Targeting Replicative Stress and DNA Repair by Combining PARP and Wee1 Kinase Inhibitors Is Synergistic in Triple Negative Breast Cancers with Cyclin E or BRCA1 Alteration
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
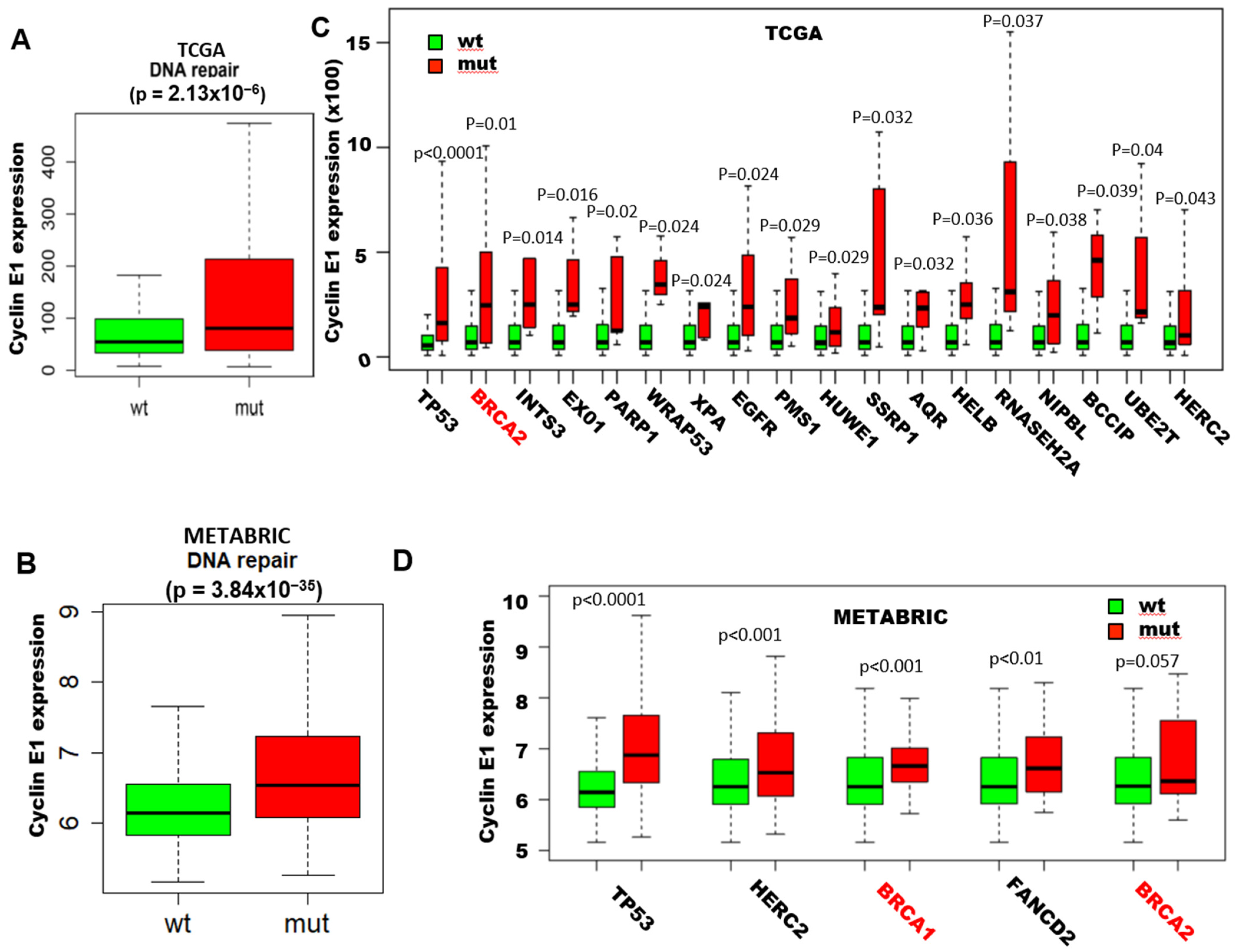
2.1. High Cyclin E RNA and Protein Levels Predict Poor Outcome in Breast Cancer Patients with BRCA1/2 Mutations
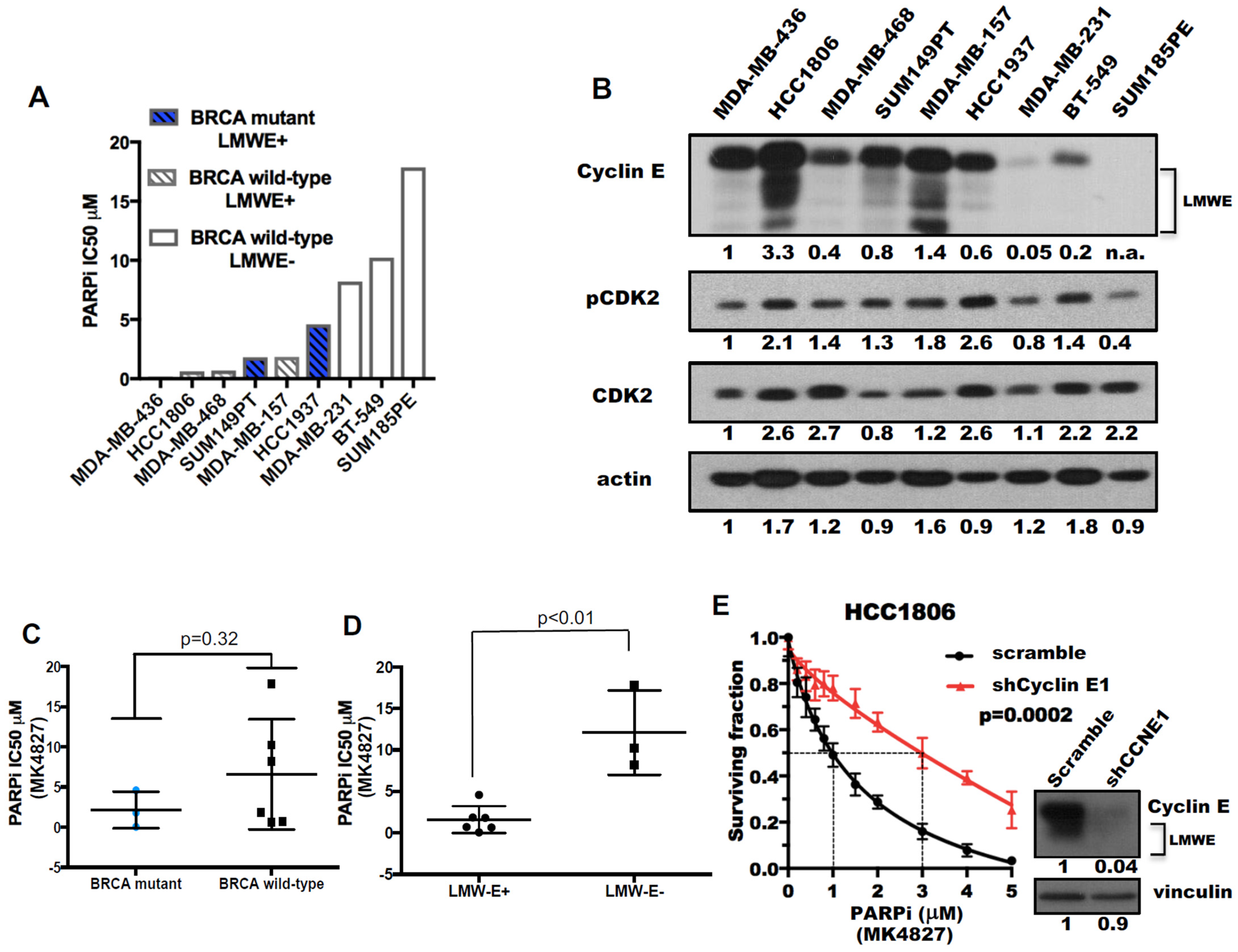
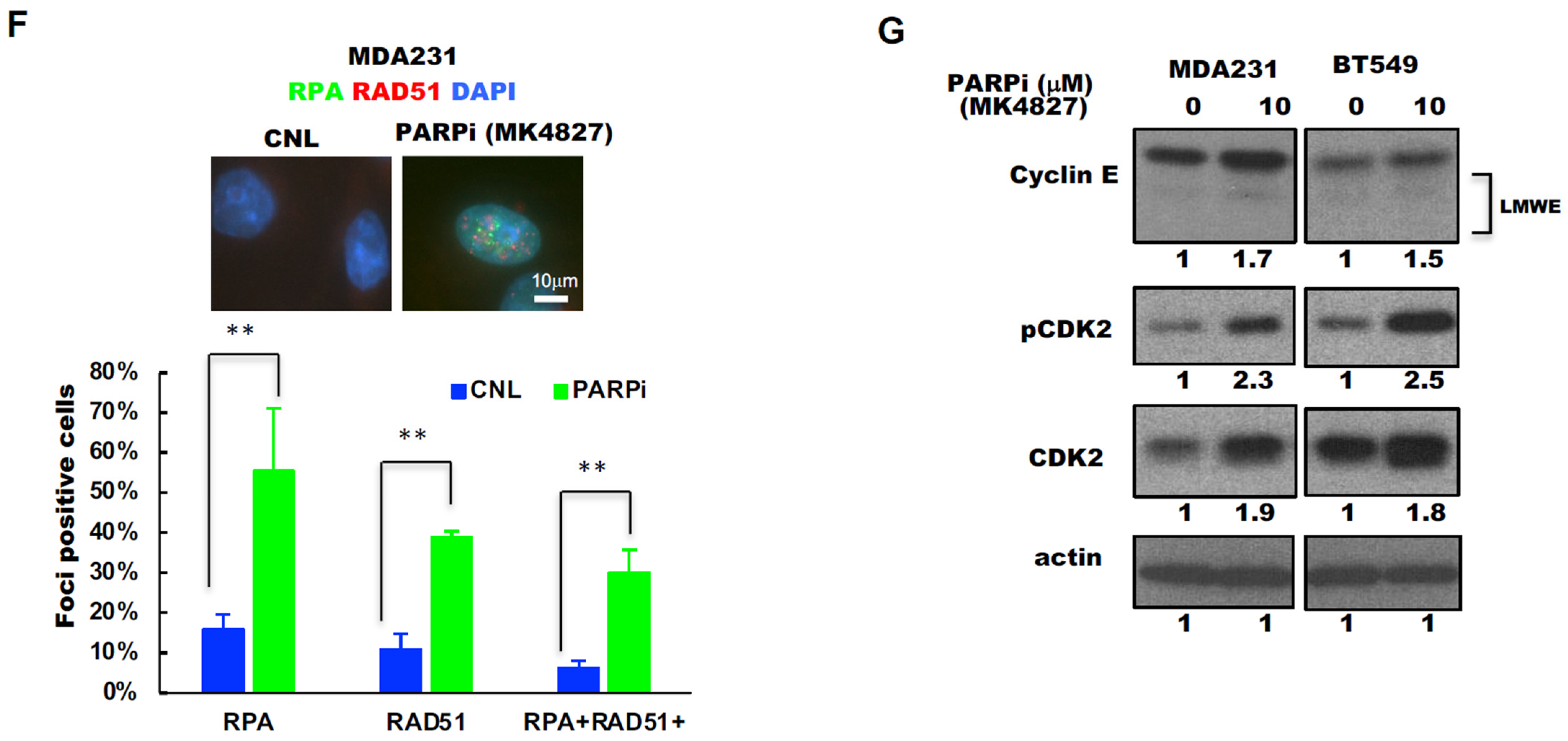
2.2. Triple-Negative Breast Cancers (TNBC) with Low-Molecular-Weight Cyclin E (LMWE) Are Sensitive to PARP Inhibition
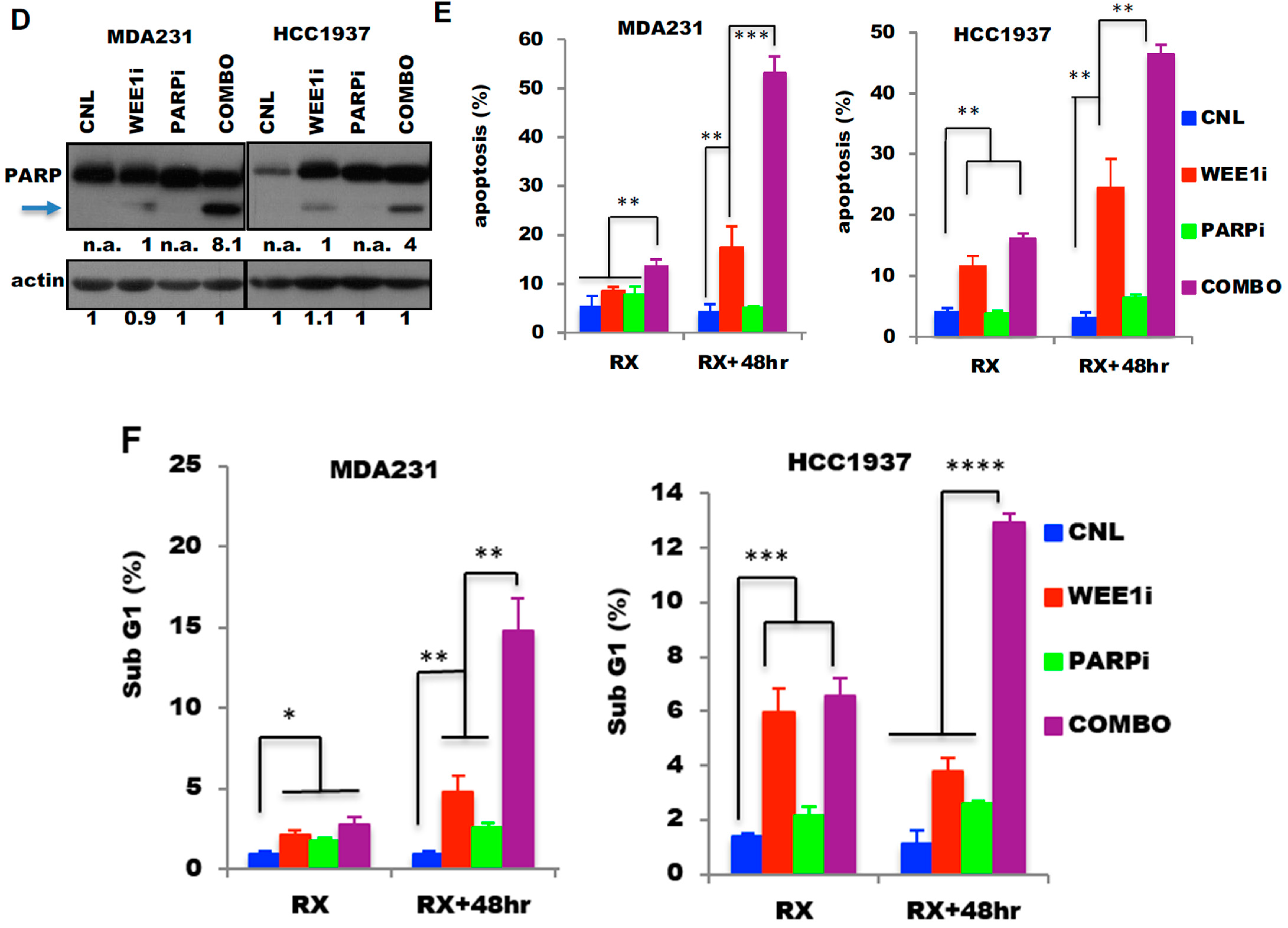
2.3. The Combination of PARP Inhibitor (PARPi) and Wee1 Kinase Inhibitor (Wee1i) Is Synergistic by Inducing Apoptosis in TNBC Cells
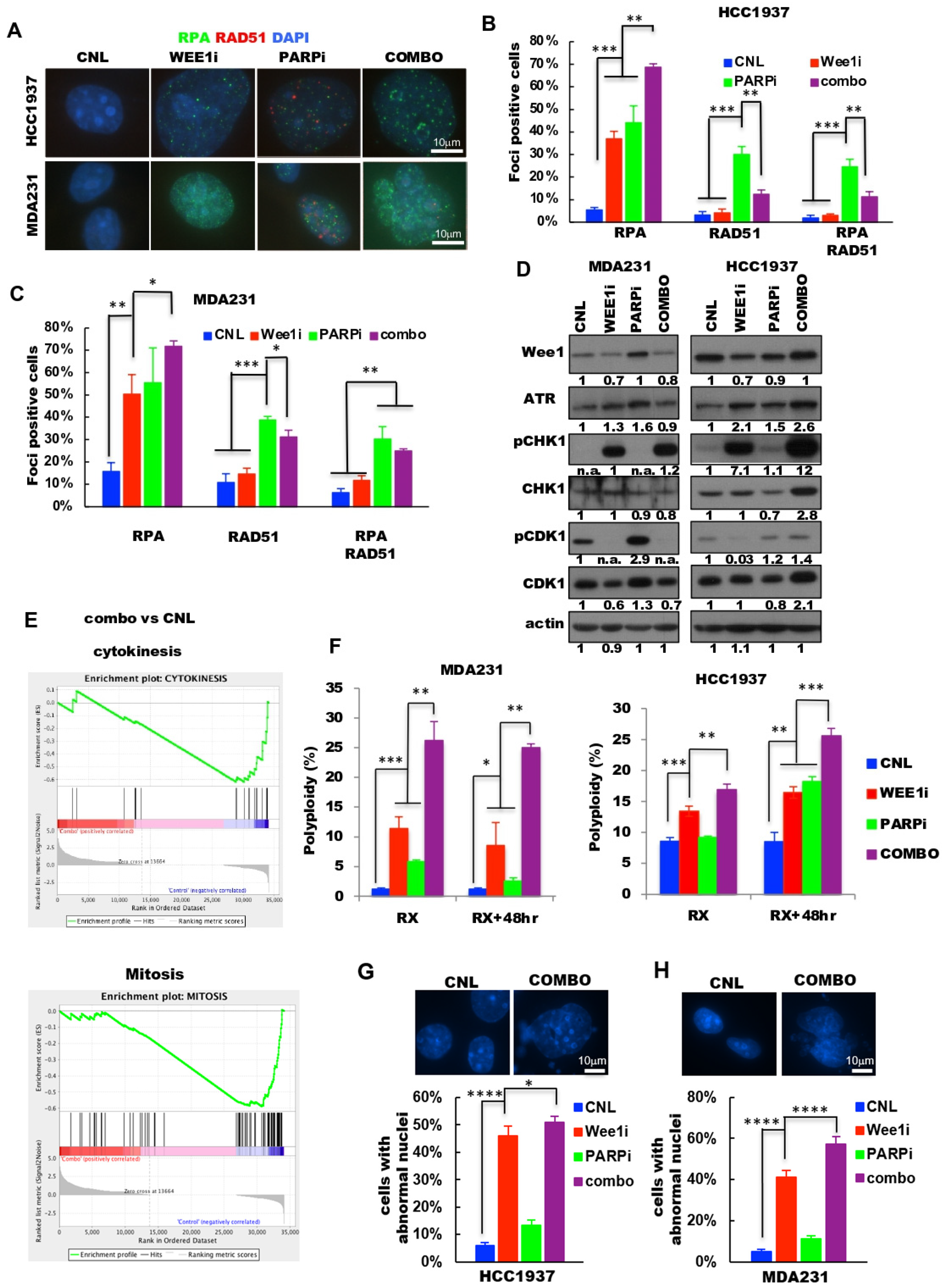
2.4. Combination of PARP and Wee1 Kinase Inhibition Increases DNA Replicative Stress and Polyploidy
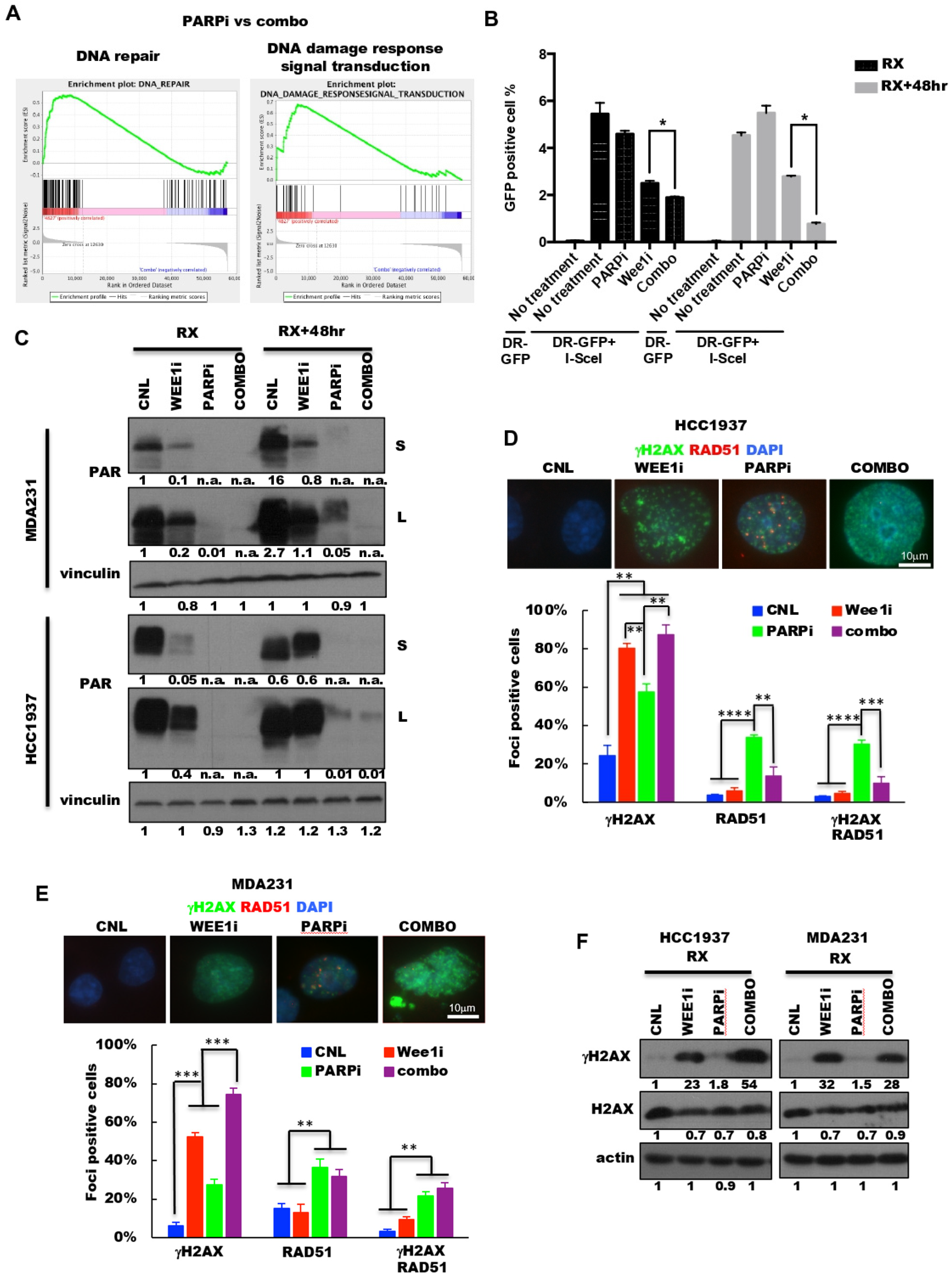
2.5. PARP and Wee1 Kinase Inhibition Deregulates DNA Repair and Increases DNA Damage
2.6. The Combination of PARP and Wee1 Kinase Inhibition Inhibited Tumor Growth and Improved Survival In Vivo
3. Discussion
4. Material and Methods
4.1. Clinical Samples
4.2. Immunohistochemistry (IHC) and Scoring of Cyclin E
4.3. Cell Lines
4.4. High-Throughput Survival Assay (HTSA)
4.5. Immunoblot Assays
4.6. ShRNA Knockdown
4.7. Immunofluorescence Staining
4.8. Cell Cycle Analysis
4.9. Annexin/Propidium Iodide Assay
4.10. Homologous Recombination (HR) Assay
4.11. Animal Studies
4.12. RNA Sequencing and Gene Set Enrichment Analysis (GSEA)
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Costa, R.L.; Gradishar, W.J. Triple-Negative Breast Cancer: Current Practice and Future Directions. J. Oncol. Pract. 2017, 13, 301–303. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; O’Shaughnessy, J.; Zhao, J.; Haiderali, A.; Cortés, J.; Ramsey, S.D.; Briggs, A.; Hu, P.; Karantza, V.; Aktan, G.; et al. Association of Pathologic Complete Response with Long-Term Survival Outcomes in Triple-Negative Breast Cancer: A Meta-Analysis. Cancer Res. 2020, 80, 5427–5434. [Google Scholar] [CrossRef]
- Chen, B.V.E.; Gillespie, E.F.; Zakeri, K.; Murphy, J.D.; Yashar, C.M.; Lu, S.; Einck, J.P. Pathologic response after neoadjuvant chemotherapy predicts locoregional control in patients with triple negative breast cancer. Adv. Radiat. Oncol. 2017, 2, 105–109. [Google Scholar] [CrossRef]
- Kwon, S.J.; Gutierrez-Barrera, A.M.; Young, D.; Sun, C.C.; Daniels, M.S.; Lu, K.H.; Arun, B. Expanding the Criteria for Brca Mutation Testing in Breast Cancer Survivors. J. Clin. Oncol. 2010, 28, 4214–4220. [Google Scholar] [CrossRef] [PubMed]
