Simple Summary
Members of the adamalysin family are multi-domain proteins involved in many cancer-related functions. In this review, we will examine the literature on the involvement of adamalysins in hepatocellular carcinoma progression and their importance in the tumor microenvironment where they regulate the inflammatory response and the epithelial–mesenchymal transition. We complete this review with an analysis of adamalysin expression in a large cohort of patients with hepatocellular carcinoma from The Cancer Genome Atlas (TCGA) database. These original results give a new insight into the involvement of all adamalysins in the primary liver cancer.
Abstract
The tumor microenvironment plays a major role in tumor growth, invasion and resistance to chemotherapy, however understanding how all actors from microenvironment interact together remains a complex issue. The tumor microenvironment is classically represented as three closely connected components including the stromal cells such as immune cells, fibroblasts, adipocytes and endothelial cells, the extracellular matrix (ECM) and the cytokine/growth factors. Within this space, proteins of the adamalysin family (ADAM for a disintegrin and metalloproteinase; ADAMTS for ADAM with thrombospondin motifs; ADAMTSL for ADAMTS-like) play critical roles by modulating cell–cell and cell–ECM communication. During last decade, the implication of adamalysins in the development of hepatocellular carcinoma (HCC) has been supported by numerous studies however the functional characterization of most of them remain unsettled. In the present review we propose both an overview of the literature and a meta-analysis of adamalysins expression in HCC using data generated by The Cancer Genome Atlas (TCGA) Research Network.
1. Adamalysins Are Multidomain Proteins with Multiple Functions
Adamalysins belong to the zinc protease superfamily and are characterized by a multidomain organization that confers multiple functions [1]. Numerous reviews have already detailed the classification and functions of ADAM and ADAMTS proteins (ADAM for a disintegrin and metalloproteinase; ADAMTS for ADAM with thrombospondin motifs; ADAMTSL for ADAMTS-like) and their implication in physiological and pathological processes [2,3]. The ADAM–TS–TSL family consists of 20 ADAMs, 19 ADAMTS and 7 ADAMTSLs (1 to 6 and papillin) in human. These proteins share a multi-domain organization including a signal peptide, a pro-peptide and metalloprotease, disintegrin and cysteine-rich domains (Figure 1). While ADAMs are membrane proteins characterized by additional epidermal growth factor (EGF)-like, transmembrane and cytoplasmic domains, ADAMTS members are secreted proteins characterized by an ancillary domain containing a thrombospondin type 1 repeat (TSR), a spacer domain and except for ADAMTS4, additional motifs including TSRs. Unlike ADAM and ADAMTS, ADAMTSL genes encode secreted proteins that lack catalytic and disintegrin domains and are likely involved in ECM assembly.
A major feature of ADAM and ADAMTS proteins resides in their catalytic properties. ADAM metalloproteinases were rapidly named sheddases since most of their substrates were membrane-bound precursors. However only 12 ADAMs including ADAM8, 9, 10, 12, 15, 17, 19, 20, 21, 28, 30 and 33 have a functional catalytic site and two of them (ADAM20 and 21) have no known substrates (reviewed in [4,5]). Most of substrates for the mentioned 12 ADAMs are growth factors, chemokines, adhesion molecules and their receptors and only very few extracellular matrix components were reported (Figure 2A). While there is no specific substrate repertoire for each ADAMs, the phylogenic neighbors ADAM10 and 17 share many substrates including Notch and its ligand Delta-like 1 (DLL1), TNF receptor (CD30), the receptor activator of nuclear factor κ-B ligand (RANKL), the receptor to IL-6 and adhesion molecules such as CD44, desmoglein and L1CAM. One important feature that characterize ADAM9, 10, 12, 15 and 17 is their ability to interfere with EGFR- and Notch-dependent signaling pathways through processing of either the receptor or the ligands [6,7,8]. The regulation of these signaling pathways can occur in an ADAM’s catalytic-independent way such as the ADAM12-dependent activation of TGF-β signaling pathway [9] and the disintegrin domain-dependent binding for integrin-mediated signaling [10].
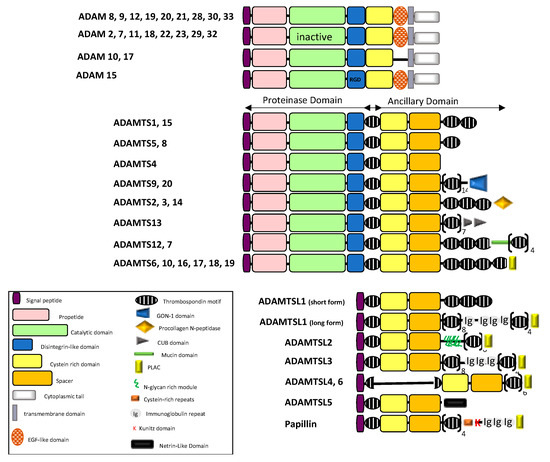
Figure 1.
Schematic diagram illustrating the domain organization of the ADAM/TS-TSL family members (adapted from [2,3,5]).
Splice variants have been reported for ADAM9 and ADAM12 giving rise to secreted forms with implication in cancer cell invasion. Similarly, ADAM11, ADAM28 and ADAM33 have short spliced variants lacking transmembrane and cytoplasmic tails while variants for ADAM15 and ADAM22 affect only cytoplasmic domain. ADAM19 and ADAM33 variants are characterized by alternative splicing within the pro-, metalloprotease and disintegrin domains.
ADAMTS proteins are proteases which have been historically classified according to their substrates: the aggrecanase and proteoglycanase (ADAMTS1, 4, 5, 8, 9, 15 and 20); the pro-collagen N-propeptidases (ADAMTS-2, 3 and 14); the COMP proteinases (ADAMTS7 and 12) and the von Willebrand factor proteinase (ADAMTS13). For a long time, the remaining ADAMTS were considered as orphan enzymes [2,3,11] however, identification of several new substrates complicated the picture. A large screening for ADAMTS2, 3, and 14 substrates identified components of TGF-β network (the latent TGF-β binding proteins LTBP1 and 2, the Transforming Growth Factor Beta Receptor 3, TGFBR3 and Decorin that binds active TGF-β) as well as extracellular matrix proteins such as fibronectin [12]. Fibronectin was also recently identified as a substrate of ADAMTS16 [13] and ADAMTS9 [14] thereby supporting evidence for the implication of ADAMTS9 and ADAMTS16 in ECM assembly and turnover. Using Terminal Amine Isotopic Labeling of Substrates (TAILS) method, Colige et al. recently identified new substrates repertoire for ADAMTS7 including LTBPs, fibronectin, and fibrillin-2 [15], the latter being also identified as a new substrate for ADAMTS10 [16]. The absence of ADAM-specific motif in the substrates led to the complex identification of ADAM substrates. Based on the recent review from [17], we built the substrates network of ADAMTS proteins giving rise to a new classification based on functional proximity (Figure 2B). An important feature of ADAMTS proteins is the non-catalytic ancillary domains which support substrate recognition [18] and share homologies with ADAMTS-like proteins. The critical contribution of ADAMTSs and ADAMTSLs in microfibril formation, stabilization and functions is now well established [19]. In accordance with this, mutations in ADAMTS2, 3, 10, 13, 17, 20 and in ADAMTSL2 and ADAMTSL4 are associated with Mendelian disorders affecting ECM assembly and regulation of growth factors bioavailability [20,21,22].
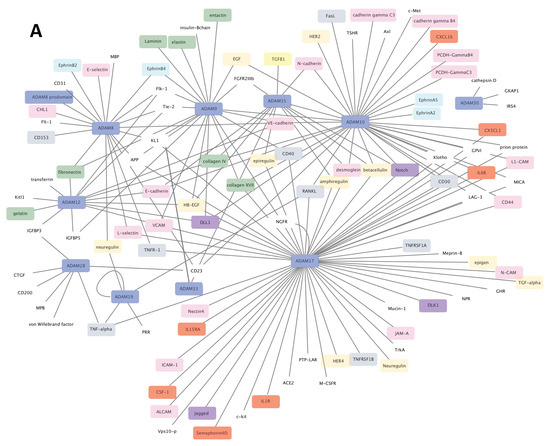
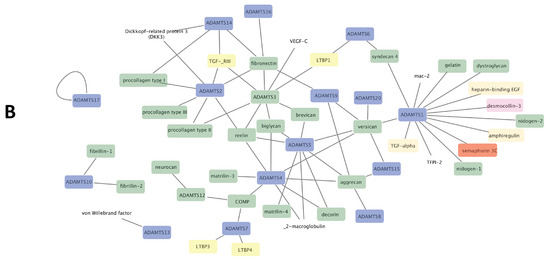
Figure 2.
Substrates networks for ADAM (A) and ADAMTS (B). Generated from data reported in [5,17] and plotted using cytoscape software platform (https://cytoscape.org/, accessed on 14 January 2021). Colors of nodes: blue, ADAM/ADAMTS; green, extracellular matrix components; red, cytokine/chemokine; pink, adhesion/junction molecules; grey, TNF superfamily components; purple, Notch signaling components; yellow, TGF-β signaling components; turquoise, ephrin signaling components. The remaining uncolored nodes correspond to miscellaneous functions.
Because of their multiple functions in regulation of cell–cell and cell–matrix communication, ADAM and ADAMTS proteins are key players in maintaining tissue homeostasis and their deregulation is associated with numerous biological processes including tissue remodeling, inflammation and cell migration. In that context, the role of ADAMs [5,23] and ADAMTSs [24,25] in cancer have been widely documented especially in angiogenesis [26], their contribution to the tumor environment [27] and their use as targets for the treatment of cancer [28]. Here we will focus on hepatocellular carcinoma, the most common primary liver cancers that mostly develop in the context of chronic liver disease.
