A Neural Network Approach to Identify Glioblastoma Progression Phenotype from Multimodal MRI
Abstract
Simple Summary
Abstract
1. Introduction
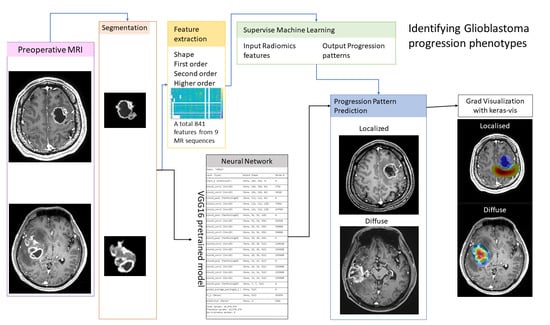
2. Materials and Methods
2.1. Patient Inclusion and Exclusion Criteria
2.2. MRI Acquisition and Imaging Processing
- Diffuse versus localized patterns: the diffuse progress pattern is defined as continuous progression ≥ 2 cm from the primary resection margin;
- Distal progression is defined as a separate progression 2 cm beyond the margin;
- Ventricular spread showed the progression of the tumor with contrast enhancement along the ventricular wall.
2.3. Radiomic Analysis and Machine Learning
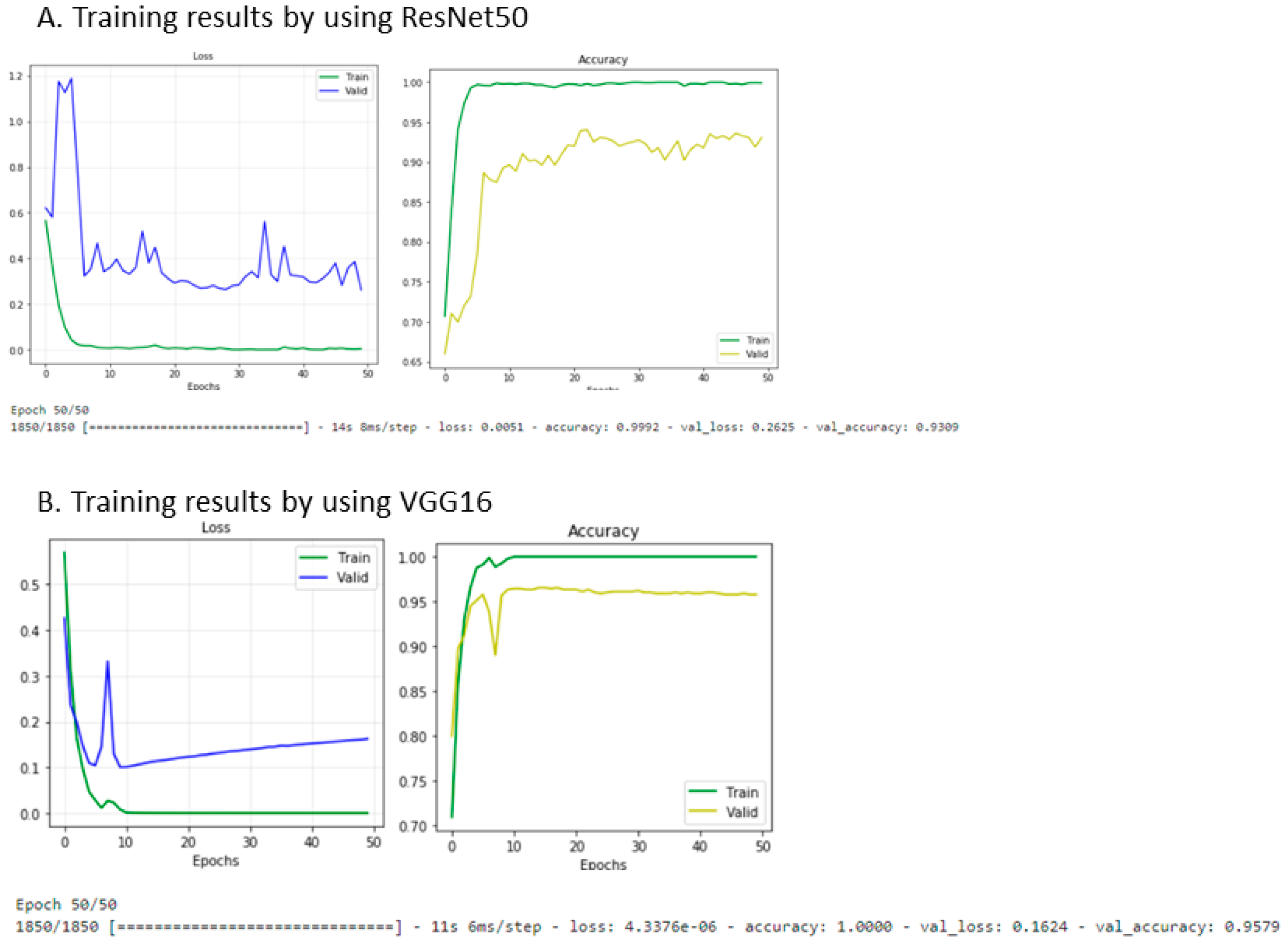
2.4. Neural Network Approach for Imaging Phenotype Prediction
3. Results
3.1. Patient Characteristics
3.2. The Clinical Impact of the GBM Progression Pattern
3.3. Radiomic Analysis and the Prediction Model of GBM Progression
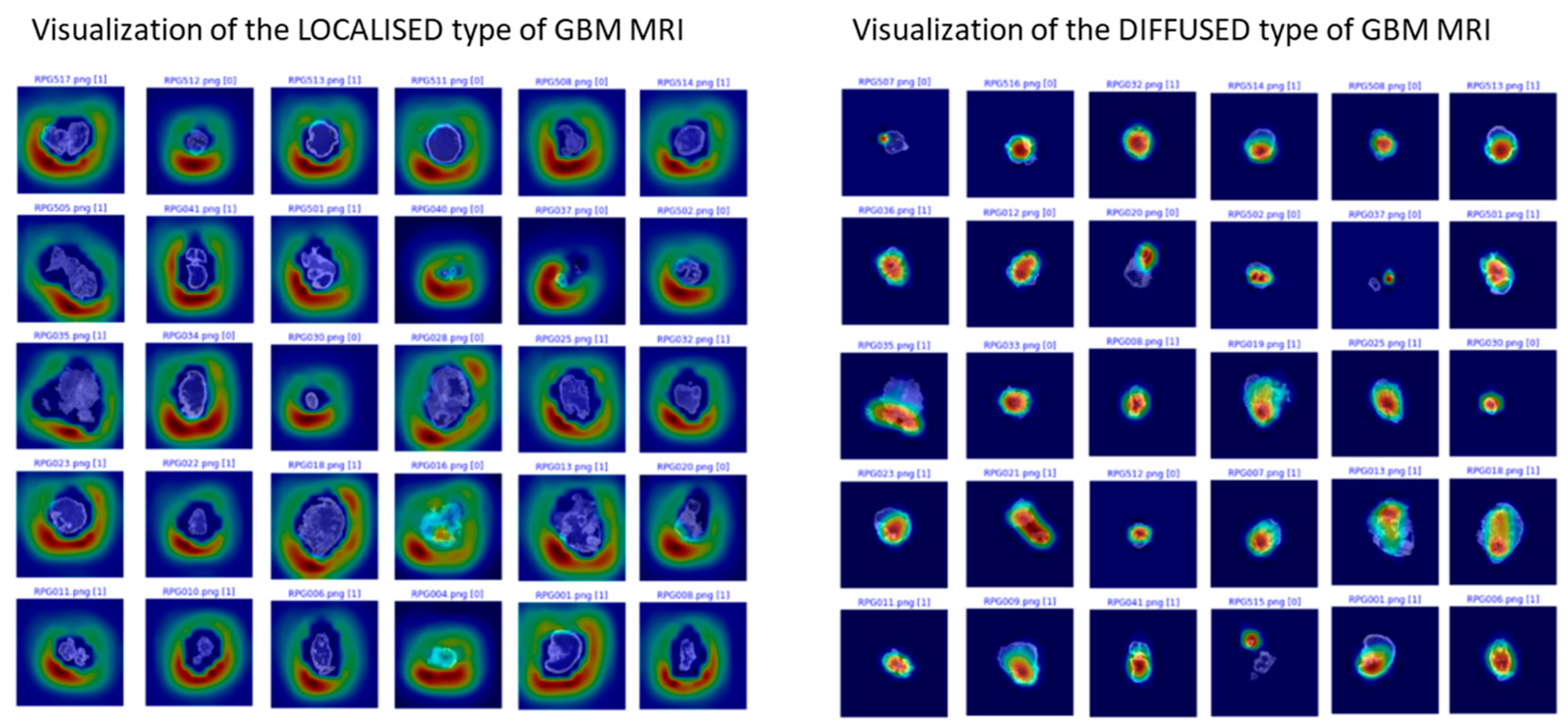
3.4. The Neural Network Approach for the Identification of the MR Progression Phenotype
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ostrom, Q.T.; Patil, N.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2013–2017. Neuro Oncol. 2020, 22, iv1–iv96. [Google Scholar] [CrossRef] [PubMed]
- Piper, R.J.; Senthil, K.K.; Yan, J.-L.; Price, S.J. Neuroimaging classification of progression patterns in glioblastoma: A systematic review. J. Neuro Oncol. 2018, 139, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Bette, S.; Barz, M.; Huber, T.; Straube, C.; Schmidt-Graf, F.; Combs, S.E.; Delbridge, C.; Gerhardt, J.; Zimmer, C.; Meyer, B.; et al. Retrospective Analysis of Radiological Recurrence Patterns in Glioblastoma, Their Prognostic Value and Association to Postoperative Infarct Volume. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Mabray, M.C.; Barajas, R.F.; Cha, S. Modern Brain Tumor Imaging. Brain Tumor Res. Treat. 2015, 3, 8–23. [Google Scholar] [CrossRef] [PubMed]