- Eskiler, G.G.; Cecener, G.; Egeli, U.; Tunca, B. Triple Negative Breast Cancer: New Therapeutic Approaches and Brca Status. APMIS 2018, 126, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Ollier, M.; Radosevic-Robin, N.; Kwiatkowski, F.; Ponelle, F.; Viala, S.; Privat, M.; Uhrhammer, N.; Bernard-Gallon, D.; Penault-Llorca, F.; Bignon, Y.J.; et al. DNA Repair Genes Implicated in Triple Negative Familial Non-Brca1/2 Breast Cancer Predisposition. Am. J. Cancer Res. 2015, 5, 2113–2126. [Google Scholar] [PubMed]
- Couch, F.J.; Hart, S.N.; Sharma, P.; Toland, A.E.; Wang, X.; Miron, P.; Olson, J.E.; Godwin, A.K.; Pankratz, V.S.; Olswold, C.; et al. Inherited Mutations in 17 Breast Cancer Susceptibility Genes Among a Large Triple-Negative Breast Cancer Cohort Unselected for Family History of Breast Cancer. J. Clin. Oncol. 2015, 33, 304–311. [Google Scholar] [CrossRef]
- Farmer, H.; McCabe, N.; Lord, C.J.; Tutt, A.N.J.; Johnson, D.A.; Richardson, T.B.; Santarosa, M.; Dillon, K.J.; Hickson, I.D.; Knights, C.; et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nat. Cell Biol. 2005, 434, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Sonnenblick, A.; De Azambuja, E.; Azim, H.A.; Piccart, M. An Update on Parp Inhibitors—Moving to the Adjuvant Setting. Nat. Rev. Clin. Oncol. 2015, 12, 27–41. [Google Scholar] [CrossRef]
- Minami, C.A.; Chung, D.U.; Chang, H.R. Management Options in Triple-Negative Breast Cancer. Breast Cancer Basic Clin. Res. 2011, 5, 175–199. [Google Scholar] [CrossRef]
- Papadimitriou, M.; Mountzios, G.; Papadimitriou, C.A. The role of PARP inhibition in triple-negative breast cancer: Unraveling the wide spectrum of synthetic lethality. Cancer Treat. Rev. 2018, 67, 34–44. [Google Scholar] [CrossRef]
- Litton, J.K.; Rugo, H.S.; Ettl, J.; Hurvitz, S.A.; Gonçalves, A.; Lee, K.-H.; Fehrenbacher, L.; Yerushalmi, R.; Mina, L.A.; Martin, M.; et al. Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. N. Engl. J. Med. 2018, 379, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, B.; Shapira-Frommer, R.; Schmutzler, R.K.; Audeh, M.W.; Friedlander, M.; Balmaña, J.; Mitchell, G.; Fried, G.; Stemmer, S.M.; Hubert, A.; et al. Olaparib Monotherapy in Patients with Advanced Cancer and a Germline Brca1/2 Mutation. J. Clin. Oncol. 2015, 33, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Tutt, A.; Robson, M.E.; Garber, J.; Domchek, S.M.; Audeh, M.W.; Weitzel, J.N.; Friedlander, M.; Arun, B.; Loman, N.; Schmutzler, R.K.; et al. Oral poly (ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: A proof-of-concept trial. Lancet 2010, 376, 235–244. [Google Scholar] [CrossRef]
- Diéras, V.; Han, H.S.; Kaufman, B.; Wildiers, H.; Friedlander, M.; Ayoub, J.-P.; Puhalla, S.L.; Bondarenko, I.; Campone, M.; Jakobsen, E.H.; et al. Veliparib with carboplatin and paclitaxel in BRCA-mutated advanced breast cancer (BROCADE3): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2020, 21, 1269–1282. [Google Scholar] [CrossRef]
