. 2016, 136, 838–846. [Google Scholar] [CrossRef]
- Chartrain, M.; Riond, J.; Stennevin, A.; Vandenberghe, I.; Gomes, B.; Lamant, L.; Meyer, N.; Gairin, J.E.; Guilbaud, N.; Annereau, J.P. Melanoma chemotherapy leads to the selection of ABCB5-expressing cells. PLoS ONE 2012, 7, e36762. [Google Scholar] [CrossRef]
- Bueno, R.C.; Canevari, R.A.; Villacis, R.A.; Domingues, M.A.; Caldeira, J.R.; Rocha, R.M.; Drigo, S.A.; Rogatto, S.R. ATM down-regulation is associated with poor prognosis in sporadic breast carcinomas. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2014, 25, 69–75. [Google Scholar] [CrossRef]
- AlDubayan, S.H.; Giannakis, M.; Moore, N.D.; Han, G.C.; Reardon, B.; Hamada, T.; Mu, X.J.; Nishihara, R.; Qian, Z.; Liu, L.; et al. Inherited DNA-Repair Defects in Colorectal Cancer. Am. J. Hum. Genet. 2018, 102, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Petersen, L.F.; Klimowicz, A.C.; Otsuka, S.; Elegbede, A.A.; Petrillo, S.K.; Williamson, T.; Williamson, C.T.; Konno, M.; Lees-Miller, S.P.; Hao, D.; et al. Loss of tumour-specific ATM protein expression is an independent prognostic factor in early resected NSCLC. Oncotarget 2017, 8, 38326–38336. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Levine, B. Autophagy in Human Diseases. N. Engl. J. Med. 2020, 383, 1564–1576. [Google Scholar] [CrossRef] [PubMed]
- Amaravadi, R.K.; Kimmelman, A.C.; Debnath, J. Targeting Autophagy in Cancer: Recent Advances and Future Directions. Cancer Discov. 2019, 9, 1167–1181. [Google Scholar] [CrossRef]
- Yun, C.W.; Jeon, J.; Go, G.; Lee, J.H.; Lee, S.H. The Dual Role of Autophagy in Cancer Development and a Therapeutic Strategy for Cancer by Targeting Autophagy. Int. J. Mol. Sci. 2020, 22, 179. [Google Scholar] [CrossRef] [PubMed]
- Rangel, M.; Kong, J.; Bhatt, V.; Khayati, K.; Guo, J.Y. Autophagy and tumorigenesis. FEBS J. 2021. [Google Scholar] [CrossRef] [PubMed]
- Chourasia, A.H.; Macleod, K.F. Tumor suppressor functions of BNIP3 and mitophagy. Autophagy 2015, 11, 1937–1938. [Google Scholar] [CrossRef]
- Ney, P.A. Mitochondrial autophagy: Origins, significance, and role of BNIP3 and NIX. Biochim. Biophys. Acta 2015, 1853, 2775–2783. [Google Scholar] [CrossRef]
- Chourasia, A.H.; Tracy, K.; Frankenberger, C.; Boland, M.L.; Sharifi, M.N.; Drake, L.E.; Sachleben, J.R.; Asara, J.M.; Locasale, J.W.; Karczmar, G.S.; et al. Mitophagy defects arising from BNip3 loss promote mammary tumor progression to metastasis. EMBO Rep. 2015, 16, 1145–1163. [Google Scholar] [CrossRef]
- Vijayalingam, S.; Pillai, S.G.; Rashmi, R.; Subramanian, T.; Sagartz, J.E.; Chinnadurai, G. Overexpression of BH3-Only Protein BNIP3 Leads to Enhanced Tumor Growth. Genes Cancer 2010, 1, 964–971. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wu, H.; Huang, S.; Li, W.; Zhang, S.; Zheng, P.; Zhou, X.; Liu, W.; Zhang, D. Expression of BNIP3 and its correlations to hypoxia-induced autophagy and clinicopathological features in salivary adenoid cystic carcinoma. Cancer Biomark. 2015, 15, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Vara-Perez, M.; Rossi, M.; Van den Haute, C.; Maes, H.; Sassano, M.L.; Venkataramani, V.; Michalke, B.; Romano, E.; Rillaerts, K.; Garg, A.D.; et al. BNIP3 promotes HIF-1alpha-driven melanoma growth by curbing intracellular iron homeostasis. EMBO J. 2021, 40, e106214. [Google Scholar] [CrossRef] [PubMed]
- Lebrero-Fernandez, C.; Wenzel, U.A.; Akeus, P.; Wang, Y.; Strid, H.; Simren, M.; Gustavsson, B.; Borjesson, L.G.; Cardell, S.L.; Ohman, L.; et al. Altered expression of Butyrophilin (BTN) and BTN-like (BTNL) genes in intestinal inflammation and colon cancer. Immun. Inflamm. Dis. 2016, 4, 191–200. [Google Scholar] [CrossRef]
- Mo, Q.; Xu, K.; Luo, C.; Zhang, Q.; Wang, L.; Ren, G. BTNL9 is frequently downregulated and inhibits proliferation and metastasis via the P53/CDC25C and P53/GADD45 pathways in breast cancer. Biochem. Biophys. Res. Commun. 2021, 553, 17–24. [Google Scholar] [CrossRef]
- Ren, H.; Li, S.; Liu, X.; Li, W.; Hao, J.; Zhao, N. Multi-omics analysis of the expression and prognostic value of the butyrophilins in breast cancer. J. Leukoc. Biol. 2021, 110, 1181–1195. [Google Scholar] [CrossRef]
- Pergaris, A.; Danas, E.; Goutas, D.; Sykaras, A.G.; Soranidis, A.; Theocharis, S. The Clinical Impact of the EPH/Ephrin System in Cancer: Unwinding the Thread. Int. J. Mol. Sci. 2021, 22, 8412. [Google Scholar] [CrossRef]
- Anderton, M.; van der Meulen, E.; Blumenthal, M.J.; Schafer, G. The Role of the Eph Receptor Family in Tumorigenesis. Cancers 2021, 13, 206. [Google Scholar] [CrossRef]
- Laussu, J.; Khuong, A.; Gautrais, J.; Davy, A. Beyond boundaries--Eph:ephrin signaling in neurogenesis. Cell Adhes. Migr. 2014, 8, 349–359. [Google Scholar] [CrossRef]
- Wilkinson, D.G. Regulation of cell differentiation by Eph receptor and ephrin signaling. Cell Adhes. Migr. 2014, 8, 339–348. [Google Scholar] [CrossRef]
- Cayuso, J.; Xu, Q.; Wilkinson, D.G. Mechanisms of boundary formation by Eph receptor and ephrin signaling. Dev. Biol. 2015, 401, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Klein, R. Eph/ephrin signalling during development. Development 2012, 139, 4105–4109. [Google Scholar] [CrossRef]
- Lodola, A.; Giorgio, C.; Incerti, M.; Zanotti, I.; Tognolini, M. Targeting Eph/ephrin system in cancer therapy. Eur. J. Med. Chem. 2017, 142, 152–162. [Google Scholar] [CrossRef]
- Tejido, C.; Pakravan, D.; Bosch, L.V.D. Potential Therapeutic Role of HDAC Inhibitors in FUS-ALS. Front. Mol. Neurosci. 2021, 14, 686995. [Google Scholar] [CrossRef]
- Klingl, Y.E.; Pakravan, D.; Van Den Bosch, L. Opportunities for histone deacetylase inhibition in amyotrophic lateral sclerosis. Br. J. Pharmacol. 2021, 178, 1353–1372. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Sun, B.; Zhong, C.; Xu, K.; Wang, Z.; Hofman, P.; Nagano, T.; Legras, A.; Breadner, D.; Ricciuti, B.; et al. Targeting histone deacetylase enhances the therapeutic effect of Erastin-induced ferroptosis in EGFR-activating mutant lung adenocarcinoma. Transl. Lung Cancer Res. 2021, 10, 1857–1872. [Google Scholar] [CrossRef] [PubMed]