2. Adamalysins in Hepatocellular Carcinoma
2.1. Adamalysin Expression and Association with Overall Survival
In 2010, Mazzocca et al. wondered about the «real pathogenic link» between ADAM proteins and tumorigenesis and progression of hepatocellular carcinoma [29]. There is no longer any doubt today that adamalysins are involved in the development of HCC. Increased expression of many Adamalysins including ADAM8 [30,31], ADAM9 [32,33], ADAM10 [34,35], ADAM12 [36] and ADAM17 [37] was reported in HCC and associated with tumor progression. Additional studies demonstrated that over-expression of these ADAMs promoted proliferation of HCC cells and was predictive of poor survival outcomes. In parallel, miRNA-dependent down-regulation of ADAM9 [38,39,40], ADAM10 [41,42,43,44,45,46] and ADAM17 [47,48,49,50] was associated with reduced tumor progression, inhibition of cell proliferation and invasion and chemotherapy sensitization of HCC cells. The liver-specific miR-122 that targets both ADAM10 and ADAM17 is decreased in patient with HCC and was recently proposed as a biomarker for hepatocellular carcinoma related to hepatitis C virus infection [51]. By contrast, the long non coding LIN0551 competes miR122 in regulating ADAM10 thereby promoting HCC cell proliferation and invasion [52]. Alternatively, spliced variants of ADAM12 and ADAM9 were identified in activated hepatic stellate cells and among those of ADAM9, the short form (ADAM9-S) promoted cancer cell invasion [36,53]. In addition to these spliced variants, single nucleotide polymorphism (SNP) variants of ADAM10 were sequenced from patients with HCC and some of them including rs514049 and rs653765 variants were associated with a higher risk of developing metastases [54]. Besides the well documented ADAM9, ADAM10 and ADAM17 genes, the over-expression of ADAM18, ADAM21 and ADAM32 was more recently associated with HCC invasion [55,56], however the molecular mechanisms that were involved remain unexplored.
To have a complete view of the expression of all adamalysins in HCC, we took advantage of RNAseq data generated by The Cancer Genome Atlas Program (TCGA) Research Network: https://www.cancer.gov/tcga, accessed on 14 January 2019. Normalized expression values were extracted via the ‘International Cancer Genome Consortium” website (https://icgc.org/, accessed on 11 July 2016). The Cancer Genome Atlas Liver Hepatocellular Carcinoma (TCGA-LIHC) data collection contains 364 LIHC samples including 48 paired hepatocellular carcinoma and adjacent non-tumor tissues. Of course, RNA sequencing data provide new valuable information although do not permit to evaluate adamalysin activity. We also analyzed the prognosis value of adamalysins expression in TCGA-LIHC samples using Kaplan–Meier plotter online tool (http://kmplot.com/analysis/, accessed on 1 September 2020). Comparison of adamalysin expression levels between tumor (T) and adjacent non-tumor (NT) tissues and survival analyses are given in Table 1, left and right panels, respectively. Note that the same TCGA-LIHC samples were recently used to propose ADAM9 as a biomarker in advanced HCC [57]. In agreement with previous reports, we observed that expression levels of ADAM9, ADAM10, ADAM12 and ADAM17 were increased in tumor versus adjacent non tumor tissues except for ADAM10 that was non-significant, all were associated with poor prognosis. Importantly, we reported for the first time an up-regulation of ADAM15, ADAM21, ADAM22, ADAM23 in tumors compared with adjacent non tumor tissues, however only ADAM15 is indicative of worse prognosis (0.007). The very high significant increase expression of ADAM15 in tumor (8.2 × 10−19) is in agreement with the up-regulation of ADAM15 previously reported in other cancers including breast, lung, prostate and bladder cancers [23]. The role of ADAM15 in invasion and metastasis involves the regulation of claudin-1 and its interaction with the tight junction proteins ZO1 and ZO2 [58]. Interestingly, these tight junction proteins also play a critical role in hepatitis C virus infection [59]. In addition, ADAM15 is known for its role in inflammation [60] suggesting its contribution to HCC development.

Table 1.
Adamalysin expression in liver cancers from The Cancer Genome Atlas (TCGA) database. nd; not detectable; ns, not significant.
The implication of ADAMTS proteins in cancer was also previously reported and both anti-tumor and pro-tumor functions were described [24,25,61]. In the liver, expression levels of ADAMTS1 were firstly reported as being lower in human HCC samples than in the underlying cirrhotic tissue [62,63]. Low expression of ADAMTS1 in HCC compared with healthy livers was next reported [62,64]. More recently, up-regulation of ADAMTS1 expression was observed in HCC cells upon stimulation by either the inflammatory cytokines IL-1β and IL-6 [65] or by hypoxia [66]. ADAMTS1 was also reported in non-parenchymal cells including endothelial cells [67] and activated hepatic stellate cells [68]. Using the NT/T paired sample data set from TCGA database, we confirmed decreased expression of ADAMTS1 in tumor samples compared with the non-tumor adjacent tissues (p = 0.000059) suggesting anti-tumor functions. By contrast, the high expression of ADAMTS1 in cirrhotic tissues [63], the up-regulation of ADAMTS1 in liver fibrosis [69] and the implication of ADAMTS1 in TGF-β activation in activated hepatic stellate cells (HCS) [68] suggest a role of ADAMTS1 in liver fibrosis that might favor tumor onset.
Low expression levels of ADAMTS5 [70] and ADAMTS8 [71] were associated with poor prognosis of patients with HCC. ADAMTS8 inhibited proliferation and favored apoptosis of HCC cells [71]. Using TCGA data, we confirmed that low expression of ADAMTS8 was associated with overall survival however, we observed the increased expression of ADAMTS5 in tumor and its association with worse survival (Table 1). We also observed a significant increase in ADAMTS9 expression in tumors compared with adjacent non tumor tissue and an association between high expression and worse prognosis. However, ADAMTS9 was previously suggested to act as a suppressor of tumor since increased expression of microRNA-32 [72] and long non-coding RNA ADAMTS9-AS1 [73] that both target ADAMTS9, were correlated with decreased patient survival, and increased proliferation and invasion, respectively. Our own analysis showed also increased expression of ADAMTS7, 14 and 18 in tumors compared with adjacent non tumor tissues and high expression in tumors was associated with worse prognosis (Table 1). In accordance with this observation, the pro-tumor role of ADAMTS7 was recently supported by its identification in a signature of cancer stem cells that contribute to the development and therapeutic resistance of HCC [74]. ADAMTS14 and 18 have not yet been documented in liver cancers. By contrast ADAMTS13 was widely studied since liver is the main source of ADAMTS13 in human and changes in ADAMTS13 activity was associated with liver disease [75]. Several reports showed a decrease of ADAMTS13 activity either in cirrhosis [76] or in plasma of patients with viral [77] or alcohol [78] hepatitis. However, Lisman et al. [79] did not report significant difference in ADAMTS13 expression levels between cirrhosis and control livers and Ikeda et al. [80] recently showed that high plasma ADAMTS13 activity was a risk factor for HCC development in patients with chronic hepatitis B and C. Ikeda et al. [80] suggested that these discrepancies might be related to the difference in hepatitis activity and wound healing in selected patients. More recently, Takaya et al. proposed to use the ratio between ADAMTS13 activity and its substrate the von Willebrand factor as a biomarker either for the response to sorafenib treatment [81] or early diagnosis of HCC [82]. In line with this controversy, Kume et al. [83] showed that apoptosis of HSCs, the major ADAMTS13-producing cells [84] contributes to decrease ADAMTS13 levels in dimethylnitrosamine-treated rats but not in (CCl4)-treated rats which are characterized by the absence of HSC apoptosis and up-regulation of ADAMTS13 [85]. Also, a novel spliced ADAMTS13 transcript was found in both hepatic stellate cells and HCC cell lines [86] suggesting that tumor cells could also participate in expression of ADAMTS13 in HCC. Using paired HCC samples from TCGA database, we observed a very significant decrease of ADAMTS13 expression in tumor compared to adjacent non tumor tissues (3.8 × 10−21), however ADAMTS13 expression was not predictive for prognosis in the whole unpaired cohort (Table 1).
The balance between pro-tumor and anti-tumor effects of ADAMTSs has been partly attributed to their dual role in angiogenesis, a critical process in cancer progression [26]. The anti- and pro-angiogenic effects of ADAMTS1 were widely documented elsewhere [87] acting either as protease or in a catalytic-independent way. In hepatocellular carcinoma, only ADAMTS5 was suggested to inhibit angiogenesis through down-regulation of VEGF in HCC cells [70]. Variants of ADAMTS14 [88] and ADAMTS5 [89] were associated with susceptibility to hepatocellular carcinoma in a Chinese Han population, however the functional effects of these polymorphisms that occur near the anti-angiogenic TSR domain were not demonstrated.
The implication of ADAMTSL proteins in cancer is still largely undocumented. Changes in expression levels of ADAMTSL3 and ADAMTSL5 were reported in hepatocellular carcinoma, however opposite roles were suggested. ADAMTSL3 was identified as a tumor suppressor gene since down-regulation of ADAMTSL3 is associated with poor overall survival and predicted poor relapse-free survival [90]. At the opposite end, high ADAMTSL5 expression was associated with hypermethylation in HCC and a shorter overall survival of patients [91]. Our own analysis of HCC samples from the TCGA database provides new information on ADAMTSL expression levels in these samples (Table 1). In agreement with previous studies, we observed decreased ADAMTSL3 expression in tumors compared with adjacent non tumor tissues and low ADAMTSL3 expression was associated with a worse prognosis. By contrast ADAMTSL5 expression was increased in tumor versus adjacent non tumor tissues. In addition to these expected observations, our analysis now revealed that low expression levels of ADAMTSL1, ADAMTSL2 and ADAMTSL4 were predictive of worse prognosis suggesting similar suppressor role for the four ADAMTSLs (1–4) in HCC. These observation in HCC are consistent with previous reports in other cancers. Decreased ADAMTSL3 expression was reported in colon carcinoma [92] and ADAMTSL5 was over-expressed in melanomas [93]. In addition, increased methylation in ADAMTSL5 was associated with chemo-resistance in Acute B Lymphoblastic Leukemia patients [94]. Very few information about ADAMTSL1, 2 and 4 is available in cancers, however two SNPs in ADAMTSL1 gene were associated with early-onset disease-free survival in breast cancers [95] and ADAMTSL1 was shown to regulate chondrosarcoma cell proliferation [96]. More recently ADAMTSL4 expression was identified in glioma stem-like cells contributing to a five gene signature associated with bad prognosis of patients with primary glioblastoma, suggesting that ADAMTSL4 might have contrasting roles depending on cell origin [97].