- Robson, M.; Im, S.-A.; Senkus, E.; Xu, B.; Domchek, S.M.; Masuda, N.; Delaloge, S.; Li, W.; Tung, N.; Armstrong, A.; et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N. Engl. J. Med. 2017, 377, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Keyomarsi, K.; Pardee, A.B. Redundant cyclin overexpression and gene amplification in breast cancer cells. Proc. Natl. Acad. Sci. USA 1993, 90, 1112–1116. [Google Scholar] [CrossRef]
- Porter, D.C.; Zhang, N.; Danes, C.; McGahren, M.J.; Harwell, R.M.; Faruki, S.; Keyomarsi, K. Tumor-Specific Proteolytic Processing of Cyclin E Generates Hyperactive Lower-Molecular-Weight Forms. Mol. Cell. Biol. 2001, 21, 6254–6269. [Google Scholar] [CrossRef] [PubMed]
- Harwell, R.M.; Porter, D.C.; Danes, C.; Keyomarsi, K. Processing of cyclin E differs between normal and tumor breast cells. Cancer Res. 2000, 60, 481–489. [Google Scholar] [PubMed]
- Mittendorf, E.A.; Alatrash, G.; Qiao, N.; Wu, Y.; Sukhumalchandra, P.; John, L.S.S.; Philips, A.V.; Xiao, H.; Zhang, M.; Ruisaard, K.; et al. Breast Cancer Cell Uptake of the Inflammatory Mediator Neutrophil Elastase Triggers an Anticancer Adaptive Immune Response. Cancer Res. 2012, 72, 3153–3162. [Google Scholar] [CrossRef]
- Wang, X.D.; Rosales, J.L.; Magliocco, A.; Gnanakumar, R.; Lee, K.-Y. Cyclin E in breast tumors is cleaved into its low molecular weight forms by calpain. Oncogene 2003, 22, 769–774. [Google Scholar] [CrossRef] [PubMed]
- Libertini, S.J.; Robinson, B.S.; Dhillon, N.K.; Glick, D.; George, M.; Dandekar, S.; Gregg, J.P.; Sawai, E.; Mudryj, M. Cyclin E Both Regulates and Is Regulated by Calpain 2, a Protease Associated with Metastatic Breast Cancer Phenotype. Cancer Res. 2005, 65, 10700–10708. [Google Scholar] [CrossRef]
- Corin, I.; Di Giacomo, M.C.; Lastella, P.; Bagnulo, R.; Guanti, G.; Simone, C. Tumor-specific hyperactive low-molecular-weight cyclin E isoform detection and characterization in non-metastatic colorectal tumors. Cancer Biol. Ther. 2006, 5, 198–203. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhou, Y.; Xie, Y.; Gu, J.; Yan, L.; Guan, G.; Liu, X. Overexpression of cyclin E isoforms correlates with poor prognosis in rectal cancer. Eur. J. Surg. Oncol. 2011, 37, 1078–1084. [Google Scholar] [CrossRef] [PubMed]
- Caruso, J.A.; Duong, M.T.; Carey, J.P.W.; Hunt, K.K.; Keyomarsi, K. Low-Molecular-Weight Cyclin E in Human Cancer: Cellular Consequences and Opportunities for Targeted Therapies. Cancer Res. 2018, 78, 5481–5491. [Google Scholar] [CrossRef]
- Karakas, C.; Francis, A.M.; Ha, M.J.; Wingate, H.F.; Meena, R.A.; Yi, M.; Rasaputra, K.S.; Barrera, A.M.G.; Arun, B.; Do, K.-A.; et al. Cytoplasmic Cyclin E Expression Predicts for Response to Neoadjuvant Chemotherapy in Breast Cancer. Ann. Surg. 2020. [Google Scholar] [CrossRef]