- Damaskos, C.; Tomos, I.; Garmpis, N.; Karakatsani, A.; Dimitroulis, D.; Garmpi, A.; Spartalis, E.; Kampolis, C.F.; Tsagkari, E.; Loukeri, A.A.; et al. Histone Deacetylase Inhibitors as a Novel Targeted Therapy Against Non-small Cell Lung Cancer: Where Are We Now and What Should We Expect? Anticancer. Res. 2018, 38, 37–43. [Google Scholar] [CrossRef]
- Strub, T.; Ballotti, R.; Bertolotto, C. The “ART” of Epigenetics in Melanoma: From histone “Alterations, to Resistance and Therapies”. Theranostics 2020, 10, 1777–1797. [Google Scholar] [CrossRef]
- Chen, I.C.; Sethy, B.; Liou, J.P. Recent Update of HDAC Inhibitors in Lymphoma. Front. Cell Dev. Biol. 2020, 8, 576391. [Google Scholar] [CrossRef] [PubMed]
- Bondarev, A.D.; Attwood, M.M.; Jonsson, J.; Chubarev, V.N.; Tarasov, V.V.; Schioth, H.B. Recent developments of HDAC inhibitors: Emerging indications and novel molecules. Br. J. Clin. Pharmacol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; Tekwani, B.L. Histone Deacetylases Inhibitors in Neurodegenerative Diseases, Neuroprotection and Neuronal Differentiation. Front. Pharmacol. 2020, 11, 537. [Google Scholar] [CrossRef] [PubMed]
- Souri, Z.; Jochemsen, A.G.; Versluis, M.; Wierenga, A.P.A.; Nemati, F.; van der Velden, P.A.; Kroes, W.G.M.; Verdijk, R.M.; Luyten, G.P.M.; Jager, M.J. HDAC Inhibition Increases HLA Class I Expression in Uveal Melanoma. Cancers 2020, 12, 3690. [Google Scholar] [CrossRef]
- Souri, Z.; Jochemsen, A.G.; Wierenga, A.P.A.; Kroes, W.G.M.; Verdijk, R.M.; van der Velden, P.A.; Luyten, G.P.M.; Jager, M.J. Expression of HDACs 1, 3 and 8 Is Upregulated in the Presence of Infiltrating Lymphocytes in Uveal Melanoma. Cancers 2021, 13, 4146. [Google Scholar] [CrossRef] [PubMed]
- Levinzon, L.; Madigan, M.; Nguyen, V.; Hasic, E.; Conway, M.; Cherepanoff, S. Tumour Expression of Histone Deacetylases in Uveal Melanoma. Ocul. Oncol. Pathol. 2019, 5, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Ny, L.; Jespersen, H.; Karlsson, J.; Alsen, S.; Filges, S.; All-Eriksson, C.; Andersson, B.; Carneiro, A.; Helgadottir, H.; Levin, M.; et al. The PEMDAC phase 2 study of pembrolizumab and entinostat in patients with metastatic uveal melanoma. Nat. Commun. 2021, 12, 5155. [Google Scholar] [CrossRef] [PubMed]
- Lendahl, U.; Zimmerman, L.B.; McKay, R.D. CNS stem cells express a new class of intermediate filament protein. Cell 1990, 60, 585–595. [Google Scholar] [CrossRef]
- Bernal, A.; Arranz, L. Nestin-expressing progenitor cells: Function, identity and therapeutic implications. Cell. Mol. Life Sci. CMLS 2018, 75, 2177–2195. [Google Scholar] [CrossRef]
- Thill, M.; Berna, M.J.; Grierson, R.; Reinhart, I.; Voelkel, T.; Piechaczek, C.; Galambos, P.; Jager, M.J.; Richard, G.; Lange, C.; et al. Expression of CD133 and other putative stem cell markers in uveal melanoma. Melanoma Res. 2011, 21, 405–416. [Google Scholar] [CrossRef]
- Neradil, J.; Veselska, R. Nestin as a marker of cancer stem cells. Cancer Sci. 2015, 106, 803–811. [Google Scholar] [CrossRef]
- Zhang, X.; Xing, C.; Guan, W.; Chen, L.; Guo, K.; Yu, A.; Xie, K. Clinicopathological and prognostic significance of nestin expression in patients with breast cancer: A systematic review and meta-analysis. Cancer Cell Int. 2020, 20, 169. [Google Scholar] [CrossRef] [PubMed]
- Szymanska-Chabowska, A.; Swiatkowski, F.; Jankowska-Polanska, B.; Mazur, G.; Chabowski, M. Nestin Expression as a Diagnostic and Prognostic Marker in Colorectal Cancer and Other Tumors. Clin. Med. Insights. Oncol. 2021, 15, 11795549211038256. [Google Scholar] [CrossRef] [PubMed]
- Sone, K.; Maeno, K.; Masaki, A.; Kunii, E.; Takakuwa, O.; Kagawa, Y.; Takeuchi, A.; Fukuda, S.; Uemura, T.; Fukumitsu, K.; et al. Nestin Expression Affects Resistance to Chemotherapy and Clinical Outcome in Small Cell Lung Cancer. Front. Oncol. 2020, 10, 1367. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Alsharif, S.; Fallatah, A.; Chung, B.M. Intermediate Filaments as Effectors of Cancer Development and Metastasis: A Focus on Keratins, Vimentin, and Nestin. Cells 2019, 8, 497. [Google Scholar] [CrossRef]
- Djirackor, L.; Kalirai, H.; Coupland, S.E.; Petrovski, G. CD166high Uveal Melanoma Cells Represent a Subpopulation With Enhanced Migratory Capacity. Investig. Ophthalmol. Vis. Sci. 2019, 60, 2696–2704. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-kappaB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef]
- Taniguchi, K.; Karin, M. NF-κB, inflammation, immunity and cancer: Coming of age. Nat. Rev. Immunol. 2018, 18, 309–324. [Google Scholar] [CrossRef]
- Zhang, Q.; Lenardo, M.J.; Baltimore, D. 30 Years of NF-kappaB: A Blossoming of Relevance to Human Pathobiology. Cell 2017, 168, 37–57. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, T.D. NF-kappaB and Human Cancer: What Have We Learned over the Past 35 Years? Biomedicines 2021, 9, 889. [Google Scholar] [CrossRef]
- Eluard, B.; Thieblemont, C.; Baud, V. NF-kappaB in the New Era of Cancer Therapy. Trends Cancer 2020, 6, 677–687. [Google Scholar] [CrossRef]
- Barnabei, L.; Laplantine, E.; Mbongo, W.; Rieux-Laucat, F.; Weil, R. NF-kappaB: At the Borders of Autoimmunity and Inflammation. Front. Immunol. 2021, 12, 716469. [Google Scholar] [CrossRef] [PubMed]
- Van Beek, L.; McClay, E.; Patel, S.; Schimpl, M.; Spagnolo, L.; Maia de Oliveira, T. PARP Power: A Structural Perspective on PARP1, PARP2, and PARP3 in DNA Damage Repair and Nucleosome Remodelling. Int. J. Mol. Sci. 2021, 22, 5112. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, A.; Merlin, M.A.; Shah, G.M. Poly (ADP-ribose) polymerase (PARP) inhibition in cancer: Potential impact in cancer stem cells and therapeutic implications. Eur. J. Pharmacol. 2021, 911, 174546. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.Y.; Brown, L.J.; Reid, L.; Joshua, A.M. PARP Inhibitors in Melanoma-An Expanding Therapeutic Option? Cancers 2021, 13, 4520. [Google Scholar] [CrossRef]
- Iams, W.T.; Porter, J.; Horn, L. Immunotherapeutic approaches for small-cell lung cancer. Nat. Rev. Clin. Oncol. 2020, 17, 300–312. [Google Scholar] [CrossRef]
- Trojaniello, C.; Luke, J.J.; Ascierto, P.A. Therapeutic Advancements Across Clinical Stages in Melanoma, With a Focus on Targeted Immunotherapy. Front. Oncol. 2021, 11, 670726. [Google Scholar] [CrossRef]