- Price, S.J.; Gillard, J.H. Imaging biomarkers of brain tumour margin and tumour invasion. Br. J. Radiol. 2011, 84, S159–S167. [Google Scholar] [CrossRef] [PubMed]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Srinivasan, G.N.; Shobha, G. Statistical Texture Analysis. Proc. World Acad. Sci. Eng. Technol. 2008, 36, 1264–1269. [Google Scholar]
- Bae, S.; Choi, Y.S.; Ahn, S.S.; Chang, J.H.; Kang, S.-G.; Kim, E.H.; Kim, S.H.; Lee, S.-K. Radiomic MRI Phenotyping of Glioblastoma: Improving Survival Prediction. Radiology 2018, 289, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Kazerooni, A.F.; Akbari, H.; Shukla, G.; Badve, C.; Rudie, J.D.; Sako, C.; Rathore, S.; Bakas, S.; Pati, S.; Singh, A.; et al. Cancer Imaging Phenomics via CaPTk: Multi-Institutional Prediction of Progression-Free Survival and Pattern of Recurrence in Glioblastoma. JCO Clin. Cancer Inf. 2020, 4, 234–244. [Google Scholar] [CrossRef]
- Yan, J.-L.; Li, C.; Van Der Hoorn, A.; Boonzaier, N.R.; Matys, T.; Price, S.J. A Neural Network Approach to Identify the Peritumoral Invasive Areas in Glioblastoma Patients by Using MR Radiomics. Sci. Rep. 2020, 10, 9748. [Google Scholar] [CrossRef] [PubMed]
- Jenkinson, M.; Beckmann, C.F.; Behrens, T.E.; Woolrich, M.W.; Smith, S.M. FSL. NeuroImage 2012, 62, 782–790. [Google Scholar] [CrossRef] [PubMed]
- Jenkinson, M.; Smith, S. A global optimisation method for robust affine registration of brain images. Med. Image Anal. 2001, 5, 143–156. [Google Scholar] [CrossRef]
- Jenkinsona, M.; Bannisterab, P.; Bradyb, M.; Smitha, S. Improved Optimization for the Robust and Accurate Linear Registration and Motion Correction of Brain Images. NeuroImage 2002, 17, 825–841. [Google Scholar] [CrossRef]
- Fedorov, A.; Beichel, R.; Kalpathy-Cramer, J.; Finet, J.; Fillion-Robin, J.-C.; Pujol, S.; Bauer, C.; Jennings, M.; Fennessy, F.; Sonka, M.; et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn. Reson. Imaging 2012, 30, 1323–1341. [Google Scholar] [CrossRef]