- Hunt, K.K.; Karakas, C.; Ha, M.J.; Biernacka, A.; Yi, M.; Sahin, A.A.; Adjapong, O.; Hortobagyi, G.N.; Bondy, M.L.; Thompson, P.A.; et al. Cytoplasmic Cyclin E Predicts Recurrence in Patients with Breast Cancer. Clin. Cancer Res. 2017, 23, 2991–3002. [Google Scholar] [CrossRef]
- Karakas, C.; Biernacka, A.; Bui, T.; Sahin, A.A.; Yi, M.; Akli, S.; Schafer, J.; Alexander, A.; Adjapong, O.; Hunt, K.K.; et al. Cytoplasmic Cyclin E and Phospho-Cyclin-Dependent Kinase 2 Are Biomarkers of Aggressive Breast Cancer. Am. J. Pathol. 2016, 186, 1900–1912. [Google Scholar] [CrossRef]
- Keyomarsi, K.; Tucker, S.L.; Buchholz, T.A.; Callister, M.; Ding, Y.; Hortobagyi, G.N.; Bedrosian, I.; Knickerbocker, C.; Toyofuku, W.; Lowe, M.; et al. Cyclin E and Survival in Patients with Breast Cancer. N. Engl. J. Med. 2002, 347, 1566–1575. [Google Scholar] [CrossRef]
- Romagnolo, A.P.; Romagnolo, D.F.; Selmin, O.I. BRCA1 as Target for Breast Cancer Prevention and Therapy. Anti Cancer Agents Med. Chem. 2015, 15, 4–14. [Google Scholar] [CrossRef] [PubMed]
- De Talhouet, S.; Peron, J.; Vuilleumier, A.; Friedlaender, A.; Viassolo, V.; Ayme, A.; Bodmer, A.; Treilleux, I.; Lang, N.; Tille, J.C.; et al. Clinical Outcome of Breast Cancer in Carriers of Brca1 and Brca2 Mutations According to Molecular Subtypes. Sci. Rep. 2020, 10, 7073. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.R.; Hashimoto, Y.; Herrador, R.; Neelsen, K.J.; Fachinetti, D.; Bermejo, R.; Cocito, A.; Costanzo, V.; Lopes, M. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat. Struct. Mol. Biol. 2012, 19, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Berti, M.; Chaudhuri, A.R.; Thangavel, S.; Gomathinayagam, S.; Kenig, S.; Vujanovic, M.; Odreman, F.; Glatter, T.; Graziano, S.; Mendoza-Maldonado, R.; et al. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat. Struct. Mol. Biol. 2013, 20, 347–354. [Google Scholar] [CrossRef]
- Parsels, L.A.; Karnak, D.; Parsels, J.D.; Zhang, Q.; Vélez-Padilla, J.; Reichert, Z.R.; Wahl, D.R.; Maybaum, J.; O’Connor, M.J.; Lawrence, T.S.; et al. PARP1 Trapping and DNA Replication Stress Enhance Radiosensitization with Combined WEE1 and PARP Inhibitors. Mol. Cancer Res. 2018, 16, 222–232. [Google Scholar] [CrossRef]
- Colicchia, V.; Petroni, M.; Guarguaglini, G.; Sardina, F.; Sahún-Roncero, M.; Carbonari, M.; Ricci, B.; Heil, C.; Capalbo, C.; Belardinilli, F.; et al. PARP inhibitors enhance replication stress and cause mitotic catastrophe in MYCN-dependent neuroblastoma. Oncogene 2017, 36, 4682–4691. [Google Scholar] [CrossRef]
- Neelsen, K.J.; Zanini, I.M.; Herrador, R.; Lopes, M. Oncogenes induce genotoxic stress by mitotic processing of unusual replication intermediates. J. Cell Biol. 2013, 200, 699–708. [Google Scholar] [CrossRef]