- Mei, Z.; Huang, J.; Qiao, B.; Lam, A.K. Immune checkpoint pathways in immunotherapy for head and neck squamous cell carcinoma. Int. J. Oral Sci. 2020, 12, 16. [Google Scholar] [CrossRef]
- Okazaki, T.; Honjo, T. PD-1 and PD-1 ligands: From discovery to clinical application. Int. Immunol. 2007, 19, 813–824. [Google Scholar] [CrossRef]
- Lucibello, G.; Mograbi, B.; Milano, G.; Hofman, P.; Brest, P. PD-L1 regulation revisited: Impact on immunotherapeutic strategies. Trends Mol. Med. 2021, 27, 868–881. [Google Scholar] [CrossRef]
- Kleffel, S.; Posch, C.; Barthel, S.R.; Mueller, H.; Schlapbach, C.; Guenova, E.; Elco, C.P.; Lee, N.; Juneja, V.R.; Zhan, Q.; et al. Melanoma Cell-Intrinsic PD-1 Receptor Functions Promote Tumor Growth. Cell 2015, 162, 1242–1256. [Google Scholar] [CrossRef]
- Algazi, A.P.; Tsai, K.K.; Shoushtari, A.N.; Munhoz, R.R.; Eroglu, Z.; Piulats, J.M.; Ott, P.A.; Johnson, D.B.; Hwang, J.; Daud, A.I.; et al. Clinical outcomes in metastatic uveal melanoma treated with PD-1 and PD-L1 antibodies. Cancer 2016, 122, 3344–3353. [Google Scholar] [CrossRef] [PubMed]
- Basile, M.S.; Mazzon, E.; Fagone, P.; Longo, A.; Russo, A.; Fallico, M.; Bonfiglio, V.; Nicoletti, F.; Avitabile, T.; Reibaldi, M. Immunobiology of Uveal Melanoma: State of the Art and Therapeutic Targets. Front. Oncol. 2019, 9, 1145. [Google Scholar] [CrossRef] [PubMed]
- Piulats, J.M.; Espinosa, E.; de la Cruz Merino, L.; Varela, M.; Alonso Carrion, L.; Martin-Algarra, S.; Lopez Castro, R.; Curiel, T.; Rodriguez-Abreu, D.; Redrado, M.; et al. Nivolumab Plus Ipilimumab for Treatment-Naive Metastatic Uveal Melanoma: An Open-Label, Multicenter, Phase II Trial by the Spanish Multidisciplinary Melanoma Group (GEM-1402). J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2021, 39, 586–598. [Google Scholar] [CrossRef]
- Zoroquiain, P.; Esposito, E.; Logan, P.; Aldrees, S.; Dias, A.B.; Mansure, J.J.; Santapau, D.; Garcia, C.; Saornil, M.A.; Belfort Neto, R.; et al. Programmed cell death ligand-1 expression in tumor and immune cells is associated with better patient outcome and decreased tumor-infiltrating lymphocytes in uveal melanoma. Mod. Pathol. Off. J. United States Can. Acad. Pathol. Inc 2018, 31, 1201–1210. [Google Scholar] [CrossRef] [PubMed]
- Iliaki, S.; Beyaert, R.; Afonina, I.S. Polo-like kinase 1 (PLK1) signaling in cancer and beyond. Biochem. Pharmacol. 2021, 193, 114747. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, C.E.; MacAuley, M.J.; Vizeacoumar, F.S.; Abuhussein, O.; Freywald, A.; Vizeacoumar, F.J. The CINs of Polo-Like Kinase 1 in Cancer. Cancers 2020, 12, 2953. [Google Scholar] [CrossRef]
- Colicino, E.G.; Hehnly, H. Regulating a key mitotic regulator, polo-like kinase 1 (PLK1). Cytoskeleton 2018, 75, 481–494. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, Z.; Liu, D.; Wang, M.; Xiao, G.; Wang, P.; Sun, X.; Ren, H.; Tang, S.C.; Du, N. Augmented expression of polo-like kinase 1 indicates poor clinical outcome for breast patients: A systematic review and meta-analysis. Oncotarget 2017, 8, 57723–57732. [Google Scholar] [CrossRef]
- Shin, S.B.; Jang, H.R.; Xu, R.; Won, J.Y.; Yim, H. Active PLK1-driven metastasis is amplified by TGF-beta signaling that forms a positive feedback loop in non-small cell lung cancer. Oncogene 2020, 39, 767–785. [Google Scholar] [CrossRef] [PubMed]
- Murga-Zamalloa, C.; Inamdar, K.V.; Wilcox, R.A. The role of aurora A and polo-like kinases in high-risk lymphomas. Blood Adv. 2019, 3, 1778–1787. [Google Scholar] [CrossRef]
- Liu, Z.; Sun, Q.; Wang, X. PLK1, A Potential Target for Cancer Therapy. Transl. Oncol. 2017, 10, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Gutteridge, R.E.; Ndiaye, M.A.; Liu, X.; Ahmad, N. Plk1 Inhibitors in Cancer Therapy: From Laboratory to Clinics. Mol. Cancer Ther. 2016, 15, 1427–1435. [Google Scholar] [CrossRef] [PubMed]
- Ismail, T.; Kim, Y.; Lee, H.; Lee, D.S.; Lee, H.S. Interplay Between Mitochondrial Peroxiredoxins and ROS in Cancer Development and Progression. Int. J. Mol. Sci. 2019, 20, 4407. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J. Knockout Mouse Models for Peroxiredoxins. Antioxidants 2020, 9, 182. [Google Scholar] [CrossRef]
- Westbrook, V.A.; Diekman, A.B.; Klotz, K.L.; Khole, V.V.; von Kap-Herr, C.; Golden, W.L.; Eddy, R.L.; Shows, T.B.; Stoler, M.H.; Lee, C.Y.; et al. Spermatid-specific expression of the novel X-linked gene product SPAN-X localized to the nucleus of human spermatozoa. Biol. Reprod. 2000, 63, 469–481. [Google Scholar] [CrossRef]
- Salemi, M.; Calogero, A.E.; Di Benedetto, D.; Cosentino, A.; Barone, N.; Rappazzo, G.; Vicari, E. Expression of SPANX proteins in human-ejaculated spermatozoa and sperm precursors. Int. J. Androl. 2004, 27, 134–139. [Google Scholar] [CrossRef]
- Urizar-Arenaza, I.; Osinalde, N.; Akimov, V.; Puglia, M.; Munoa-Hoyos, I.; Gianzo, M.; Rodriguez, J.A.; Ganzabal, T.; Blagoev, B.; Kratchmarova, I.; et al. SPANX-A/D protein subfamily plays a key role in nuclear organisation, metabolism and flagellar motility of human spermatozoa. Sci. Rep. 2020, 10, 5625. [Google Scholar] [CrossRef]
- Maine, E.A.; Westcott, J.M.; Prechtl, A.M.; Dang, T.T.; Whitehurst, A.W.; Pearson, G.W. The cancer-testis antigens SPANX-A/C/D and CTAG2 promote breast cancer invasion. Oncotarget 2016, 7, 14708–14726. [Google Scholar] [CrossRef]
- Lazar, I.; Fabre, B.; Feng, Y.; Khateb, A.; Turko, P.; Martinez Gomez, J.M.; Frederick, D.T.; Levesque, M.P.; Feld, L.; Zhang, G.; et al. SPANX Control of Lamin A/C Modulates Nuclear Architecture and Promotes Melanoma Growth. Mol. Cancer Res. MCR 2020, 18, 1560–1573. [Google Scholar] [CrossRef]
- Urizar-Arenaza, I.; Benedicto, A.; Perez-Valle, A.; Osinalde, N.; Akimov, V.; Munoa-Hoyos, I.; Rodriguez, J.A.; Asumendi, A.; Boyano, M.D.; Blagoev, B.; et al. The multifunctional role of SPANX-A/D protein subfamily in the promotion of pro-tumoural processes in human melanoma. Sci. Rep. 2021, 11, 3583. [Google Scholar] [CrossRef]