2.2. Implication of Adamalysins in Inflammation and HCC Progression
To further characterize biological processes associated with adamalysins expression in hepatocellular carcinoma, we used the molecular and clinical feature previously described for TCGA HCC samples in [98]. As shown in Figure 3, the main molecular signatures associated with adamalysin expression are related to inflammation (leukocyte estimate), recurrence (SNUR, RS65.Sscore) and poor differentiation state (NCIHS, NCIPHS, cholangiocarcinoma-like). Because of their shedding activity that targets molecular actors of the immune system, the regulatory role of ADAM proteins in inflammatory processes have been widely documented [99]. Such activities particularly make sense in HCC since this cancer mainly develops in the context of chronic liver inflammation leading to fibrosis which in turn promotes the initiation and progression of tumors [100].
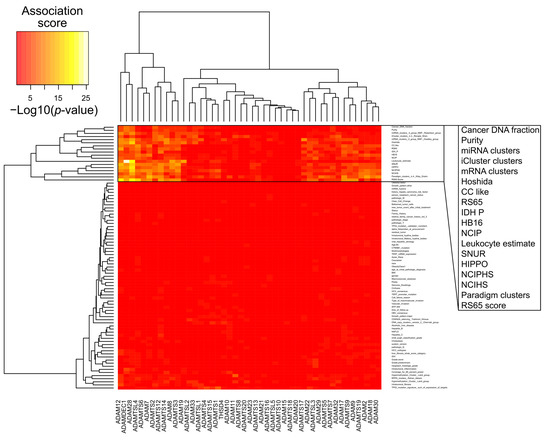
Figure 3.
Association between expression levels of Adamalysin genes and molecular features of hepatocellular carcinoma (HCC) samples (TCGA-LIHC primary tumors, 294 samples) extracted from the Cancer Genome Atlas Research Network [98]. Heat map shows association scores expressed as −Log10(p-values). For quantitative features, p-values result from a Pearson’s correlation test between feature values in HCC samples and the corresponding expression values for a given gene. For qualitative values (HCC subtypes), p-values result from a non-parametric distribution test (Kruskal-Wallis) of the expression values for a given gene among HCC subtypes (clusters).
2.2.1. Regulation of Pro-Tumorigenic Cytokines by Adamalysins in HCC
Among the targets of ADAMs, inflammatory cytokines such as tumor necrosis factor-α (TNF-a) and their receptors such as interleukin-6 receptor play critical roles in liver inflammation and HCC development [101]. Shedding of membrane-bound TNFα releases a soluble TNFa that binds to TNFRs to induce different signaling pathways leading either to cell survival and pro-inflammatory gene expression or to apoptosis and cell death [102]. In addition, the proteolytic cleavage of TNFR by ADAMs generates a soluble receptor that regulates TNFα bioavailability. In the liver, ADAM17 was directly involved in regulation of TNFα system in inflammation associated with different hepatic processes such as regeneration [103] and severe Alcoholic Hepatitis [104]. A link between inflammatory signals and pro-tumorigenic mechanisms in liver cells is supported by the ability of the inflammatory cytokine TNFα to induce activation of EGFR signaling. Argast et al. [105] firstly suggested the implication of ADAM17 in the proliferative effect of TNFα through the release of TGFα that activates EGFR in mouse hepatocyte cell lines. Similar mechanism was reported in human HCC cells where TNFα treatment induced increase of the EGFR ligand amphiregulin (AR) and this effect was abrogated by the ADAM17 inhibitor GM6001 [106]. ADAM17 was also involved in the regulation of IL-6-dependent signaling pathway that links chronic inflammation to HCC progression. Besides the classical pathway induced by IL-6 binding on its receptor, the proteolytic release of the soluble IL-6 receptor (sIL-6R) by ADAM17 induces a trans-signaling pathway, a mechanism allowing to sensitize cells that do not express IL-6R [107,108]. Increase of IL-6 and sIL-6R has been reported in cirrhosis and HCC [109,110] and Bergmann et al. demonstrated that the IL-6 trans-signaling mechanism is essential for the development of HCC [111]. Notch signaling is another pathway tightly regulated by adamalysins either through shedding of Notch receptor or its ligand [7,112]. The role of Notch signaling in the development of liver cancers has been recently reviewed [113,114] however, the direct implication of adamalysins in this pathway remains poorly documented, except for ADAM17. ADAM17-dependent Notch activation was involved in the induction of IL10 producing CD4 + T cells mediated by hepatocytes from Con A-pretreated mice contributing to the regulation of immune response [115]. Notch signaling plays also a regulatory role in maintenance of cancer stem-like cells (CSC) phenotype and Notch proteolytic activation by ADAM17 was implicated in promotion of liver CSCs upon iNOS over-expression [116]. Proteolytic activity of ADAM10 and 17 was also involved in the shedding of the receptor tyrosine kinase Met in HCC cells and hepatic stellate cells [117]. The authors further demonstrated a correlation between the soluble form of Met (sMet) and liver damage and inflammation. While sMet has been proposed as biomarker of cancer aggressiveness and bad prognostic [118,119,120], it was shown to down-regulate HGF signaling by trapping this growth factor and by preventing c-Met receptor dimerization [121].
2.2.2. Role of Adamalysins in Tumor Escape from Immune Surveillance
ADAM-dependent shedding of the major histocompatibility complex class I-related chain A (MICA) expressed by tumor cells prevents recognition by the NKG2D receptor expressed by natural killer (NK) cells thereby contributing to evading anti-tumor immunity [122]. According with this, the multi kinase inhibitors sorafenib and regorafenib enhance natural killer (NK) cells cytotoxicity against HCC cells by reducing ADAM9 and ADAM10 expression [123,124,125]. The contribution of ADAM17 in MICA shedding in HCC cells is more controversial. Although ADAM17 was firstly unrelated to MICA shedding [124], more recent data showed that ADAM17 contributed to MICA shedding in HEPG2 and HEP3B cells [123,126]. At the opposite, ADAM28 was recently proposed to play a protective role against the dissemination of cancer cells by promoting the T cell immune response [127]. However, proliferation of liver cancer cells was inhibited by miR-574-3p through ADAM28 targeting thereby suggesting that ADAM28 promotes liver cancer [128]. The ambivalent functions of ADAM28 in cancer were recently reviewed in [129] where expression of ADAM28 in cancer cells contribute to their proliferation and migration while stromal expression of ADAM28 contributes to protective effects.
2.2.3. Adamalysins Regulate Invasion in HCC
Among shedding activities associated with cancer, the importance of ADAM-dependent shedding of EGF-like ligands in proliferation and invasion of cells is widely documented [130,131]. In hepatocellular carcinoma, Itabashi et al. [132] firstly reported the suppression of angiotensin II-induced cell proliferation by targeting the AngII–EGFR cross-talk signaling mediated by ADAM9 and ADAM17. ADAM17 was further involved in the hypoxia-induced drug resistance of HCC cells through activation of EGFR/PI3K/Akt pathway [133]. The contribution of ADAM9 and ADAM17 to HCC proliferation and invasiveness was also supported by the suppressive effects of miRNAs that specifically target them [39,48,49,50]. More specifically, several adamalysins were associated with the epithelio–mesenchymal transition (EMT), a mechanism that favors proliferation, stemness, invasiveness, immune escape and resistance to anti-cancer treatments [134,135]. Different molecular mechanisms have been reported in ADAM-mediated EMT in liver. ADAM9 was involved in the IL-6 dependent EMT of HCC cell lines through interaction with the NADPH oxydase, thereby favoring ROS production [136]. This effect is linked to an increased expression of Snail, a major transcriptional driver of EMT, while the expression of other EMT-promoting factors such as Slug, Twist or Zeb were not affected suggesting that ADAM9 promotes a partial EMT. The contribution of ADAM17 in promoting EMT of HCC cells involves the activation of Notch signaling pathway, by increasing the proteolytic cleavage and release of the active Notch intracellular domain (NICD) [49]. In this study, pro-EMT effects of ADAM17 were antagonized by a specific micro-RNA (miR-3163) whose anti-EMT effects were associated with a decreased expression of EMT-promoting transcription factors (ZEB1, SNAIL and TWIST) and EMT markers (N-cadherin and Vimentin), while E-Cadherin (an epithelial marker) was increased. In agreement with this, ADAM17 inhibitors such as ZLDI-8 were shown to prevent EMT in HCC cells by decreasing the release of Notch NICD, thereby improving the effects of anti-cancer drugs [137,138]. ADAM17 was also involved in transactivation of Notch signaling in liver cancer stem cells, contributing to an aggressive phenotype [116,139]. In addition, Notch signaling was implicated in the ADAM17-dependent activation of integrin β1 thereby promoting the migration and invasion of HCC cells [140]. In that context, interaction of ADAM17 with G-protein-coupled receptor 50 was suggested to drive the ADAM17-induced Notch signaling toward HCC progression [141]. While ADAM10 also activates Notch signaling, no report involving a Notch-ADAM10 signaling pathway to promote HCC progression has been reported yet, may be due to its ligand-dependency mechanism while ADAM17 activates Notch signaling by a ligand-independent mechanism [7]. However, ADAM10 promotes the proliferation, invasion and migration of HCC cells [34] and contributes to HCC progression through the shedding of CD147 (EMMPRIN or basigin), a transmembrane glycoprotein involved in metabolic adaptation of cancer cells, chemo-resistance and EMT [142].