- Hills, A.S.; Diffley, J.F. DNA Replication and Oncogene-Induced Replicative Stress. Curr. Biol. 2014, 24, R435–R444. [Google Scholar] [CrossRef]
- Chen, X.; Low, K.-H.; Alexander, A.; Jiang, Y.; Karakas, C.; Hess, K.R.; Carey, J.P.; Bui, T.N.; Vijayaraghavan, S.; Evans, K.W.; et al. Cyclin E Overexpression Sensitizes Triple-Negative Breast Cancer to Wee1 Kinase Inhibition. Clin. Cancer Res. 2018, 24, 6594–6610. [Google Scholar] [CrossRef]
- Chow, J.P.H.; Poon, R.Y.C. The CDK1 inhibitory kinase MYT1 in DNA damage checkpoint recovery. Oncogene 2013, 32, 4778–4788. [Google Scholar] [CrossRef]
- Russell, P.; Nurse, P. Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell 1987, 49, 559–567. [Google Scholar] [CrossRef]
- Aarts, M.; Sharpe, R.; Garcia-Murillas, I.; Gevensleben, H.; Hurd, M.S.; Shumway, S.D.; Toniatti, C.; Ashworth, A.; Turner, N.C. Forced Mitotic Entry of S-Phase Cells as a Therapeutic Strategy Induced by Inhibition of WEE1. Cancer Discov. 2012, 2, 524–539. [Google Scholar] [CrossRef] [PubMed]
- Mir, S.E.; Hamer, P.C.D.W.; Krawczyk, P.M.; Balaj, L.; Claes, A.; Niers, J.M.; Van Tilborg, A.A.; Zwinderman, A.H.; Geerts, D.; Kaspers, G.J.; et al. In Silico Analysis of Kinase Expression Identifies WEE1 as a Gatekeeper against Mitotic Catastrophe in Glioblastoma. Cancer Cell 2010, 18, 244–257. [Google Scholar] [CrossRef]
- Krajewska, M.; Heijink, A.M.; Bisselink, Y.J.W.M.I.; Seinstra, R.; Sillje, H.H.W.; De Vries, E.G.E.; Van Vugt, M.A.T.M. Forced activation of Cdk1 via wee1 inhibition impairs homologous recombination. Oncogene 2013, 32, 3001–3008. [Google Scholar] [CrossRef]
- Domínguez-Kelly, R.; Martín, Y.; Koundrioukoff, S.; Tanenbaum, M.E.; Smits, V.A.; Medema, R.H.; Debatisse, M.; Freire, R. Wee1 controls genomic stability during replication by regulating the Mus81-Eme1 endonuclease. J. Cell Biol. 2011, 194, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Kogiso, T.; Nagahara, H.; Hashimoto, E.; Ariizumi, S.; Yamamoto, M.; Shiratori, K. Efficient Induction of Apoptosis by Wee1 Kinase Inhibition in Hepatocellular Carcinoma Cells. PLoS ONE 2014, 9, e100495. [Google Scholar] [CrossRef]
- Harris, P.S.; Venkataraman, S.; Alimova, I.; Birks, D.K.; Balakrishnan, I.; Cristiano, B.; Donson, A.M.; Dubuc, A.M.; Taylor, M.D.; Foreman, N.K.; et al. Integrated genomic analysis identifies the mitotic checkpoint kinase WEE1 as a novel therapeutic target in medulloblastoma. Mol. Cancer 2014, 13, 72. [Google Scholar] [CrossRef]
- Garcia, T.B.; Snedeker, J.C.; Baturin, D.; Gardner, L.; Fosmire, S.P.; Zhou, C.; Jordan, C.T.; Venkataraman, S.; Vibhakar, R.; Porter, C.C. A Small-Molecule Inhibitor of WEE1, AZD1775, Synergizes with Olaparib by Impairing Homologous Recombination and Enhancing DNA Damage and Apoptosis in Acute Leukemia. Mol. Cancer Ther. 2017, 16, 2058–2068. [Google Scholar] [CrossRef]