- Jang, G.F.; Crabb, J.S.; Hu, B.; Willard, B.; Kalirai, H.; Singh, A.D.; Coupland, S.E.; Crabb, J.W. Proteomics of Primary Uveal Melanoma: Insights into Metastasis and Protein Biomarkers. Cancers 2021, 13, 3520. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Zhang, Z.; Chan, D.W. Detection of Uveal Melanoma by Multiplex Immunoassays of Serum Biomarkers. Methods Mol. Biol. 2021, 2265, 447–459. [Google Scholar] [CrossRef]
- Song, J.; Merbs, S.L.; Sokoll, L.J.; Chan, D.W.; Zhang, Z. A multiplex immunoassay of serum biomarkers for the detection of uveal melanoma. Clin. Proteom. 2019, 16, 10. [Google Scholar] [CrossRef] [PubMed]
- Velez, G.; Nguyen, H.V.; Chemudupati, T.; Ludwig, C.A.; Toral, M.; Reddy, S.; Mruthyunjaya, P.; Mahajan, V.B. Liquid biopsy proteomics of uveal melanoma reveals biomarkers associated with metastatic risk. Mol. Cancer 2021, 20, 39. [Google Scholar] [CrossRef]
- Honavar, S.G. Is Collaborative Ocular Melanoma Study (COMS) still relevant? Indian J. Ophthalmol. 2018, 66, 1385–1387. [Google Scholar] [CrossRef] [PubMed]
- Damato, B.; Eleuteri, A.; Hussain, R.; Kalirai, H.; Thornton, S.; Taktak, A.; Heimann, H.; Coupland, S.E. Parsimonious Models for Predicting Mortality from Choroidal Melanoma. Investig. Ophthalmol. Vis. Sci. 2020, 61, 35. [Google Scholar] [CrossRef]
- Vaquero-Garcia, J.; Lalonde, E.; Ewens, K.G.; Ebrahimzadeh, J.; Richard-Yutz, J.; Shields, C.L.; Barrera, A.; Green, C.J.; Barash, Y.; Ganguly, A. PRiMeUM: A Model for Predicting Risk of Metastasis in Uveal Melanoma. Investig. Ophthalmol. Vis. Sci. 2017, 58, 4096–4105. [Google Scholar] [CrossRef]
- Martel, A.; Baillif, S.; Nahon-Esteve, S.; Gastaud, L.; Bertolotto, C.; Romeo, B.; Mograbi, B.; Lassalle, S.; Hofman, P. Liquid Biopsy for Solid Ophthalmic Malignancies: An Updated Review and Perspectives. Cancers 2020, 12, 3284. [Google Scholar] [CrossRef]
- Ignatiadis, M.; Sledge, G.W.; Jeffrey, S.S. Liquid biopsy enters the clinic—Implementation issues and future challenges. Nat. Rev. Clin. Oncol. 2021, 18, 297–312. [Google Scholar] [CrossRef]
- Heitzer, E.; Haque, I.S.; Roberts, C.E.S.; Speicher, M.R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 2019, 20, 71–88. [Google Scholar] [CrossRef]
- Im, Y.R.; Tsui, D.W.Y.; Diaz, L.A., Jr.; Wan, J.C.M. Next-Generation Liquid Biopsies: Embracing Data Science in Oncology. Trends Cancer 2021, 7, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Heeke, S.; Hofman, V.; Ilie, M.; Allegra, M.; Lespinet, V.; Bordone, O.; Benzaquen, J.; Boutros, J.; Poudenx, M.; Lalvee, S.; et al. Prospective evaluation of NGS-based liquid biopsy in untreated late stage non-squamous lung carcinoma in a single institution. J. Transl. Med. 2020, 18, 87. [Google Scholar] [CrossRef] [PubMed]
- Heeke, S.; Benzaquen, J.; Hofman, V.; Ilie, M.; Allegra, M.; Long-Mira, E.; Lassalle, S.; Tanga, V.; Salacroup, C.; Bonnetaud, C.; et al. Critical Assessment in Routine Clinical Practice of Liquid Biopsy for EGFR Status Testing in Non-Small-Cell Lung Cancer: A Single-Laboratory Experience (LPCE, Nice, France). Clin. Lung Cancer 2020, 21, 56–65.e8. [Google Scholar] [CrossRef] [PubMed]
- Hofman, P. Liquid Biopsy and Therapeutic Targets: Present and Future Issues in Thoracic Oncology. Cancers 2017, 9, 154. [Google Scholar] [CrossRef] [PubMed]
- Hofman, P. Next-Generation Sequencing with Liquid Biopsies from Treatment-Naive Non-Small Cell Lung Carcinoma Patients. Cancers 2021, 13, 2049. [Google Scholar] [CrossRef]
- Hofman, P. Detecting Resistance to Therapeutic ALK Inhibitors in Tumor Tissue and Liquid Biopsy Markers: An Update to a Clinical Routine Practice. Cells 2021, 10, 168. [Google Scholar] [CrossRef] [PubMed]
- Jin, E.; Burnier, J.V. Liquid Biopsy in Uveal Melanoma: Are We There Yet? Ocul. Oncol. Pathol. 2021, 7, 1–16. [Google Scholar] [CrossRef]
- Beasley, A.; Isaacs, T.; Khattak, M.A.; Freeman, J.B.; Allcock, R.; Chen, F.K.; Pereira, M.R.; Yau, K.; Bentel, J.; Vermeulen, T.; et al. Clinical Application of Circulating Tumor Cells and Circulating Tumor DNA in Uveal Melanoma. JCO Precis. Oncol. 2018, 2, 1–12. [Google Scholar] [CrossRef]
- Park, J.J.; Diefenbach, R.J.; Byrne, N.; Long, G.V.; Scolyer, R.A.; Gray, E.S.; Carlino, M.S.; Rizos, H. Circulating Tumor DNA Reflects Uveal Melanoma Responses to Protein Kinase C Inhibition. Cancers 2021, 13, 1740. [Google Scholar] [CrossRef]
- Le Guin, C.H.D.; Bornfeld, N.; Bechrakis, N.E.; Jabbarli, L.; Richly, H.; Lohmann, D.R.; Zeschnigk, M. Early detection of metastatic uveal melanoma by the analysis of tumor-specific mutations in cell-free plasma DNA. Cancer Med. 2021, 10, 5974–5982. [Google Scholar] [CrossRef]
- Krishna, Y.; McCarthy, C.; Kalirai, H.; Coupland, S.E. Inflammatory cell infiltrates in advanced metastatic uveal melanoma. Hum. Pathol. 2017, 66, 159–166. [Google Scholar] [CrossRef]
- Wierenga, A.P.A.; Cao, J.; Mouthaan, H.; van Weeghel, C.; Verdijk, R.M.; van Duinen, S.G.; Kroes, W.G.M.; Dogrusoz, M.; Marinkovic, M.; van der Burg, S.S.H.; et al. Aqueous Humor Biomarkers Identify Three Prognostic Groups in Uveal Melanoma. Investig. Ophthalmol. Vis. Sci. 2019, 60, 4740–4747. [Google Scholar] [CrossRef] [PubMed]
- Berry, J.L.; Xu, L.; Murphree, A.L.; Krishnan, S.; Stachelek, K.; Zolfaghari, E.; McGovern, K.; Lee, T.C.; Carlsson, A.; Kuhn, P.; et al. Potential of Aqueous Humor as a Surrogate Tumor Biopsy for Retinoblastoma. JAMA Ophthalmol. 2017, 135, 1221–1230. [Google Scholar] [CrossRef]
- Gerrish, A.; Stone, E.; Clokie, S.; Ainsworth, J.R.; Jenkinson, H.; McCalla, M.; Hitchcott, C.; Colmenero, I.; Allen, S.; Parulekar, M.; et al. Non-invasive diagnosis of retinoblastoma using cell-free DNA from aqueous humour. Br. J. Ophthalmol. 2019, 103, 721–724. [Google Scholar] [CrossRef]
- Kuiper, J.; Ten Dam-van Loon, N.; Domanian, A.; Schellekens, P.; Nierkens, S.; Radstake, T.; de Boer, J. Correlation between measurement of IL-10 and IL-6 in paired aqueous humour and vitreous fluid in primary vitreoretinal lymphoma. Acta Ophthalmol. 2015, 93, e680–e681. [Google Scholar] [CrossRef] [PubMed]
- Pochat-Cotilloux, C.; Bienvenu, J.; Nguyen, A.M.; Ohanessian, R.; Ghesquieres, H.; Seve, P.; Garnier, L.; Kodjikian, L. Use of a Threshold of Interleukin-10 and Il-10/Il-6 Ratio in Ocular Samples for the Screening of Vitreoretinal Lymphoma. Retina 2018, 38, 773–781. [Google Scholar] [CrossRef] [PubMed]