- Burger, P.C.; Dubois, P.J.; Schold, S.C.; Smith, K.R.; Odom, G.L.; Crafts, D.C.; Giangaspero, F. Computerized tomographic and pathologic studies of the untreated, quiescent, and recurrent glioblastoma multiforme. J. Neurosurg. 1983, 58, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Massey, V.; Wallner, K.E. Patterns of second recurrence of malignant astrocytomas. Int. J. Radiat. Oncol. 1990, 18, 395–398. [Google Scholar] [CrossRef]
- De Bonis, P.; Anile, C.; Pompucci, A.; Fiorentino, A.; Balducci, M.; Chiesa, S.; Lauriola, L.; Maira, G.; Mangiola, A. The influence of surgery on recurrence pattern of glioblastoma. Clin. Neurol. Neurosurg. 2013, 115, 37–43. [Google Scholar] [CrossRef]
- Dörner, L.; Mustafa, A.; Rohr, A.; Mehdorn, H.M.; Nabavi, A. Growth pattern of tumor recurrence following bis-chloroethylnitrosourea (BCNU) wafer implantation in malignant glioma. J. Clin. Neurosci. 2013, 20, 429–434. [Google Scholar] [CrossRef]
- Brandes, A.A.; Tosoni, A.; Franceschi, E.; Sotti, G.; Frezza, G.; Amistà, P.; Morandi, L.; Spagnolli, F.; Ermani, M. Recurrence Pattern After Temozolomide Concomitant with and Adjuvant to Radiotherapy in Newly Diagnosed Patients With Glioblastoma: Correlation With MGMT Promoter Methylation Status. J. Clin. Oncol. 2009, 27, 1275–1279. [Google Scholar] [CrossRef]
- Sherriff, J.; Tamangani, J.; Senthil, L.; Cruickshank, G.; Spooner, D.; Jones, B.; Brookes, C.; Sanghera, P. Patterns of relapse in glioblastoma multiforme following concomitant chemoradiotherapy with temozolomide. Br. J. Radiol. 2013, 86, 20120414. [Google Scholar] [CrossRef] [PubMed]