- Karnak, D.; Engelke, C.G.; Parsels, L.A.; Kausar, T.; Wei, D.; Robertson, K.B., Jr.; Marsh, M.A.; Davis, L.; Zhao, J.; Maybaum, T.S.; et al. Combined Inhibition of Wee1 and Parp1/2 for Radiosensitization in Pancreatic Cancer. Clin. Cancer Res. 2014, 20, 5085–5096. [Google Scholar] [CrossRef]
- Fang, Y.; McGrail, D.J.; Sun, C.; Labrie, M.; Chen, X.; Zhang, D.; Ju, Z.; Vellano, C.P.; Lu, Y.; Li, Y.; et al. Sequential Therapy with PARP and WEE1 Inhibitors Minimizes Toxicity while Maintaining Efficacy. Cancer Cell 2019, 35, 851–867.e7. [Google Scholar] [CrossRef] [PubMed]
- McAlpine, J.N.; Porter, H.; Köbel, M.; Nelson, B.H.; Prentice, L.M.; E Kalloger, S.; Senz, J.; Milne, K.; Ding, J.; Shah, S.P.; et al. BRCA1 and BRCA2 mutations correlate with TP53 abnormalities and presence of immune cell infiltrates in ovarian high-grade serous carcinoma. Mod. Pathol. 2012, 25, 740–750. [Google Scholar] [CrossRef] [PubMed]
- Greenblatt, M.S.O.; Chappuis, P.; Bond, J.P.; Hamel, N.; Foulkes, W.D. TP53 mutations in breast cancer associated with BRCA1 or BRCA2 germ-line mutations: Distinctive spectrum and structural distribution. Cancer Res. 2001, 61, 4092–4097. [Google Scholar] [PubMed]
- Martin, A.-M.A.; Kanetsky, P.; Amirimani, B.A.; Colligon, T.; Athanasiadis, G.A.; Shih, H.; Gerrero, M.R.; Calzone, K.; Rebbeck, T.R.; Weber, B.L. Germline TP53 mutations in breast cancer families with multiple primary cancers: Is TP53 a modifier of BRCA1? J. Med. Genet. 2003, 40, e34. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jabbour-Leung, N.A.; Chen, X.; Bui, T.; Jiang, Y.; Yang, D.; Vijayaraghavan, S.; McArthur, M.J.; Hunt, K.K.; Keyomarsi, K. Sequential Combination Therapy of CDK Inhibition and Doxorubicin Is Synthetically Lethal in p53-Mutant Triple-Negative Breast Cancer. Mol. Cancer Ther. 2016, 15, 593–607. [Google Scholar] [CrossRef]
- Gibson, B.A.; Kraus, W.L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012, 13, 411–424. [Google Scholar] [CrossRef]
- Elstrodt, F.; Hollestelle, A.; Nagel, J.H.; Gorin, M.; Wasielewski, M.; Ouweland, A.V.D.; Merajver, S.D.; Ethier, S.P.; Schutte, M. BRCA1 Mutation Analysis of 41 Human Breast Cancer Cell Lines Reveals Three New Deleterious Mutants. Cancer Res. 2006, 66, 41–45. [Google Scholar] [CrossRef]
- Keung, M.Y.; Wu, Y.; Badar, F.; Vadgama, J.V. Response of Breast Cancer Cells to PARP Inhibitors Is Independent of BRCA Status. J. Clin. Med. 2020, 9, 940. [Google Scholar] [CrossRef]
- Etemadmoghadam, D.; Weir, B.A.; Au-Yeung, G.; Alsop, K.; Mitchell, G.; George, J.; Davis, S.; D’Andrea, A.D.; Simpson, K.; Hahn, W.C.; et al. Synthetic lethality between CCNE1 amplification and loss of BRCA1. Proc. Natl. Acad. Sci. USA 2013, 110, 19489–19494. [Google Scholar] [CrossRef]