- Midena, E.; Parrozzani, R.; Midena, G.; Trainiti, S.; Marchione, G.; Cosmo, E.; Londei, D.; Frizziero, L. In vivo intraocular biomarkers: Changes of aqueous humor cytokines and chemokines in patients affected by uveal melanoma. Medicine 2020, 99, e22091. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.; Gullerova, M. Home and Away: The Role of Non-Coding RNA in Intracellular and Intercellular DNA Damage Response. Genes 2021, 12, 1475. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Acha-Sagredo, A.; Uko, B.; Pantazi, P.; Bediaga, N.G.; Moschandrea, C.; Rainbow, L.; Marcus, M.W.; Davies, M.P.A.; Field, J.K.; Liloglou, T. Long non-coding RNA dysregulation is a frequent event in non-small cell lung carcinoma pathogenesis. Br. J. Cancer 2020, 122, 1050–1058. [Google Scholar] [CrossRef]
- Wozniak, M.; Czyz, M. The Functional Role of Long Non-Coding RNAs in Melanoma. Cancers 2021, 13, 4848. [Google Scholar] [CrossRef] [PubMed]
- Bande, M.; Fernandez-Diaz, D.; Fernandez-Marta, B.; Rodriguez-Vidal, C.; Lago-Baameiro, N.; Silva-Rodriguez, P.; Paniagua, L.; Blanco-Teijeiro, M.J.; Pardo, M.; Pineiro, A. The Role of Non-Coding RNAs in Uveal Melanoma. Cancers 2020, 12, 2944. [Google Scholar] [CrossRef] [PubMed]
- Milan-Rois, P.; Quan, A.; Slack, F.J.; Somoza, A. The Role of LncRNAs in Uveal Melanoma. Cancers 2021, 13, 4041. [Google Scholar] [CrossRef]
- Aughton, K.; Kalirai, H.; Coupland, S.E. MicroRNAs and Uveal Melanoma: Understanding the Diverse Role of These Small Molecular Regulators. Int. J. Mol. Sci. 2020, 21, 5648. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Biswas, A.; Liu, H.; Sen, S.; Paruchuri, A.; Katsonis, P.; Lichtarge, O.; Chand Dakal, T.; Maulik, U.; Gromiha, M.M.; et al. Mutational Landscape of the BAP1 Locus Reveals an Intrinsic Control to Regulate the miRNA Network and the Binding of Protein Complexes in Uveal Melanoma. Cancers 2019, 11, 1600. [Google Scholar] [CrossRef]
- Yang, C.; Wang, Y.; Hardy, P. Emerging roles of microRNAs and their implications in uveal melanoma. Cell. Mol. Life Sci. CMLS 2021, 78, 545–559. [Google Scholar] [CrossRef]
- Wroblewska, J.P.; Lach, M.S.; Ustaszewski, A.; Kulcenty, K.; Ibbs, M.; Jagiello, I.; Suchorska, W.M.; Marszalek, A. The Potential Role of Selected miRNA in Uveal Melanoma Primary Tumors as Early Biomarkers of Disease Progression. Genes 2020, 11, 271. [Google Scholar] [CrossRef]
- Yang, C.; Wang, R.; Hardy, P. Potential of miRNA-Based Nanotherapeutics for Uveal Melanoma. Cancers 2021, 13, 5192. [Google Scholar] [CrossRef]
- Krishna, Y.; Acha-Sagredo, A.; Sabat-Pospiech, D.; Kipling, N.; Clarke, K.; Figueiredo, C.R.; Kalirai, H.; Coupland, S.E. Transcriptome Profiling Reveals New Insights into the Immune Microenvironment and Upregulation of Novel Biomarkers in Metastatic Uveal Melanoma. Cancers 2020, 12, 2832. [Google Scholar] [CrossRef]
- Kelly, D.; Rose, A.A.N.; Muniz, T.P.; Hogg, D.; Butler, M.O.; Saibil, S.D.; King, I.; Kamil, Z.S.; Ghazarian, D.; Ross, K.; et al. Development of a Metastatic Uveal Melanoma Prognostic Score (MUMPS) for Use in Patients Receiving Immune Checkpoint Inhibitors. Cancers 2021, 13, 3640. [Google Scholar] [CrossRef]
- Bol, K.F.; Ellebaek, E.; Hoejberg, L.; Bagger, M.M.; Larsen, M.S.; Klausen, T.W.; Kohler, U.H.; Schmidt, H.; Bastholt, L.; Kiilgaard, J.F.; et al. Real-World Impact of Immune Checkpoint Inhibitors in Metastatic Uveal Melanoma. Cancers 2019, 11, 1489. [Google Scholar] [CrossRef]
- Figueiredo, C.R.; Kalirai, H.; Sacco, J.J.; Azevedo, R.A.; Duckworth, A.; Slupsky, J.R.; Coulson, J.M.; Coupland, S.E. Loss of BAP1 expression is associated with an immunosuppressive microenvironment in uveal melanoma, with implications for immunotherapy development. J. Pathol. 2020, 250, 420–439. [Google Scholar] [CrossRef] [PubMed]
- Stalhammar, G.; Seregard, S.; Grossniklaus, H.E. Expression of immune checkpoint receptors Indoleamine 2,3-dioxygenase and T cell Ig and ITIM domain in metastatic versus nonmetastatic choroidal melanoma. Cancer Med. 2019, 8, 2784–2792. [Google Scholar] [CrossRef] [PubMed]
- Durante, M.A.; Rodriguez, D.A.; Kurtenbach, S.; Kuznetsov, J.N.; Sanchez, M.I.; Decatur, C.L.; Snyder, H.; Feun, L.G.; Livingstone, A.S.; Harbour, J.W. Single-cell analysis reveals new evolutionary complexity in uveal melanoma. Nat. Commun. 2020, 11, 496. [Google Scholar] [CrossRef] [PubMed]
- Souri, Z.; Wierenga, A.P.A.; Kroes, W.G.M.; van der Velden, P.A.; Verdijk, R.M.; Eikmans, M.; Luyten, G.P.M.; Jager, M.J. LAG3 and Its Ligands Show Increased Expression in High-Risk Uveal Melanoma. Cancers 2021, 13, 4445. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, Y.; Gao, J.; Fu, Y.; Hua, P.; Jing, Y.; Cai, M.; Wang, H.; Tong, T. The role of CD47-SIRPalpha immune checkpoint in tumor immune evasion and innate immunotherapy. Life Sci. 2021, 273, 119150. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Yan, B.; Tian, X.; Liu, Q.; Jin, J.; Shi, J.; Hou, Y. CD47/SIRPalpha pathway mediates cancer immune escape and immunotherapy. Int. J. Biol. Sci. 2021, 17, 3281–3287. [Google Scholar] [CrossRef]
- Sun, J.; Chen, Y.; Lubben, B.; Adebayo, O.; Muz, B.; Azab, A.K. CD47-targeting antibodies as a novel therapeutic strategy in hematologic malignancies. Leuk. Res. Rep. 2021, 16, 100268. [Google Scholar] [CrossRef]
- Xu, Y.; Li, J.; Tong, B.; Chen, M.; Liu, X.; Zhong, W.; Zhao, J.; Wang, M. Positive tumour CD47 expression is an independent prognostic factor for recurrence in resected non-small cell lung cancer. ESMO Open 2020, 5, e000823. [Google Scholar] [CrossRef]
- Kosaka, A.; Ishibashi, K.; Nagato, T.; Kitamura, H.; Fujiwara, Y.; Yasuda, S.; Nagata, M.; Harabuchi, S.; Hayashi, R.; Yajima, Y.; et al. CD47 blockade enhances the efficacy of intratumoral STING-targeting therapy by activating phagocytes. J. Exp. Med. 2021, 218, e20200792. [Google Scholar] [CrossRef]
- Petralia, M.C.; Mazzon, E.; Fagone, P.; Russo, A.; Longo, A.; Avitabile, T.; Nicoletti, F.; Reibaldi, M.; Basile, M.S. Characterization of the Pathophysiological Role of CD47 in Uveal Melanoma. Molecules 2019, 24, 2450. [Google Scholar] [CrossRef]