The interplay between adamalysins and TGF-β-dependent signal might be a paradigm of the complex contribution of adamalysins activities in hepatocellular carcinoma. TGF-β plays a central role in chronic liver diseases, EMT and HCC progression [143,144]. We previously showed the association between ADAM12 expression and HCC aggressiveness and its regulation by TGF-β in hepatic stellate cells [36,145]. We next demonstrated that ADAM12 interacts with and stabilizes TGFBRII receptor, thereby promoting TGF-β signaling pathways through increase of receptor trafficking [9,146]. Using dedicated cellular models, we showed that ADAM12 promotes TGF-β-mediated EMT in a proteolytic independent manner [147]. ADAM12 also acts as a downstream player of EMT by regulating invadopodia and focal adhesion structures [148,149]. As an actor of TGF-β and Notch [8] signaling pathways, which are both involved in EMT, ADAM12 might enhance EMT by synergizing these pathways, although such a mechanism has not yet been addressed in the context of HCC progression. Beside ADAM12, we demonstrate that ADAMTS1 expression is increased in HCC and also contributes to amplify the TGF-β signal by stimulating the conversion of the latent-TGF-β into its active form in hepatic stellate cells [68,150]. Since HSCs are major cellular components of HCC stroma, where they modulate the proliferation and invasiveness of cancer cells [151], adamalysins expressed by HSCs are likely to play a pivotal role in these processes.
To conclude, deregulation of the expression of many adamalysins is associated with hepatocellular carcinoma. These have been implicated at all stages of HCC progression including inflammation, fibrosis, angiogenesis, proliferation, epithelial–mesenchymal transition and invasion offering broad clinical perspectives. However, their spatial and temporal dynamics of expression and their mechanisms of action are still insufficiently characterized. Future work should focus on better characterizing the contribution of the different hepatic cell types in the expression and activity of these new regulators of HCC progression.
Author Contributions
All authors have read and agreed to the published version of the manuscript.
Funding
Inserm and University of Rennes 1.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Seals, D.F.; Courtneidge, S.A. The ADAMs family of metalloproteases: Multidomain proteins with multiple functions. Genes Dev. 2003, 17, 7–30. [Google Scholar] [CrossRef]
- Apte, S.S. A Disintegrin-like and Metalloprotease (Reprolysin-Type) with Thrombospondin Type 1 Motif (ADAMTS) Superfamily: Functions and Mechanisms. J. Biol. Chem. 2009, 284, 31493–31497. [Google Scholar] [CrossRef] [PubMed]
- Kelwick, R.; Desanlis, I.; Wheeler, G.N.; Edwards, D.R. The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin Motifs) Family. Genome Biol. 2015, 16, 113. [Google Scholar] [CrossRef]
- Huovila, A.-P.J.; Turner, A.J.; Pelto-Huikko, M.; Kärkkäinen, I.; Ortiz, R.M. Shedding Light on ADAM Metalloproteinases. Trends Biochem. Sci. 2005, 30, 413–422. [Google Scholar] [CrossRef]
- Zadka, L.; Kulus, M.J.; Piatek, K. ADAM Protein Family—Its Role in Tumorigenesis, Mechanisms of Chemoresistance and Potential as Diagnostic and Prognostic Factors. Neoplasma 2018, 65, 823–839. [Google Scholar] [CrossRef] [PubMed]
- Blobel, C.P. ADAMs: Key Components in EGFR signalling and development. Nat. Rev. Mol. Cell Biol. 2005, 6, 32–43. [Google Scholar] [CrossRef]
- Christian, L.M. The ADAM family: Insights into notch proteolysis. Fly 2012, 6, 30–34. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zolkiewska, A. ADAM proteases: Ligand processing and modulation of the notch pathway. Cell Mol. Life Sci. 2008, 65, 2056–2068. [Google Scholar] [CrossRef]
- Atfi, A.; Dumont, E.; Colland, F.; Bonnier, D.; L’helgoualc’h, A.; Prunier, C.; Ferrand, N.; Clément, B.; Wewer, U.M.; Théret, N. The disintegrin and metalloproteinase ADAM12 contributes to TGF-Beta signaling through interaction with the Type II receptor. J. Cell Biol. 2007, 178, 201–208. [Google Scholar] [CrossRef]
- Reiss, K.; Ludwig, A.; Saftig, P. Breaking up the Tie: Disintegrin-like metalloproteinases as regulators of cell migration in inflammation and invasion. Pharmacol. Ther 2006, 111, 985–1006. [Google Scholar] [CrossRef] [PubMed]
- Dancevic, C.M.; McCulloch, D.R.; Ward, A.C. The ADAMTS hyalectanase family: Biological insights from diverse species. Biochem. J. 2016, 473, 2011–2022. [Google Scholar] [CrossRef]
- Bekhouche, M.; Leduc, C.; Dupont, L.; Janssen, L.; Delolme, F.; Vadon-Le Goff, S.; Smargiasso, N.; Baiwir, D.; Mazzucchelli, G.; Zanella-Cleon, I.; et al. Determination of the Substrate Repertoire of ADAMTS2, 3, and 14 significantly broadens their functions and identifies extracellular matrix organization and TGF-β signaling as primary targets. FASEB J. 2016, 30, 1741–1756. [Google Scholar] [CrossRef]
- Schnellmann, R.; Sack, R.; Hess, D.; Annis, D.S.; Mosher, D.F.; Apte, S.S.; Chiquet-Ehrismann, R. A selective extracellular matrix proteomics approach identifies fibronectin proteolysis by a disintegrin-like and metalloprotease domain with thrombospondin Type 1 Motifs (ADAMTS16) and its impact on spheroid morphogenesis. Mol. Cell Proteom. 2018, 17, 1410–1425. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.W.; Nandadasa, S.; Annis, D.S.; Dubail, J.; Mosher, D.F.; Willard, B.B.; Apte, S.S. A disintegrin-like and metalloproteinase domain with thrombospondin Type 1 Motif 9 (ADAMTS9) regulates fibronectin fibrillogenesis and turnover. J. Biol. Chem. 2019, 294, 9924–9936. [Google Scholar] [CrossRef]
- Colige, A.; Monseur, C.; Crawley, J.T.B.; Santamaria, S.; de Groot, R. Proteomic discovery of substrates of the cardiovascular protease ADAMTS. J. Biol. Chem. 2019, 294, 8037–8045. [Google Scholar] [CrossRef]
- Wang, L.W.; Kutz, W.E.; Mead, T.J.; Beene, L.C.; Singh, S.; Jenkins, M.W.; Reinhardt, D.P.; Apte, S.S. Adamts10 inactivation in mice leads to persistence of ocular microfibrils subsequent to reduced Fibrillin-2 cleavage. Matrix Biol. 2019, 77, 117–128. [Google Scholar] [CrossRef]
- Satz-Jacobowitz, B.; Hubmacher, D. The Quest for Substrates and Binding Partners: A Critical Barrier for Understanding the Role of ADAMTS Proteases in Musculoskeletal Development and Disease. Dev. Dyn. 2020. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S. Three-dimensional domain architecture of the ADAM family proteinases. Semin. Cell Dev. Biol. 2009, 20, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Hubmacher, D.; Apte, S.S. ADAMTS proteins as modulators of microfibril formation and function. Matrix Biol. 2015, 47, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Hynes, R.O. The extracellular matrix: Not just pretty fibrils. Science 2009, 326, 1216–1219. [Google Scholar] [CrossRef] [PubMed]
- Le Goff, C.; Cormier-Daire, V. The ADAMTS(L) family and human genetic disorders. Hum. Mol. Genet. 2011, 20, R163–R167. [Google Scholar] [CrossRef] [PubMed]
- Mead, T.J.; Apte, S.S. ADAMTS proteins in human disorders. Matrix Biol. 2018, 71–72, 225–239. [Google Scholar] [CrossRef]
- Duffy, M.J.; McKiernan, E.; O’Donovan, N.; McGowan, P.M. Role of ADAMs in cancer formation and progression. Clin. Cancer Res. 2009, 15, 1140–1144. [Google Scholar] [CrossRef]
- Binder, M.J.; McCoombe, S.; Williams, E.D.; McCulloch, D.R.; Ward, A.C. The extracellular matrix in cancer progression: Role of hyalectan proteoglycans and ADAMTS enzymes. Cancer Lett. 2017, 385, 55–64. [Google Scholar] [CrossRef]
- Cal, S.; López-Otín, C. ADAMTS proteases and cancer. Matrix Biol. 2015, 44–46, 77–85. [Google Scholar] [CrossRef]
- Sun, Y.; Huang, J.; Yang, Z. The roles of ADAMTS in angiogenesis and cancer. Tumour Biol. 2015, 36, 4039–4051. [Google Scholar] [CrossRef] [PubMed]