- Gorski, J.W.; Ueland, F.R.; Kolesar, J.M. CCNE1 Amplification as a Predictive Biomarker of Chemotherapy Resistance in Epithelial Ovarian Cancer. Diagnostics 2020, 10, 279. [Google Scholar] [CrossRef]
- Chappuis, P.O.; Donato, E.; Goffin, J.R.; Wong, N.; Begin, L.R.; Kapusta, L.R.; Brunet, J.S.; Porter, P.; Foulkes, W.D. Cyclin E Expression in Breast Cancer: Predicting Germline Brca1 Mutations, Prognosis and Response to Treatment. Ann. Oncol. 2005, 16, 735–742. [Google Scholar] [CrossRef]
- Aziz, D.; Etemadmoghadam, D.; Caldon, C.E.; Au-Yeung, G.; Deng, N.; Hutchinson, R.; Bowtell, D.; Waring, P. 19q12 amplified and non-amplified subsets of high grade serous ovarian cancer with overexpression of cyclin E1 differ in their molecular drivers and clinical outcomes. Gynecol. Oncol. 2018, 151, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Aziz, D.; Portman, K.J.N.; Fernandez, C.; Lee, S.; Alexandrou, A.; Llop-Guevara, A.; Yong, A.; Wilkinson, C.M.; Sergio, D.; Ferraro, D.; et al. Cyclin E1 Protein Is Stabilized in Brca1 Mutated Breast Cancers Leading to Synergy between Cdk2 and Parp Inhibitors. bioRxiv 2020. [Google Scholar] [CrossRef]
- Siu, K.T.; Rosner, M.R.; Minella, A.C. An integrated view of cyclin E function and regulation. Cell Cycle 2012, 11, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Delk, N.A.; Hunt, K.K.; Keyomarsi, K. Altered Subcellular Localization of Tumor-Specific Cyclin E Isoforms Affects Cyclin-Dependent Kinase 2 Complex Formation and Proteasomal Regulation. Cancer Res. 2009, 69, 2817–2825. [Google Scholar] [CrossRef]
- Beck, H.; Nähse-Kumpf, V.; Larsen, M.S.Y.; O’Hanlon, K.A.; Patzke, S.; Holmberg, C.; Mejlvang, J.; Groth, A.; Nielsen, O.; Syljuåsen, R.G.; et al. Cyclin-Dependent Kinase Suppression by WEE1 Kinase Protects the Genome through Control of Replication Initiation and Nucleotide Consumption. Mol. Cell. Biol. 2012, 32, 4226–4236. [Google Scholar] [CrossRef]
- Toledo, L.; Neelsen, K.J.; Lukas, J. Replication Catastrophe: When a Checkpoint Fails because of Exhaustion. Mol. Cell 2017, 66, 735–749. [Google Scholar] [CrossRef]
- Dreyer, S.B.; Upstill-Goddard, R.; Paulus-Hock, V.; Paris, C.; Lampraki, E.-M.; Dray, E.; Serrels, B.; Caligiuri, G.; Rebus, S.; Plenker, D.; et al. Targeting DNA Damage Response and Replication Stress in Pancreatic Cancer. Gastroenterology 2021, 160, 362–377.e13. [Google Scholar] [CrossRef]
- Lallo, A.; Frese, K.K.; Morrow, C.J.; Sloane, R.S.; Gulati, S.; Schenk, M.W.; Trapani, F.; Simms, N.; Galvin, M.; Brown, S.; et al. The Combination of the PARP Inhibitor Olaparib and the WEE1 Inhibitor AZD1775 as a New Therapeutic Option for Small Cell Lung Cancer. Clin. Cancer Res. 2018, 24, 5153–5164. [Google Scholar] [CrossRef]