- Van Dijken, B.R.J.; Van Laar, P.J.; Li, C.; Yan, J.-L.; Boonzaier, N.R.; Price, S.J.; Van Der Hoorn, A. Fcrs Ventricle contact is associated with lower survival and increased peritumoral perfusion in glioblastoma. J. Neurosurg. 2019, 131, 717–723. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.; Grinband, J.; Weinberg, B.D.; Bardis, M.; Khy, M.; Cadena, G.; Su, M.-Y.; Cha, S.; Filippi, C.G.; Bota, D.; et al. Deep-Learning Convolutional Neural Networks Accurately Classify Genetic Mutations in Gliomas. Am. J. Neuroradiol. 2018, 39, 1201–1207. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, Y.; Yu, J.; Guo, Y.; Cao, W. Deep Learning based Radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Chang, K.; Beers, A.L.; Bai, H.X.; Brown, J.M.; Ly, K.I.; Li, X.; Senders, J.T.; Kavouridis, V.K.; Boaro, A.; Su, C.; et al. Automatic assessment of glioma burden: A deep learning algorithm for fully automated volumetric and bidimensional measurement. Neuro Oncol. 2019, 21, 1412–1422. [Google Scholar] [CrossRef] [PubMed]
- Kickingereder, P.; Isensee, F.; Tursunova, I.; Petersen, J.; Neuberger, U.; Bonekamp, D.; Brugnara, G.; Schell, M.; Kessler, T.; Foltyn, M.; et al. Automated quantitative tumour response assessment of MRI in neuro-oncology with artificial neural networks: A multicentre, retrospective study. Lancet Oncol. 2019, 20, 728–740. [Google Scholar] [CrossRef]
| Characteristics | Training | Validation Group | p-Value |
|---|---|---|---|
| Total number of patients | 41 | 18 | - |
| Males/females | 33/8 | 12/6 | 0.32 |
| Age (years) | 57.4 ± 13.4 | 56.8 ± 11.9 | 0.87 |
| Pre-OP tumor size (mL) | 42.7 ± 24.4 | 36.19 ± 19.46 | 0.42 |
| * GTR/STR | 26/15 | 11/7 | 1.00 |
| PFS (median, days) | 182 | 169.5 | 0.11 |
| OS (median, days) | 463 | 362.5 * (8 died) | 0.68 |
| MGMT unmethylated | 1 | 8 | - |
| methylated | 3 | 2 | - |
| IDH-1 wild type | 20 | 17 | 0.25 |
| mutated | 3 | 0 | - |
| Progression Pattern | Number | Overall Survival (Median, Days) | Progression Free Survival (Median, Days) | ||
|---|---|---|---|---|---|
| Diffuse | 39 | 363 | p = 0.032 | 189.5 | p = 0.02 |
| Local | 20 | 668 | - | 136 | - |
| Ventricular spread | 22 | 354 | p = 0.05 | 190 | p = 0.12 |