- Shimoda, M.; Ohtsuka, T.; Okada, Y.; Kanai, Y. Stromal metalloproteinases: Crucial contributors to the tumor microenvironment. Pathol. Int. 2020. [Google Scholar] [CrossRef] [PubMed]
- Mullooly, M.; McGowan, P.M.; Crown, J.; Duffy, M.J. The ADAMs family of proteases as targets for the treatment of cancer. Cancer Biol. Ther. 2016, 17, 870–880. [Google Scholar] [CrossRef]
- Mazzocca, A.; Giannelli, G.; Antonaci, S. Involvement of ADAMs in tumorigenesis and progression of hepatocellular carcinoma: Is it merely fortuitous or a real pathogenic link? Biochim. Biophys. Acta 2010, 1806, 74–81. [Google Scholar] [CrossRef]
- Jiang, C.; Zhang, Y.; Yu, H.-F.; Yu, X.-T.; Zhou, S.-J.; Tan, Y.-F. Expression of ADAM8 and its clinical values in diagnosis and prognosis of hepatocellular carcinoma. Tumour Biol. 2012, 33, 2167–2172. [Google Scholar] [CrossRef]
- Zhang, Y.; Zha, T.-Z.; Hu, B.-S.; Jiang, C.; Ge, Z.-J.; Zhang, K.; Tan, Y.-F. High expression of ADAM8 correlates with poor prognosis in hepatocellular carcinoma. Surgeon 2013, 11, 67–71. [Google Scholar] [CrossRef]
- Tannapfel, A.; Anhalt, K.; Häusermann, P.; Sommerer, F.; Benicke, M.; Uhlmann, D.; Witzigmann, H.; Hauss, J.; Wittekind, C. Identification of novel proteins associated with hepatocellular carcinomas using protein microarrays. J. Pathol. 2003, 201, 238–249. [Google Scholar] [CrossRef]
- Tao, K.; Qian, N.; Tang, Y.; Ti, Z.; Song, W.; Cao, D.; Dou, K. Increased expression of a disintegrin and metalloprotease-9 in hepatocellular carcinoma: Implications for tumor progression and prognosis. Jpn. J. Clin. Oncol. 2010, 40, 645–651. [Google Scholar] [CrossRef]
- Yuan, S.; Lei, S.; Wu, S. ADAM10 is overexpressed in human hepatocellular carcinoma and contributes to the proliferation, invasion and migration of HepG2 cells. Oncol. Rep. 2013, 30, 1715–1722. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, S.; Liu, K.; Wang, Y.; Ji, B.; Zhang, X.; Liu, Y. A Disintegrin and Metalloprotease (ADAM)10 is highly expressed in Hepatocellular Carcinoma and is associated with tumour progression. J. Int. Med. Res. 2014, 42, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Le Pabic, H.; Bonnier, D.; Wewer, U.M.; Coutand, A.; Musso, O.; Baffet, G.; Clément, B.; Théret, N. ADAM12 in human liver cancers: TGF-Beta-Regulated expression in stellate cells is associated with matrix remodeling. Hepatology 2003, 37, 1056–1066. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Yang, L.-Y.; Huang, G.-W.; Wang, W.; Lu, W.-Q. ADAM17 MRNA expression and pathological features of hepatocellular carcinoma. World J. Gastroenterol. 2004, 10, 2735–2739. [Google Scholar] [CrossRef] [PubMed]
- Wan, D.; Shen, S.; Fu, S.; Preston, B.; Brandon, C.; He, S.; Shen, C.; Wu, J.; Wang, S.; Xie, W.; et al. MiR-203 Suppresses the proliferation and metastasis of Hepatocellular Carcinoma by targeting Oncogene ADAM9 and Oncogenic Long Non-Coding RNA HULC. Anticancer Agents Med. Chem. 2016, 16, 414–423. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.-Y.; Ou, H.-H.; Liu, X.-C.; Chen, Z.-J.; Li, X.-H.; Huang, Y.; Yang, D.-H. Loss of Tumor Suppressor MiR-126 Contributes to the development of Hepatitis B Virus-Related Hepatocellular Carcinoma metastasis through the upregulation of ADAM. Tumour Biol. 2017, 39, 1010428317709128. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Liu, J.; Li, Y.; Liu, L.; Zhang, X.; Ma, C.; Hua, S.; Yang, M.; Yuan, Q. MicroRNA-1274a, a Modulator of Sorafenib induced a disintegrin and Metalloproteinase 9 (ADAM9) down-regulation in Hepatocellular Carcinoma. FEBS Lett. 2011, 585, 1828–1834. [Google Scholar] [CrossRef]
- Bai, S.; Nasser, M.W.; Wang, B.; Hsu, S.-H.; Datta, J.; Kutay, H.; Yadav, A.; Nuovo, G.; Kumar, P.; Ghoshal, K. MicroRNA-122 inhibits tumorigenic properties of Hepatocellular Carcinoma cells and sensitizes these cells to Sorafenib. J. Biol. Chem. 2009, 284, 32015–32027. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, K.; Zhang, W.; Wang, Y.; Jin, Z.; Jia, B.; Liu, Y. MiR-449a inhibits proliferation and invasion by regulating ADAM10 in Hepatocellular Carcinoma. Am. J. Transl. Res. 2016, 8, 2609–2619. [Google Scholar]
- Liu, Y.; Zhang, W.; Liu, S.; Liu, K.; Ji, B.; Wang, Y. MiR-365 Targets ADAM10 and suppresses the cell growth and metastasis of Hepatocellular Carcinoma. Oncol. Rep. 2017, 37, 1857–1864. [Google Scholar] [CrossRef][Green Version]
- Nakao, K.; Miyaaki, H.; Ichikawa, T. Antitumor function of MicroRNA-122 against Hepatocellular Carcinoma. J. Gastroenterol. 2014, 49, 589–593. [Google Scholar] [CrossRef]
- Wu, G.; Zheng, K.; Xia, S.; Wang, Y.; Meng, X.; Qin, X.; Cheng, Y. MicroRNA-655-3p functions as a tumor suppressor by regulating ADAM10 and β-Catenin pathway in Hepatocellular Carcinoma. J. Exp. Clin. Cancer Res. 2016, 35, 89. [Google Scholar] [CrossRef]
- Xu, Y.; Cao, L.; Chen, G.; Chen, L.; Li, Y.; Lai, Y.; Weng, H.; Chen, T.; Wang, L.; Ye, Y. Human umbilical cord mesenchymal stem cells-derived exosomal MicroRNA-451a represses epithelial-mesenchymal transition of hepatocellular carcinoma cells by inhibiting ADAM. RNA Biol. 2020. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, C.; Wang, Y.; Wen, S.; Wang, J.; Chen, Z.; He, Q.; Feng, D. MicroRNA-145 inhibits cell proliferation by directly targeting ADAM17 in Hepatocellular Carcinoma. Oncol. Rep. 2014, 32, 1923–1930. [Google Scholar] [CrossRef] [PubMed]
- Tsai, W.-C.; Hsu, P.W.-C.; Lai, T.-C.; Chau, G.-Y.; Lin, C.-W.; Chen, C.-M.; Lin, C.-D.; Liao, Y.-L.; Wang, J.-L.; Chau, Y.-P.; et al. MicroRNA-122, a tumor suppressor MicroRNA that regulates intrahepatic metastasis of Hepatocellular Carcinoma. Hepatology 2009, 49, 1571–1582. [Google Scholar] [CrossRef]
- Yang, B.; Wang, C.; Xie, H.; Wang, Y.; Huang, J.; Rong, Y.; Zhang, H.; Kong, H.; Yang, Y.; Lu, Y. MicroRNA-3163 Targets ADAM-17 and enhances the sensitivity of Hepatocellular Carcinoma cells to molecular targeted agents. Cell Death Dis. 2019, 10, 784. [Google Scholar] [CrossRef]
- Yang, X.-W.; Zhang, L.-J.; Huang, X.-H.; Chen, L.-Z.; Su, Q.; Zeng, W.-T.; Li, W.; Wang, Q. MiR-145 Suppresses cell invasion in Hepatocellular Carcinoma Cells: MiR-145 targets ADAM. Hepatol. Res. 2014, 44, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.-Y.; Ding, J.; Tian, W.-G.; Yu, Y.-C. MicroRNA-122 as a diagnostic biomarker for Hepatocellular Carcinoma related to Hepatitis C Virus: A meta-analysis and systematic review. J. Int. Med. Res. 2020, 48, 300060520941634. [Google Scholar] [CrossRef]
- Gao, J.; Yin, X.; Yu, X.; Dai, C.; Zhou, F. Long Noncoding LINC01551 promotes hepatocellular carcinoma cell proliferation, migration, and invasion by acting as a competing endogenous RNA of MicroRNA-122-5p to regulate ADAM10 expression. J. Cell Biochem. 2019, 120, 16393–16407. [Google Scholar] [CrossRef]
- Mazzocca, A.; Coppari, R.; De Franco, R.; Cho, J.-Y.; Libermann, T.A.; Pinzani, M.; Toker, A. A secreted form of ADAM9 promotes Carcinoma invasion through Tumor-Stromal Interactions. Cancer Res. 2005, 65, 4728–4738. [Google Scholar] [CrossRef]
- Shiu, J.-S.; Hsieh, M.-J.; Chiou, H.-L.; Wang, H.-L.; Yeh, C.-B.; Yang, S.-F.; Chou, Y.-E. Impact of ADAM10 gene polymorphisms on Hepatocellular Carcinoma development and clinical characteristics. Int. J. Med. Sci. 2018, 15, 1334–1340. [Google Scholar] [CrossRef]
- Honda, H.; Takamura, M.; Yamagiwa, S.; Genda, T.; Horigome, R.; Kimura, N.; Setsu, T.; Tominaga, K.; Kamimura, H.; Matsuda, Y.; et al. Overexpression of a disintegrin and Metalloproteinase 21 is associated with motility, metastasis, and poor prognosis in Hepatocellular Carcinoma. Sci. Rep. 2017, 7, 15485. [Google Scholar] [CrossRef] [PubMed]
- Zou, B.; Liu, X.; Gong, Y.; Cai, C.; Li, P.; Xing, S.; Pokhrel, B.; Zhang, B.; Li, J. A Novel 12-Marker panel of cancer-associated fibroblasts involved in progression of Hepatocellular Carcinoma. Cancer Manag. Res. 2018, 10, 5303–5311. [Google Scholar] [CrossRef]