- Nanos-Webb, A.; Jabbour, N.A.; Multani, A.S.; Wingate, H.; Oumata, N.; Galons, H.; Joseph, B.; Meijer, L.; Hunt, K.K.; Keyomarsi, K. Targeting low molecular weight cyclin E (LMW-E) in breast cancer. Breast Cancer Res. Treat. 2012, 132, 575–588. [Google Scholar] [CrossRef]
- Carey, J.P.; Karakas, C.; Bui, T.; Chen, X.; Vijayaraghavan, S.; Zhao, Y.; Wang, J.; Mikule, K.; Litton, J.K.; Hunt, K.K.; et al. Synthetic Lethality of PARP Inhibitors in Combination with MYC Blockade Is Independent of BRCA Status in Triple-Negative Breast Cancer. Cancer Res. 2018, 78, 742–757. [Google Scholar] [CrossRef]
- Duong, M.T.; Akli, S.; Wei, C.; Wingate, H.F.; Liu, W.; Lu, Y.; Yi, M.; Mills, G.B.; Hunt, K.K.; Keyomarsi, K. Lmw-E/Cdk2 Deregulates Acinar Morphogenesis, Induces Tumorigenesis, and Associates with the Activated B-Raf-Erk1/2-Mtor Pathway in Breast Cancer Patients. PLoS Genet. 2012, 8, e1002538. [Google Scholar] [CrossRef] [PubMed]
- Seluanov, A.; Mao, Z.; Gorbunova, V. Analysis of DNA Double-strand Break (DSB) Repair in Mammalian Cells. J. Vis. Exp. 2010, 43, e2002. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Yang, D.; Carey, J.P.W.; Karakas, C.; Albarracin, C.; Sahin, A.A.; Arun, B.K.; Guray Durak, M.; Li, M.; Kohansal, M.; et al. Targeting Replicative Stress and DNA Repair by Combining PARP and Wee1 Kinase Inhibitors Is Synergistic in Triple Negative Breast Cancers with Cyclin E or BRCA1 Alteration. Cancers 2021, 13, 1656. https://doi.org/10.3390/cancers13071656
Chen X, Yang D, Carey JPW, Karakas C, Albarracin C, Sahin AA, Arun BK, Guray Durak M, Li M, Kohansal M, et al. Targeting Replicative Stress and DNA Repair by Combining PARP and Wee1 Kinase Inhibitors Is Synergistic in Triple Negative Breast Cancers with Cyclin E or BRCA1 Alteration. Cancers. 2021; 13(7):1656. https://doi.org/10.3390/cancers13071656
Chicago/Turabian StyleChen, Xian, Dong Yang, Jason P. W. Carey, Cansu Karakas, Constance Albarracin, Aysegul A. Sahin, Banu K. Arun, Merih Guray Durak, Mi Li, Mehrnoosh Kohansal, and et al. 2021. "Targeting Replicative Stress and DNA Repair by Combining PARP and Wee1 Kinase Inhibitors Is Synergistic in Triple Negative Breast Cancers with Cyclin E or BRCA1 Alteration" Cancers 13, no. 7: 1656. https://doi.org/10.3390/cancers13071656
APA StyleChen, X., Yang, D., Carey, J. P. W., Karakas, C., Albarracin, C., Sahin, A. A., Arun, B. K., Guray Durak, M., Li, M., Kohansal, M., Bui, T. N., Ha, M.-J., Hunt, K. K., & Keyomarsi, K. (2021). Targeting Replicative Stress and DNA Repair by Combining PARP and Wee1 Kinase Inhibitors Is Synergistic in Triple Negative Breast Cancers with Cyclin E or BRCA1 Alteration. Cancers, 13(7), 1656. https://doi.org/10.3390/cancers13071656