| No ventricular spread | 37 | 180 | - | 182 | - |
| Uni-direction | 20 | 490 | p = 0.66 | 185 | p = 0.98 |
| Multidirection | 39 | 449.5 | - | 173 | - |
| Distal | 10 | 558 | p = 0.01 | 185 | p = 0.19 |
| No distal progression | 49 | 449.5 | - | 173 | - |
| Train Model | Overall Accuracy | Sensitivity | Specificity | AUC |
|---|---|---|---|---|
| Linear SVM | 77.5% | 84.6% | 64.3% | 0.89 |
| Regression | 82.5% | 85.7% | 75% | 0.84 |
| KNN | 82.5% | 85.7% | 75% | 0.88 |
| Boosted trees | 80.0% | 82.8% | 72.2% | 0.83 |
| Machine Learning Models | Results | Accuracy | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| True | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | Ground Truth |
| Logistic regression | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 55.6% |
| SVM | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 61.1% |
| Tree | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 72.2% |
| KNN | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 61.1% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, J.-L.; Toh, C.-H.; Ko, L.; Wei, K.-C.; Chen, P.-Y. A Neural Network Approach to Identify Glioblastoma Progression Phenotype from Multimodal MRI. Cancers 2021, 13, 2006. https://doi.org/10.3390/cancers13092006
Yan J-L, Toh C-H, Ko L, Wei K-C, Chen P-Y. A Neural Network Approach to Identify Glioblastoma Progression Phenotype from Multimodal MRI. Cancers. 2021; 13(9):2006. https://doi.org/10.3390/cancers13092006
Chicago/Turabian StyleYan, Jiun-Lin, Cheng-Hong Toh, Li Ko, Kuo-Chen Wei, and Pin-Yuan Chen. 2021. "A Neural Network Approach to Identify Glioblastoma Progression Phenotype from Multimodal MRI" Cancers 13, no. 9: 2006. https://doi.org/10.3390/cancers13092006
APA StyleYan, J.-L., Toh, C.-H., Ko, L., Wei, K.-C., & Chen, P.-Y. (2021). A Neural Network Approach to Identify Glioblastoma Progression Phenotype from Multimodal MRI. Cancers, 13(9), 2006. https://doi.org/10.3390/cancers13092006