- Lu, Q.; Chen, X.; Wang, S.; Lu, Y.; Yang, C.; Jiang, G. Potential New Cancer Immunotherapy: Anti-CD47-SIRPalpha Antibodies. OncoTargets Ther. 2020, 13, 9323–9331. [Google Scholar] [CrossRef] [PubMed]
- Marin-Acevedo, J.A.; Kimbrough, E.O.; Lou, Y. Next generation of immune checkpoint inhibitors and beyond. J. Hematol. Oncol. 2021, 14, 45. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.B.; Ha, S.J.; Kim, H.R. Clinical Insights Into Novel Immune Checkpoint Inhibitors. Front. Pharmacol. 2021, 12, 681320. [Google Scholar] [CrossRef] [PubMed]
- Pelster, M.S.; Gruschkus, S.K.; Bassett, R.; Gombos, D.S.; Shephard, M.; Posada, L.; Glover, M.S.; Simien, R.; Diab, A.; Hwu, P.; et al. Nivolumab and Ipilimumab in Metastatic Uveal Melanoma: Results From a Single-Arm Phase II Study. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2021, 39, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Hofman, P.; Badoual, C.; Henderson, F.; Berland, L.; Hamila, M.; Long-Mira, E.; Lassalle, S.; Roussel, H.; Hofman, V.; Tartour, E.; et al. Multiplexed Immunohistochemistry for Molecular and Immune Profiling in Lung Cancer-Just About Ready for Prime-Time? Cancers 2019, 11, 283. [Google Scholar] [CrossRef]
- Wharton, K.A., Jr.; Wood, D.; Manesse, M.; Maclean, K.H.; Leiss, F.; Zuraw, A. Tissue Multiplex Analyte Detection in Anatomic Pathology—Pathways to Clinical Implementation. Front. Mol. Biosci. 2021, 8, 672531. [Google Scholar] [CrossRef]
- Grossniklaus, H.E.; Zhang, Q.; You, S.; McCarthy, C.; Heegaard, S.; Coupland, S.E. Metastatic ocular melanoma to the liver exhibits infiltrative and nodular growth patterns. Hum. Pathol. 2016, 57, 165–175. [Google Scholar] [CrossRef]
- Grossniklaus, H.E. Understanding Uveal Melanoma Metastasis to the Liver: The Zimmerman Effect and the Zimmerman Hypothesis. Ophthalmology 2019, 126, 483–487. [Google Scholar] [CrossRef]
- Bustamante, P.; Piquet, L.; Landreville, S.; Burnier, J.V. Uveal melanoma pathobiology: Metastasis to the liver. Semin. Cancer Biol. 2021, 71, 65–85. [Google Scholar] [CrossRef]
- Lin, W.; Beasley, A.B.; Ardakani, N.M.; Denisenko, E.; Calapre, L.; Jones, M.; Wood, B.A.; Warburton, L.; Forrest, A.R.R.; Gray, E.S. Intra- and inter-tumoral heterogeneity of liver metastases in a patient with uveal melanoma revealed by single-cell RNA sequencing. Cold Spring Harb. Mol. Case Stud. 2021, 7, a006111. [Google Scholar] [CrossRef] [PubMed]
- Pandiani, C.; Strub, T.; Nottet, N.; Cheli, Y.; Gambi, G.; Bille, K.; Husser, C.; Dalmasso, M.; Beranger, G.; Lassalle, S.; et al. Single-cell RNA sequencing reveals intratumoral heterogeneity in primary uveal melanomas and identifies HES6 as a driver of the metastatic disease. Cell Death Differ. 2021, 28, 1990–2000. [Google Scholar] [CrossRef] [PubMed]
- Strub, T.; Martel, A.; Nahon-Esteve, S.; Baillif, S.; Ballotti, R.; Bertolotto, C. Translation of single-cell transcriptomic analysis of uveal melanomas to clinical oncology. Prog. Retin. Eye Res. 2021, 85, 100968. [Google Scholar] [CrossRef] [PubMed]
- Simian, M.; Bissell, M.J. Organoids: A historical perspective of thinking in three dimensions. J. Cell Biol. 2017, 216, 31–40. [Google Scholar] [CrossRef]
- Lamas, N.J.; Roybon, L. Harnessing the potential of human pluripotent stem cell-derived motor neurons for drug discovery in Amyotrophic Lateral Sclerosis (ALS): From the clinic to the laboratory and back to the patient. Front. Drug Discov. 2021, 1, 1–26. [Google Scholar] [CrossRef]
- Lucey, B.P.; Nelson-Rees, W.A.; Hutchins, G.M. Henrietta Lacks, HeLa cells, and cell culture contamination. Arch. Pathol. Lab. Med. 2009, 133, 1463–1467. [Google Scholar] [CrossRef]
- Jager, M.J.; Magner, J.A.; Ksander, B.R.; Dubovy, S.R. Uveal Melanoma Cell Lines: Where do they come from? (An American Ophthalmological Society Thesis). Trans. Am. Ophthalmol. Soc. 2016, 114, T5. [Google Scholar]
- Angi, M.; Versluis, M.; Kalirai, H. Culturing Uveal Melanoma Cells. Ocul. Oncol. Pathol. 2015, 1, 126–132. [Google Scholar] [CrossRef]
- Fiorentzis, M.; Viestenz, A.; Siebolts, U.; Seitz, B.; Coupland, S.E.; Heinzelmann, J. The Potential Use of Electrochemotherapy in the Treatment of Uveal Melanoma: In Vitro Results in 3D Tumor Cultures and In Vivo Results in a Chick Embryo Model. Cancers 2019, 11, 1344. [Google Scholar] [CrossRef]
- Aughton, K.; Shahidipour, H.; Djirackor, L.; Coupland, S.E.; Kalirai, H. Characterization of Uveal Melanoma Cell Lines and Primary Tumor Samples in 3D Culture. Transl. Vis. Sci. Technol. 2020, 9, 39. [Google Scholar] [CrossRef]
- Richards, J.R.; Yoo, J.H.; Shin, D.; Odelberg, S.J. Mouse models of uveal melanoma: Strengths, weaknesses, and future directions. Pigment. Cell Melanoma Res. 2020, 33, 264–278. [Google Scholar] [CrossRef] [PubMed]
- Stei, M.M.; Loeffler, K.U.; Holz, F.G.; Herwig, M.C. Animal Models of Uveal Melanoma: Methods, Applicability, and Limitations. BioMed Res. Int. 2016, 2016, 4521807. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, S. Pluripotent Stem Cell-Based Cell Therapy-Promise and Challenges. Cell Stem Cell 2020, 27, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Bharathan, S.P.; Ferrario, A.; Stepanian, K.; Fernandez, G.E.; Reid, M.W.; Kim, J.S.; Hutchens, C.; Harutyunyan, N.; Marks, C.; Thornton, M.E.; et al. Characterization and staging of outer plexiform layer development in human retina and retinal organoids. Development 2021, 148, dev199551. [Google Scholar] [CrossRef]
- Castro-Perez, E.; Rodriguez, C.I.; Mikheil, D.; Siddique, S.; McCarthy, A.; Newton, M.A.; Setaluri, V. Melanoma Progression Inhibits Pluripotency and Differentiation of Melanoma-Derived iPSCs Produces Cells with Neural-like Mixed Dysplastic Phenotype. Stem Cell Rep. 2019, 13, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Karagiannis, P.; Kim, S.I. iPSC-Derived Natural Killer Cells for Cancer Immunotherapy. Mol. Cells 2021, 44, 541–548. [Google Scholar] [CrossRef]
- Bhan, A.; Ansari, K.; Chen, M.Y.; Jandial, R. Human induced pluripotent stem cell-derived platelets loaded with lapatinib effectively target HER2+ breast cancer metastasis to the brain. Sci. Rep. 2021, 11, 16866. [Google Scholar] [CrossRef] [PubMed]