- Oh, S.; Park, Y.; Lee, H.-J.; Lee, J.; Lee, S.-H.; Baek, Y.-S.; Chun, S.-K.; Lee, S.-M.; Kim, M.; Chon, Y.-E.; et al. A Disintegrin and Metalloproteinase 9 (ADAM9) in advanced Hepatocellular Carcinoma and their role as a Biomarker during Hepatocellular Carcinoma immunotherapy. Cancers 2020, 12, 745. [Google Scholar] [CrossRef] [PubMed]
- Mattern, J.; Roghi, C.S.; Hurtz, M.; Knäuper, V.; Edwards, D.R.; Poghosyan, Z. ADAM15 mediates upregulation of claudin-1 expression in breast cancer cells. Sci. Rep. 2019, 9, 12540. [Google Scholar] [CrossRef]
- Mailly, L.; Baumert, T.F. Hepatitis C Virus infection and tight junction proteins: The ties that bind. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183296. [Google Scholar] [CrossRef] [PubMed]
- Charrier-Hisamuddin, L.; Laboisse, C.L.; Merlin, D. ADAM-15: A Metalloprotease that mediates inflammation. FASEB J. 2008, 22, 641–653. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, L.; Kelwick, R.; Decock, J.; Edwards, D.R. The Roles of ADAMTS Metalloproteinases in tumorigenesis and metastasis. Front. Biosci. 2011, 16, 1861–1872. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Cheung, S.T.; So, S.; Fan, S.T.; Barry, C.; Higgins, J.; Lai, K.-M.; Ji, J.; Dudoit, S.; Ng, I.O.L.; et al. Gene Expression Patterns in Human Liver Cancers. Mol. Biol. Cell 2002, 13, 1929–1939. [Google Scholar] [CrossRef] [PubMed]
- Masui, T.; Hosotani, R.; Tsuji, S.; Miyamoto, Y.; Yasuda, S.; Ida, J.; Nakajima, S.; Kawaguchi, M.; Kobayashi, H.; Koizumi, M.; et al. Expression of METH-1 and METH-2 in pancreatic cancer. Clin. Cancer Res. 2001, 7, 3437–3443. [Google Scholar]
- Braconi, C.; Meng, F.; Swenson, E.; Khrapenko, L.; Huang, N.; Patel, T. Candidate therapeutic agents for Hepatocellular Cancer can be identified from phenotype-associated gene expression signatures. Cancer 2009, 115, 3738–3748. [Google Scholar] [CrossRef]
- Turner, S.L.; Mangnall, D.; Bird, N.C.; Bunning, R.A.D.; Blair-Zajdel, M.E. Expression of ADAMTS-1, ADAMTS-4, ADAMTS-5 and TIMP3 by Hepatocellular Carcinoma cell lines. Int. J. Oncol. 2012, 41, 1043–1049. [Google Scholar] [CrossRef][Green Version]
- Turkoglu, S.A.; Kockar, F. SP1 and USF differentially regulate ADAMTS1 gene expression under normoxic and hypoxic conditions in Hepatoma Cells. Gene 2016, 575, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Diamantis, I.; Lüthi, M.; Hösli, M.; Reichen, J. Cloning of the Rat ADAMTS-1 Gene and its down regulation in endothelial cells in cirrhotic rats. Liver 2000, 20, 165–172. [Google Scholar] [CrossRef]
- Bourd-Boittin, K.; Bonnier, D.; Leyme, A.; Mari, B.; Tuffery, P.; Samson, M.; Ezan, F.; Baffet, G.; Theret, N. Protease Profiling of Liver Fibrosis Reveals the ADAM Metallopeptidase with Thrombospondin Type 1 Motif, 1 as a Central Activator of Transforming Growth Factor Beta. Hepatology 2011, 54, 2173–2184. [Google Scholar] [CrossRef] [PubMed]
- Schwettmann, L.; Wehmeier, M.; Jokovic, D.; Aleksandrova, K.; Brand, K.; Manns, M.P.; Lichtinghagen, R.; Bahr, M.J. Hepatic Expression of A Disintegrin and Metalloproteinase (ADAM) and ADAMs with Thrombospondin Motives (ADAM-TS) Enzymes in patients with chronic liver diseases. J. Hepatol. 2008, 49, 243–250. [Google Scholar] [CrossRef]
- Li, C.; Xiong, Y.; Yang, X.; Wang, L.; Zhang, S.; Dai, N.; Li, M.; Ren, T.; Yang, Y.; Zhou, S.-F.; et al. Lost expression of ADAMTS5 protein associates with progression and poor prognosis of Hepatocellular Carcinoma. Drug Des. Dev. Ther. 2015, 9, 1773–1783. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yang, C.; Wu, J.; Nan, Y. ADAMTS8 Targets ERK to suppress cell proliferation, invasion, and metastasis of Hepatocellular Carcinoma. Onco Targets Ther. 2018, 11, 7569–7578. [Google Scholar] [CrossRef]
- Li, S.; Li, T.; Li, X.; Yao, Y.; Jiang, X.; Zhao, L.; Guo, W. MicroRNA-32 regulates development and progression of hepatocellular carcinoma by targeting ADAMTS9 and affects its prognosis. Med. Sci. Monit. Basic Res. 2018, 24, 177–187. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, H.; Hu, Y.; Wang, F. Long Non-Coding RNA ADAMTS9-AS1 exacerbates cell proliferation, migration, and invasion via triggering of the PI3K/AKT/MTOR Pathway in Hepatocellular Carcinoma Cells. Am. J. Transl. Res. 2020, 12, 5696–5707. [Google Scholar]
- Bai, K.-H.; He, S.-Y.; Shu, L.-L.; Wang, W.-D.; Lin, S.-Y.; Zhang, Q.-Y.; Li, L.; Cheng, L.; Dai, Y.-J. Identification of Cancer Stem Cell Characteristics in liver Hepatocellular Carcinoma by WGCNA analysis of transcriptome stemness index. Cancer Med. 2020, 9, 4290–4298. [Google Scholar] [CrossRef]
- Uemura, M.; Fujimura, Y.; Ko, S.; Matsumoto, M.; Nakajima, Y.; Fukui, H. Pivotal Role of ADAMTS13 Function in liver diseases. Int. J. Hematol. 2010, 91, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Uemura, M.; Fujimura, Y.; Matsumoto, M.; Ishizashi, H.; Kato, S.; Matsuyama, T.; Isonishi, A.; Ishikawa, M.; Yagita, M.; Morioka, C.; et al. Comprehensive analysis of ADAMTS13 in patients with liver cirrhosis. Thromb. Haemost. 2008, 99, 1019–1029. [Google Scholar] [CrossRef] [PubMed]
- Kavakli, K.; Canciani, M.T.; Mannucci, P.M. Plasma levels of the von willebrand factor-cleaving protease in physiological and pathological conditions in children. Pediatric Hematol. Oncol. 2002, 19, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Uemura, M.; Matsuyama, T.; Ishikawa, M.; Fujimoto, M.; Kojima, H.; Sakurai, S.; Ishii, S.; Toyohara, M.; Yamazaki, M.; Yoshiji, H.; et al. Decreased Activity of Plasma ADAMTS13 May Contribute to the Development of Liver Disturbance and Multiorgan Failure in Patients with Alcoholic Hepatitis. Alcohol. Clin. Exp. Res. 2005, 29, 264S–271S. [Google Scholar] [CrossRef] [PubMed]
- Lisman, T.; Bongers, T.N.; Adelmeijer, J.; Janssen, H.L.A.; de Maat, M.P.M.; de Groot, P.G.; Leebeek, F.W.G. elevated levels of von willebrand factor in cirrhosis support platelet adhesion despite reduced functional capacity. Hepatology 2006, 44, 53–61. [Google Scholar] [CrossRef]
- Ikeda, H.; Tateishi, R.; Enooku, K.; Yoshida, H.; Nakagawa, H.; Masuzaki, R.; Kondo, Y.; Goto, T.; Shiina, S.; Kume, Y.; et al. Prediction of Hepatocellular Carcinoma development by plasma ADAMTS13 in chronic Hepatitis B and C. Cancer Epidemiol. Biomark. Prev. 2011, 20, 2204–2211. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Takaya, H.; Namisaki, T.; Shimozato, N.; Kaji, K.; Kitade, M.; Moriya, K.; Sato, S.; Kawaratani, H.; Akahane, T.; Matsumoto, M.; et al. ADAMTS13 and von willebrand factor are useful biomarkers for sorafenib treatment efficiency in patients with Hepatocellular Carcinoma. World J. Gastrointest. Oncol. 2019, 11, 424–435. [Google Scholar] [CrossRef] [PubMed]
- Takaya, H.; Namisaki, T.; Kitade, M.; Kaji, K.; Nakanishi, K.; Tsuji, Y.; Shimozato, N.; Moriya, K.; Seki, K.; Sawada, Y.; et al. VWF/ADAMTS13 ratio as a potential biomarker for early detection of Hepatocellular Carcinoma. BMC Gastroenterol. 2019, 19, 167. [Google Scholar] [CrossRef]
- Kume, Y.; Ikeda, H.; Inoue, M.; Tejima, K.; Tomiya, T.; Nishikawa, T.; Watanabe, N.; Ichikawa, T.; Kaneko, M.; Okubo, S.; et al. Hepatic stellate cell damage may lead to decreased plasma ADAMTS13 activity in rats. FEBS Lett. 2007, 581, 1631–1634. [Google Scholar] [CrossRef][Green Version]
- Uemura, M.; Tatsumi, K.; Matsumoto, M.; Fujimoto, M.; Matsuyama, T.; Ishikawa, M.; Iwamoto, T.-A.; Mori, T.; Wanaka, A.; Fukui, H.; et al. Localization of ADAMTS13 to the stellate cells of human liver. Blood 2005, 106, 922–924. [Google Scholar] [CrossRef] [PubMed]
- Niiya, M.; Uemura, M.; Zheng, X.W.; Pollak, E.S.; Dockal, M.; Scheiflinger, F.; Wells, R.G.; Zheng, X.L. Increased ADAMTS-13 proteolytic activity in rat hepatic stellate cells upon activation in vitro and in vivo. J. Thromb. Haemost. 2006, 4, 1063–1070. [Google Scholar] [CrossRef]