- Lamas, N.J.; Johnson-Kerner, B.; Roybon, L.; Kim, Y.A.; Garcia-Diaz, A.; Wichterle, H.; Henderson, C.E. Neurotrophic requirements of human motor neurons defined using amplified and purified stem cell-derived cultures. PLoS ONE 2014, 9, e110324. [Google Scholar] [CrossRef]
- Li, H.; Kuwajima, T.; Oakley, D.; Nikulina, E.; Hou, J.; Yang, W.S.; Lowry, E.R.; Lamas, N.J.; Amoroso, M.W.; Croft, G.F.; et al. Protein Prenylation Constitutes an Endogenous Brake on Axonal Growth. Cell Rep. 2016, 16, 545–558. [Google Scholar] [CrossRef]
- Rodrigues, T.; Kundu, B.; Silva-Correia, J.; Kundu, S.C.; Oliveira, J.M.; Reis, R.L.; Correlo, V.M. Emerging tumor spheroids technologies for 3D in vitro cancer modeling. Pharmacol. Ther. 2018, 184, 201–211. [Google Scholar] [CrossRef]
- Monteiro, M.V.; Zhang, Y.S.; Gaspar, V.M.; Mano, J.F. 3D-bioprinted cancer-on-a-chip: Level-up organotypic in vitro models. Trends Biotechnol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Fang, J.; Huang, S.; Wu, X.; Xie, X.; Wang, J.; Liu, F.; Zhang, M.; Peng, Z.; Hu, N. Tumor-on-a-chip: From bioinspired design to biomedical application. Microsyst. Nanoeng. 2021, 7, 50. [Google Scholar] [CrossRef]
- Marton, R.M.; Pasca, S.P. Organoid and Assembloid Technologies for Investigating Cellular Crosstalk in Human Brain Development and Disease. Trends Cell Biol. 2020, 30, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.; Revah, O.; Miura, Y.; Thom, N.; Amin, N.D.; Kelley, K.W.; Singh, M.; Chen, X.; Thete, M.V.; Walczak, E.M.; et al. Generation of Functional Human 3D Cortico-Motor Assembloids. Cell 2020, 183, 1913–1929.e26. [Google Scholar] [CrossRef] [PubMed]
- Panoutsopoulos, A.A. Organoids, Assembloids, and Novel Biotechnology: Steps Forward in Developmental and Disease-Related Neuroscience. Neuroscientist 2020, 27, 463–472. [Google Scholar] [CrossRef]
- Nathan, P.; Hassel, J.C.; Rutkowski, P.; Baurain, J.F.; Butler, M.O.; Schlaak, M.; Sullivan, R.J.; Ochsenreither, S.; Dummer, R.; Kirkwood, J.M.; et al. Overall Survival Benefit with Tebentafusp in Metastatic Uveal Melanoma. N. Engl. J. Med. 2021, 385, 1196–1206. [Google Scholar] [CrossRef]
- Killock, D. Tebentafusp for uveal melanoma. Nat. Rev. Clin. Oncol. 2021, 18, 747. [Google Scholar] [CrossRef]
- Damato, B.E.; Dukes, J.; Goodall, H.; Carvajal, R.D. Tebentafusp: T Cell Redirection for the Treatment of Metastatic Uveal Melanoma. Cancers 2019, 11, 971. [Google Scholar] [CrossRef]
- Goebeler, M.E.; Bargou, R.C. T cell-engaging therapies—BiTEs and beyond. Nat. Rev. Clin. Oncol. 2020, 17, 418–434. [Google Scholar] [CrossRef]
- Zhao, Q.; Jiang, Y.; Xiang, S.; Kaboli, P.J.; Shen, J.; Zhao, Y.; Wu, X.; Du, F.; Li, M.; Cho, C.H.; et al. Engineered TCR-T Cell Immunotherapy in Anticancer Precision Medicine: Pros and Cons. Front. Immunol. 2021, 12, 658753. [Google Scholar] [CrossRef]
- Han, A.; Purwin, T.J.; Bechtel, N.; Liao, C.; Chua, V.; Seifert, E.; Sato, T.; Schug, Z.T.; Speicher, D.W.; William Harbour, J.; et al. BAP1 mutant uveal melanoma is stratified by metabolic phenotypes with distinct vulnerability to metabolic inhibitors. Oncogene 2021, 40, 618–632. [Google Scholar] [CrossRef] [PubMed]
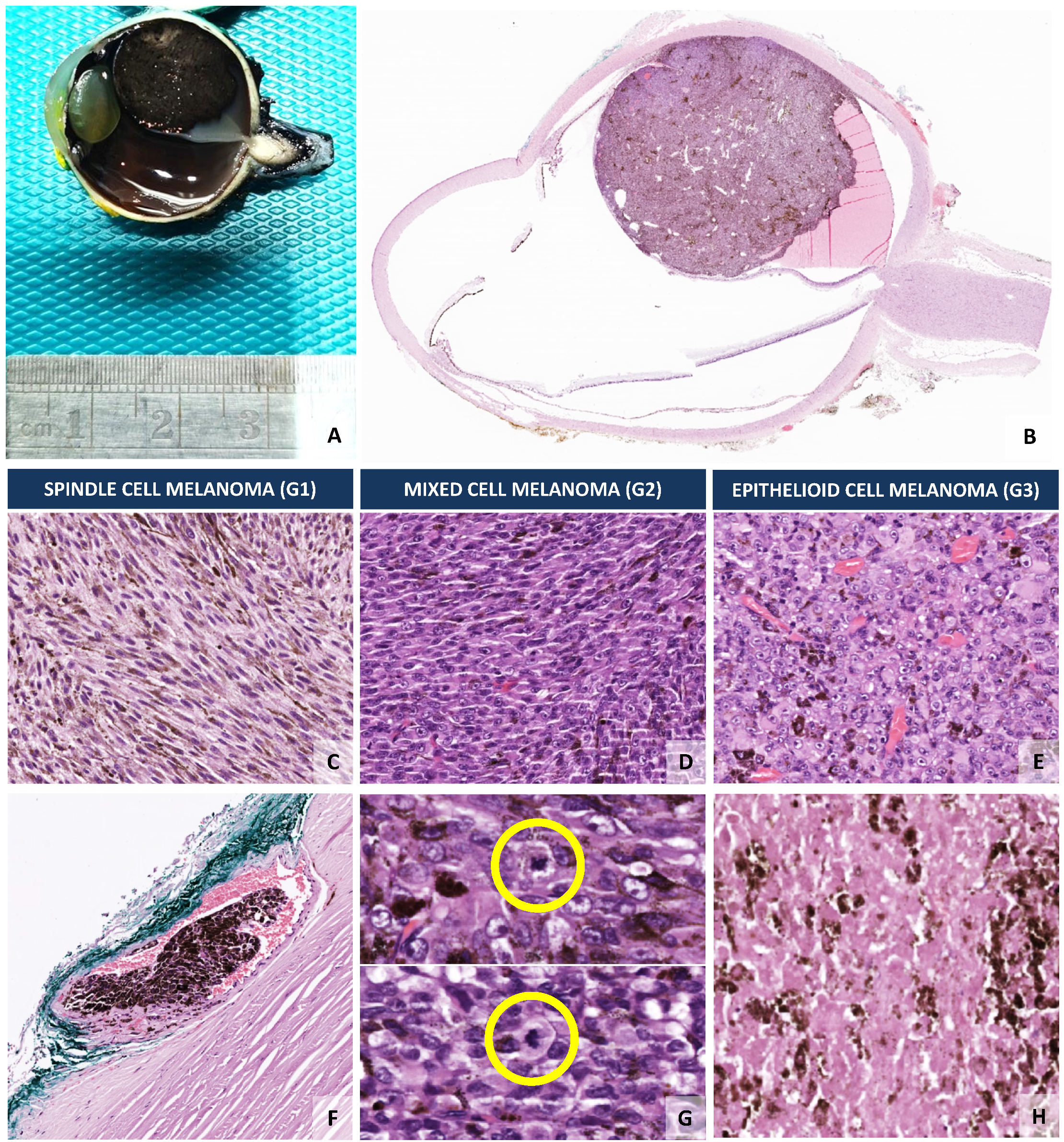
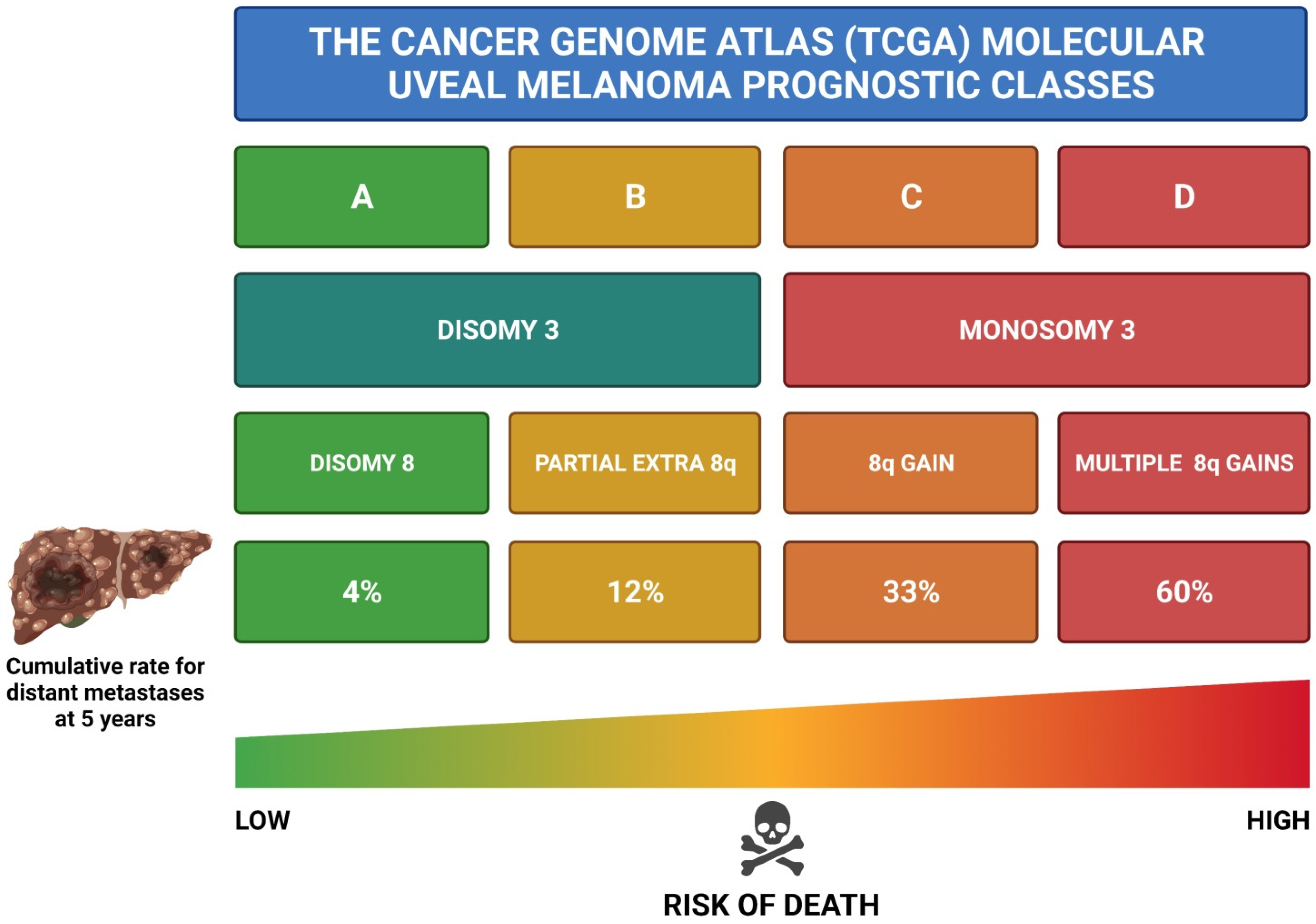
| Anatomical Site | % of Cases |
|---|---|
| Liver | 85 |
| Lung | 29 |
| Bones | 16 |
| Subcutaneous tissue | 12 |
| Lymph Nodes | 11 |
| Brain | 5 |
| Other sites | 13 |
| Multiple sites | 32 |
| Study | Gene Signature | Relevance of the Study |
|---|---|---|
| Wan et al., 2018 [148] | ABTB1, ADPRHL1, NTRK2 and SLC17A7 are hub genes in UM | Important diagnostic and prognostic markers for UM recurrence detection |
| Xue et al., 2019 [149] | AC010442.3, AC023790.2, AC092821.1, AL137784.1, CA12, FABP5P1, FAM189A2, GRIN2A, MGLL, MIR4655, MMP9, PARP8, RNF208, S100A13, SIRT3, TCTN1, ZBED1 and ZNF497 | Early identification of UM patients with poor and good prognosis |
| Ni et al., 2019 [150] | ABHD3, APOM, CALHM2, CENPV, CHAC1, HTR2B, HTRA3, LZTFL1, UBE2W, VCPIP1, ZNF391, ZNF415, ZNF667-AS1 and ZNF835 | Gene signature that allowed prediction of overall survival (OS) and recurrence-free survival (RFS) |
| Choi et al., 2020 [151] | CTNNB1, CYC1, NDUFB9 and NDUFV2 are hub genes in UM | Lower expression of CTNNB1 and increased expression of NDUFB9, NDUFV2 and CYC1 are associated with decreased survival of UM patients |
| Luo et al., 2020 [152] | ANXA2P2, CA12, HMCES, POMGNT2, RNF208, SIRT3, SLC44A3, STPG1, TCTN1 and ULBP1 | High expression of ANXA2P2, CA12 and ULBP1 and a low expression of HMCES, POMGNT2, RNF208, SIRT3, SLC44A3, STPG1 and TCTN1 are associated with higher metastatic risk and a shorter survival |
| Wan et al., 2020 [153] | CREG1, HIST1H4E, LZTS1, NIPA1, SH2D3A and TMEM201 | Low expression of LZTS1 and TMEM201 plus high expression of CREG1, HIST1H4E, NIPA1, SH2D3A are associated with decreased survival of UM patients |
| Tang and Cai, 2021 [154] | EIF1B, MEGF10, PHLDA1, RPL10A, RPL15, SLC25A38, TFDP2 and TIPARP | Robust prediction model of metastases-free survival |
| Jun Liu et al., 2021 [155] | ARPC1B, BTBD6, GUSB, KRTCAP2, RHBDD3 and SLC39A4 | Robust prediction model of OS for UM patients |
| Protein | Function | Relevant Conclusions of the Study |
|---|---|---|
| ABCB5 (ATP-binding cassette sub-family B member 5) [184] | P-glycoprotein involved in the transport of molecules across membranes Cancer stem cell marker | Higher expression of ABCB5 is associated with metastases development and worse prognosis |
| Adiponectin [185] | Anti-carcinogenic and insulin-sensitizing actions | Expression of Adiponectin and its receptor Adipor1 was decreased in cases of UM with M3, suggesting that the lower levels of adiponectin could boost the metastatic potential of UM |
| ATR (ataxia telangiectasia and Rad3-related) [186] | Member of the DNA damage response (DDR) protein machinery | Loss of nuclear ATR is associated with well-established markers of poor prognosis in UM (epithelioid cell morphology, increased tumour thickness, higher number of mitotic figures and BAP1 loss) |
| ATM (ataxia-telangiectasia mutated) [187] | Member of the DNA damage response (DDR) protein machinery | Loss of nuclear ATM is associated with well-established markers of poor prognosis in UM (epithelioid cell morphology, large tumour diameter above 10 mm, TILs and nuclear BAP1 loss) and a significant shorter DFS |
| Beclin-1 [188] | Protein involved in autophagy | Higher expression of Beclin-1 was correlated with a decreased risk of metastases and increased DFS times |
| BNIP3 (BCL2 19 kD protein-interacting protein 3) [189] | Mitochondrial protein involved in regulation of cell death, autophagy and cellular protection | Higher expression of BNIP3 was correlated with a shorter survival |
| BTNL9 (Butyrophilin-like protein 9) [190] | Modulator of T-cell mediated immune function | Higher expression of BTNL9 was significantly correlated with a better OS |
| c-Rel [191] | Member of the NF-κB pathway, which regulates a large array of genes implicated in cell survival, inflammatory disorders, response to infection, autoimmune disorders and cancer, among other processes | Nuclear expression of c-Rel expression was significantly associated with inferior survival |
| EphA1 (Eph-A1 receptor, erythropoietin-producing human hepatocellular receptor A1) [192] | Member of the Ephrin receptors, which are receptor tyrosine kinases (RTKs) that play a myriad of roles during the embryonic development (for example, in axon guidance, cell migration, segmentation and formation of tissue boundaries) and adulthood (for example, in angiogenesis, stem cell differentiation, regulation of the immune system and in cancer development) | Lower expression of EphA1 is associated with a worse prognosis |
| EphA5 (Eph-A5 receptor, erythropoietin-producing human hepatocellular receptor A5) [192] | Member of the Ephrin receptors, which are receptor tyrosine kinases (RTKs) that play a myriad of roles during the embryonic development (for example, in axon guidance, cell migration, segmentation and formation of tissue boundaries) and adulthood (for example, in angiogenesis, stem cell differentiation, regulation of the immune system and in cancer development) | Lower expression of EphA5 is associated with a worse prognosis |
| HDAC-2 (Histone Deacetylase 2) [193] | Regulation cellular proliferation, differentiation, angiogenesis and cell death, being implicated in neurodegeneration and cancer | Higher expression of HDAC-2 is an independent factor of better survival in UM |
| Nestin [194] | Intermediate filament protein marker of stem cells in the central nervous system and a cancer stem cell marker | Correlation between nestin positivity and well-established markers of bad prognosis (epithelioid cell morphology, higher mitotic counts, M3 and chromosome 8q gain) Nestin positivity in UM is associated with a worse prognosis |
| p50 [195] | Member of the NF-κB pathway, which regulates a large array of genes implicated in cell survival, inflammatory disorders, response to infection, autoimmune disorders and cancer, among other processes | Nuclear immunoreactivity of p50 significantly correlated with metastases development |
| p52 [196] | Member of the NF-κB pathway, which regulates a large array of genes implicated in cell survival, inflammatory disorders, response to infection, autoimmune disorders and cancer, among other processes | Expression of p52 was associated with BAP1 loss Higher p52 expression was associated with worse MFS and OS |
| p65 [195] | Member of the NF-κB pathway, which regulates a large array of genes implicated in cell survival, inflammatory disorders, response to infection, autoimmune disorders and cancer, among other processes | Nuclear immunoreactivity of p65 significantly correlated with metastases development |
| PARP1 [Poly(ADP-ribose) polymerase 1)] [197] | Involved in DNA repair and programmed cell death | Higher expression of PARP-1 is associated with decreased DFS and OS |
| PD-1 (Programmed cell death receptor-1) [198] | Involved in immune regulation | High expression of PD-1 in UM cells is associated with decreased DFS and OS |
| PD-L1 (Programmed cell death-ligand 1) [199] | Involved in immune regulation | PD-L1 immunoexpression was a significant prognostic factor of a reduced DFS |
| PLK-1 (Polio-like kinase 1) [200] | Kinase involved in the regulation of cell cycle | Low expression of PLK-1 was correlated with a higher TNM staging and a significantly decreased OS |
| PRDX3 (thioredoxin-dependent peroxidase reductase) [201] | Mitochondria protein with a fundamental role in the antioxidant defence of the cell | High PRDX3 expression is correlated with metastatic disease development and reduced OS |
| RelB [196] | Member of the NF-κB pathway, which regulates a large array of genes implicated in cell survival, inflammatory disorders, response to infection, autoimmune disorders and cancer, among other processes | Expression of RelB was associated with BAP1 loss and with inferior MFS |
| SPANX-C (Sperm protein associated with the nucleus on the X chromosome protein C) [202] | Belongs to a family of proteins expressed in the testis during spermatogenesis | Higher expression of SPANX-C in primary UM is associated with a decreased MFS |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lamas, N.J.; Martel, A.; Nahon-Estève, S.; Goffinet, S.; Macocco, A.; Bertolotto, C.; Lassalle, S.; Hofman, P. Prognostic Biomarkers in Uveal Melanoma: The Status Quo, Recent Advances and Future Directions. Cancers 2022, 14, 96. https://doi.org/10.3390/cancers14010096
Lamas NJ, Martel A, Nahon-Estève S, Goffinet S, Macocco A, Bertolotto C, Lassalle S, Hofman P. Prognostic Biomarkers in Uveal Melanoma: The Status Quo, Recent Advances and Future Directions. Cancers. 2022; 14(1):96. https://doi.org/10.3390/cancers14010096
Chicago/Turabian StyleLamas, Nuno Jorge, Arnaud Martel, Sacha Nahon-Estève, Samantha Goffinet, Adam Macocco, Corine Bertolotto, Sandra Lassalle, and Paul Hofman. 2022. "Prognostic Biomarkers in Uveal Melanoma: The Status Quo, Recent Advances and Future Directions" Cancers 14, no. 1: 96. https://doi.org/10.3390/cancers14010096
APA StyleLamas, N. J., Martel, A., Nahon-Estève, S., Goffinet, S., Macocco, A., Bertolotto, C., Lassalle, S., & Hofman, P. (2022). Prognostic Biomarkers in Uveal Melanoma: The Status Quo, Recent Advances and Future Directions. Cancers, 14(1), 96. https://doi.org/10.3390/cancers14010096