- Shomron, N.; Hamasaki-Katagiri, N.; Hunt, R.; Hershko, K.; Pommier, E.; Geetha, S.; Blaisdell, A.; Dobkin, A.; Marple, A.; Roma, I.; et al. A splice variant of ADAMTS13 is expressed in human hepatic stellate cells and cancerous tissues. Thromb. Haemost. 2010, 104, 531–535. [Google Scholar] [CrossRef] [PubMed]
- De Tan, I.A.; Ricciardelli, C.; Russell, D.L. The Metalloproteinase ADAMTS1: A Comprehensive review of its role in tumorigenic and Metastatic pathways. Int. J. Cancer 2013, 133, 2263–2276. [Google Scholar] [CrossRef] [PubMed]
- Sheu, M.-J.; Hsieh, M.-J.; Chou, Y.-E.; Wang, P.-H.; Yeh, C.-B.; Yang, S.-F.; Lee, H.-L.; Liu, Y.-F. Effects of ADAMTS14 genetic polymorphism and cigarette smoking on the clinicopathologic development of Hepatocellular Carcinoma. PLoS ONE 2017, 12, e0172506. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Xiong, Y.; Zhong, Z.; Zhang, S.; Peng, Y.; Wang, L.; Dai, N.; Li, M.; Ren, T.; Gan, L.; et al. association between a variant in ADAMTS5 and the susceptibility to hepatocellular carcinoma in a chinese han population. Cell Biochem. Biophys. 2015, 72, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, R.; Jing, R.; Zuo, B.; Zheng, Q. Genome-Wide CRISPR Knockout Screens Identify ADAMTSL3 and PTEN Genes as suppressors of HCC proliferation and metastasis, respectively. J. Cancer Res. Clin. Oncol. 2020, 146, 1509–1521. [Google Scholar] [CrossRef]
- Arechederra, M.; Bazai, S.K.; Abdouni, A.; Sequera, C.; Mead, T.J.; Richelme, S.; Daian, F.; Audebert, S.; Dono, R.; Lozano, A.; et al. ADAMTSL5 is an epigenetically activated gene underlying tumorigenesis and drug resistance in Hepatocellular Carcinoma. J. Hepatol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Koo, B.-H.; Hurskainen, T.; Mielke, K.; Aung, P.P.; Casey, G.; Autio-Harmainen, H.; Apte, S.S. ADAMTSL3/Punctin-2, a Gene frequently mutated in colorectal tumors, is widely expressed in normal and malignant epithelial cells, vascular endothelial cells and other cell types, and its MRNA is reduced in colon cancer. Int. J. Cancer 2007, 121, 1710–1716. [Google Scholar] [CrossRef]
- Nonomura, Y.; Otsuka, A.; Ohtsuka, M.; Yamamoto, T.; Dummer, R.; Kabashima, K. ADAMTSL5 is upregulated in melanoma tissues in patients with idiopathic psoriasis vulgaris induced by Nivolumab. J. Eur. Acad. Dermatol. Venereol. 2017, 31, e100–e101. [Google Scholar] [CrossRef]
- Abdullah, M.; Choo, C.W.; Alias, H.; Abdul Rahman, E.J.; Mohd Ibrahim, H.; Jamal, R.; Hussin, N.H. ADAMTSL5 and CDH11: Putative epigenetic markers for therapeutic resistance in acute Lymphoblastic Leukemia. Hematology 2017, 22, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Kadalayil, L.; Khan, S.; Nevanlinna, H.; Fasching, P.A.; Couch, F.J.; Hopper, J.L.; Liu, J.; Maishman, T.; Durcan, L.; Gerty, S.; et al. Germline variation in ADAMTSL1 is associated with prognosis following breast cancer treatment in young women. Nat. Commun. 2017, 8, 1632. [Google Scholar] [CrossRef]
- Campbell, V.T.; Nadesan, P.; Ali, S.A.; Wang, C.Y.Y.; Whetstone, H.; Poon, R.; Wei, Q.; Keilty, J.; Proctor, J.; Wang, L.W.; et al. Hedgehog pathway inhibition in chondrosarcoma using the smoothened inhibitor IPI-926 directly inhibits sarcoma cell growth. Mol. Cancer Ther. 2014, 13, 1259–1269. [Google Scholar] [CrossRef] [PubMed]
- Chai, R.; Zhang, K.; Wang, K.; Li, G.; Huang, R.; Zhao, Z.; Liu, Y.; Chen, J. A Novel gene signature based on five glioblastoma Stem-like cell relevant genes predicts the survival of primary glioblastoma. J. Cancer Res. Clin. Oncol. 2018, 144, 439–447. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive and integrative genomic characterization of Hepatocellular Carcinoma. Cell 2017, 169, 1327–1341. [Google Scholar] [CrossRef]
- Lambrecht, B.N.; Vanderkerken, M.; Hammad, H. The emerging role of ADAM metalloproteinases in immunity. Nat. Rev. Immunol. 2018, 18, 745–758. [Google Scholar] [CrossRef]
- Ringelhan, M.; Pfister, D.; O’Connor, T.; Pikarsky, E.; Heikenwalder, M. The Immunology of Hepatocellular Carcinoma. Nat. Immunol. 2018, 19, 222–232. [Google Scholar] [CrossRef]
- Yang, Y.M.; Kim, S.Y.; Seki, E. Inflammation and liver cancer: Molecular mechanisms and therapeutic targets. Semin. Liver Dis. 2019, 39, 26–42. [Google Scholar] [CrossRef]
- Zelová, H.; Hošek, J. TNF-α Signalling and inflammation: Interactions between old acquaintances. Inflamm. Res. 2013, 62, 641–651. [Google Scholar] [CrossRef]
- McMahan, R.S.; Riehle, K.J.; Fausto, N.; Campbell, J.S. A Disintegrin and Metalloproteinase 17 Regulates TNF and TNFR1 levels in inflammation and liver regeneration in mice. Am. J. Physiol. Gastrointest Liver Physiol. 2013, 305, G25–G34. [Google Scholar] [CrossRef] [PubMed]
- Maras, J.S.; Das, S.; Sharma, S.; Sukriti, S.; Kumar, J.; Vyas, A.K.; Kumar, D.; Bhat, A.; Yadav, G.; Choudhary, M.C.; et al. Iron-Overload triggers ADAM-17 mediated inflammation in severe alcoholic hepatitis. Sci. Rep. 2018, 8, 10264. [Google Scholar] [CrossRef]
- Argast, G.M.; Campbell, J.S.; Brooling, J.T.; Fausto, N. Epidermal growth factor receptor transactivation mediates tumor necrosis factor-induced hepatocyte replication. J. Biol. Chem. 2004, 279, 34530–34536. [Google Scholar] [CrossRef]
- Berasain, C.; Nicou, A.; Garcia-Irigoyen, O.; Latasa, M.U.; Urtasun, R.; Elizalde, M.; Salis, F.; Perugorría, M.J.; Prieto, J.; Recio, J.A.; et al. Epidermal growth factor receptor signaling in Hepatocellular Carcinoma: Inflammatory activation and a new intracellular regulatory mechanism. Dig. Dis. 2012, 30, 524–531. [Google Scholar] [CrossRef] [PubMed]
- Lokau, J.; Schoeder, V.; Haybaeck, J.; Garbers, C. Jak-Stat signaling induced by Interleukin-6 family cytokines in Hepatocellular Carcinoma. Cancers 2019, 11, 1704. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Arras, D.; Rose-John, S. Regulation of fibrotic processes in the liver by ADAM proteases. Cells 2019, 8, 1226. [Google Scholar] [CrossRef]
- Giannitrapani, L.; Cervello, M.; Soresi, M.; Notarbartolo, M.; La Rosa, M.; Virruso, L.; D’Alessandro, N.; Montalto, G. Circulating IL-6 and SIL-6R in patients with Hepatocellular Carcinoma. Ann. N. Y. Acad. Sci. 2002, 963, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Soresi, M.; Giannitrapani, L.; D’Antona, F.; Florena, A.-M.; La Spada, E.; Terranova, A.; Cervello, M.; D’Alessandro, N.; Montalto, G. Interleukin-6 and its soluble receptor in patients with liver cirrhosis and Hepatocellular Carcinoma. World J. Gastroenterol. 2006, 12, 2563–2568. [Google Scholar] [CrossRef]
- Bergmann, J.; Müller, M.; Baumann, N.; Reichert, M.; Heneweer, C.; Bolik, J.; Lücke, K.; Gruber, S.; Carambia, A.; Boretius, S.; et al. IL-6 Trans-signaling is essential for the development of hepatocellular carcinoma in mice. Hepatology 2017, 65, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Dyczynska, E.; Sun, D.; Yi, H.; Sehara-Fujisawa, A.; Blobel, C.P.; Zolkiewska, A. Proteolytic processing of Delta-like 1 by ADAM proteases. J. Biol. Chem. 2007, 282, 436–444. [Google Scholar] [CrossRef]
- Huang, Q.; Li, J.; Zheng, J.; Wei, A. The carcinogenic role of the notch signaling pathway in the development of Hepatocellular Carcinoma. J. Cancer 2019, 10, 1570–1579. [Google Scholar] [CrossRef]
- Lu, J.; Xia, Y.; Chen, K.; Zheng, Y.; Wang, J.; Lu, W.; Yin, Q.; Wang, F.; Zhou, Y.; Guo, C. Oncogenic role of the notch pathway in primary liver cancer. Oncol. Lett. 2016, 12, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Burghardt, S.; Erhardt, A.; Claass, B.; Huber, S.; Adler, G.; Jacobs, T.; Chalaris, A.; Schmidt-Arras, D.; Rose-John, S.; Karimi, K.; et al. Hepatocytes contribute to immune regulation in the liver by activation of the notch signaling pathway in T cells. J. Immunol. 2013, 191, 5574–5582. [Google Scholar] [CrossRef]
- Wang, R.; Li, Y.; Tsung, A.; Huang, H.; Du, Q.; Yang, M.; Deng, M.; Xiong, S.; Wang, X.; Zhang, L.; et al. INOS promotes CD24+CD133+ liver cancer stem cell phenotype through a TACE/ADAM17-Dependent notch signaling pathway. Proc. Natl. Acad. Sci. USA 2018, 115, E10127–E10136. [Google Scholar] [CrossRef]
- Chalupský, K.; Kanchev, I.; Žbodáková, O.; Buryová, H.; Jiroušková, M.; Kořínek, V.; Gregor, M.; Sedláček, R. ADAM10/17-dependent release of soluble c-Met correlates with hepatocellular damage. Folia Biol. 2013, 59, 76–86. [Google Scholar]
- Barisione, G.; Fabbi, M.; Gino, A.; Queirolo, P.; Orgiano, L.; Spano, L.; Picasso, V.; Pfeffer, U.; Mosci, C.; Jager, M.J.; et al. Potential Role of Soluble C-Met as a new candidate biomarker of metastatic uveal melanoma. JAMA Ophthalmol. 2015, 133, 1013–1021. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.-F.; Li, A.-N.; Yang, J.-J.; Chen, Z.-H.; Xie, Z.; Zhang, X.-C.; Su, J.; Lou, N.-N.; Yan, H.-H.; Han, J.-F.; et al. Soluble C-Met levels correlated with tissue c-Met protein expression in patients with advanced non-small-cell lung cancer. Clin. Lung Cancer 2017, 18, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.J.; Yang, J.H.; Kim, J.; Ma, S.H.; Cho, L.Y.; Ko, K.-P.; Shin, A.; Choi, B.Y.; Kim, H.J.; Han, D.S.; et al. Soluble C-Met protein as a susceptible biomarker for gastric cancer risk: A nested case-control study within the korean multicenter cancer cohort. Int. J. Cancer 2013, 132, 2148–2156. [Google Scholar] [CrossRef]
- Moosavi, F.; Giovannetti, E.; Saso, L.; Firuzi, O. HGF/MET pathway aberrations as diagnostic, prognostic, and predictive biomarkers in human cancers. Crit. Rev. Clin. Lab. Sci. 2019, 56, 533–566. [Google Scholar] [CrossRef]
- Xing, S.; Ferrari de Andrade, L. NKG2D and MICA/B shedding: A “tag Game” between NK cells and Malignant cells. Clin. Transl. Immunol. 2020, 9, e1230. [Google Scholar] [CrossRef]
- Arai, J.; Goto, K.; Tanoue, Y.; Ito, S.; Muroyama, R.; Matsubara, Y.; Nakagawa, R.; Kaise, Y.; Lim, L.A.; Yoshida, H.; et al. Enzymatic inhibition of MICA sheddase ADAM17 by lomofungin in Hepatocellular Carcinoma Cells. Int. J. Cancer 2018, 143, 2575–2583. [Google Scholar] [CrossRef] [PubMed]
- Kohga, K.; Takehara, T.; Tatsumi, T.; Miyagi, T.; Ishida, H.; Ohkawa, K.; Kanto, T.; Hiramatsu, N.; Hayashi, N. Anticancer chemotherapy inhibits MHC Class I-Related chain a ectodomain shedding by downregulating ADAM10 expression in Hepatocellular Carcinoma. Cancer Res. 2009, 69, 8050–8057. [Google Scholar] [CrossRef]
- Kohga, K.; Takehara, T.; Tatsumi, T.; Ishida, H.; Miyagi, T.; Hosui, A.; Hayashi, N. Sorafenib Inhibits the Shedding of Major Histocompatibility Complex Class I-Related chain a on hepatocellular carcinoma cells by down-regulating a disintegrin and metalloproteinase. Hepatology 2010, 51, 1264–1273. [Google Scholar] [CrossRef] [PubMed]
- Arai, J.; Goto, K.; Stephanou, A.; Tanoue, Y.; Ito, S.; Muroyama, R.; Matsubara, Y.; Nakagawa, R.; Morimoto, S.; Kaise, Y.; et al. Predominance of regorafenib over sorafenib: Restoration of membrane-bound MICA in hepatocellular carcinoma cells. J. Gastroenterol. Hepatol. 2018, 33, 1075–1081. [Google Scholar] [CrossRef]
- Gérard, C.; Hubeau, C.; Carnet, O.; Bellefroid, M.; Sounni, N.E.; Blacher, S.; Bendavid, G.; Moser, M.; Fässler, R.; Noel, A.; et al. Microenvironment-Derived ADAM28 prevents cancer dissemination. Oncotarget 2018, 9, 37185–37199. [Google Scholar] [CrossRef]
- Zha, Z.; Jia, F.; Hu, P.; Mai, E.; Lei, T. MicroRNA-574-3p inhibits the malignant behavior of liver cancer cells by targeting ADAM. Oncol. Lett. 2020, 20, 3015–3023. [Google Scholar] [CrossRef] [PubMed]
- Hubeau, C.; Rocks, N.; Cataldo, D. ADAM28: Another ambivalent protease in cancer. Cancer Lett. 2020, 494, 18–26. [Google Scholar] [CrossRef]
- Higashiyama, S.; Nanba, D. ADAM-Mediated Ectodomain shedding of HB-EGF in receptor Cross-Talk. Biochim. Biophys. Acta 2005, 1751, 110–117. [Google Scholar] [CrossRef]
- Sanderson, M.P.; Dempsey, P.J.; Dunbar, A.J. Control of ErbB signaling through metalloprotease mediated ectodomain shedding of EGF-like factors. Growth Factors 2006, 24, 121–136. [Google Scholar] [CrossRef]
- Itabashi, H.; Maesawa, C.; Oikawa, H.; Kotani, K.; Sakurai, E.; Kato, K.; Komatsu, H.; Nitta, H.; Kawamura, H.; Wakabayashi, G.; et al. Angiotensin II and epidermal growth factor receptor cross-talk mediated by a disintegrin and metalloprotease accelerates tumor cell proliferation of hepatocellular carcinoma cell lines. Hepatol. Res. 2008, 38, 601–613. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-J.; Feng, C.-W.; Li, M. ADAM17 mediates hypoxia-induced drug resistance in hepatocellular carcinoma cells through activation of EGFR/PI3K/Akt pathway. Mol. Cell Biochem. 2013, 380, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Giannelli, G.; Koudelkova, P.; Dituri, F.; Mikulits, W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J. Hepatol. 2016, 65, 798–808. [Google Scholar] [CrossRef]
- Pinzani, M. Epithelial-Mesenchymal transition in chronic liver disease: Fibrogenesis or escape from death? J. Hepatol. 2011, 55, 459–465. [Google Scholar] [CrossRef]
- Dong, Y.; Wu, Z.; He, M.; Chen, Y.; Chen, Y.; Shen, X.; Zhao, X.; Zhang, L.; Yuan, B.; Zeng, Z. ADAM9 mediates the Interleukin-6-Induced epithelial-mesenchymal transition and metastasis through ROS production in hepatoma cells. Cancer Lett. 2018, 421, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.-Y.; Chu, H.-X.; Tan, Y.-X.; Qin, X.-C.; Liu, M.-Y.; Li, J.-D.; Ren, T.-S.; Zhang, Y.-S.; Zhao, Q.-C. Novel ADAM-17 inhibitor ZLDI-8 inhibits the metastasis of hepatocellular carcinoma by reversing epithelial-mesenchymal transition in vitro and in vivo. Life Sci. 2020, 244, 117343. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, D.; Jiang, Q.; Cao, S.; Sun, H.; Chai, Y.; Li, X.; Ren, T.; Yang, R.; Feng, F.; et al. Novel ADAM-17 inhibitor ZLDI-8 enhances the in vitro and in vivo chemotherapeutic effects of sorafenib on hepatocellular carcinoma cells. Cell Death Dis. 2018, 9, 743. [Google Scholar] [CrossRef]
- Hong, S.W.; Hur, W.; Choi, J.E.; Kim, J.-H.; Hwang, D.; Yoon, S.K. Role of ADAM17 in invasion and migration of CD133-expressing liver cancer stem cells after irradiation. Oncotarget 2016, 7, 23482–23497. [Google Scholar] [CrossRef]
- Li, Y.; Ren, Z.; Wang, Y.; Dang, Y.-Z.; Meng, B.-X.; Wang, G.-D.; Zhang, J.; Wu, J.; Wen, N. ADAM17 promotes cell migration and invasion through the integrin β1 pathway in hepatocellular carcinoma. Exp. Cell Res. 2018, 370, 373–382. [Google Scholar] [CrossRef]
- Saha, S.K.; Choi, H.Y.; Yang, G.-M.; Biswas, P.K.; Kim, K.; Kang, G.-H.; Gil, M.; Cho, S.-G. GPR50 promotes hepatocellular carcinoma progression via the notch signaling pathway through direct interaction with ADAM. Mol. Ther. Oncolytics 2020, 17, 332–349. [Google Scholar] [CrossRef]
- Wu, B.; Cui, J.; Yang, X.-M.; Liu, Z.-Y.; Song, F.; Li, L.; Jiang, J.-L.; Chen, Z.-N. Cytoplasmic fragment of CD147 generated by regulated intramembrane proteolysis contributes to HCC by promoting autophagy. Cell Death Dis. 2017, 8, e2925. [Google Scholar] [CrossRef]
- Fabregat, I.; Moreno-Càceres, J.; Sánchez, A.; Dooley, S.; Dewidar, B.; Giannelli, G.; Ten Dijke, P. IT-LIVER Consortium TGF-β signalling and liver disease. FEBS J. 2016, 283, 2219–2232. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Murata, M.; Yamaguchi, T.; Matsuzaki, K. TGF-β/Smad signaling during hepatic fibro-carcinogenesis (Review). Int. J. Oncol. 2014, 45, 1363–1371. [Google Scholar] [CrossRef]
- Le Pabic, H.; L’Helgoualc’h, A.; Coutant, A.; Wewer, U.M.; Baffet, G.; Clément, B.; Théret, N. Involvement of the serine/threonine P70S6 kinase in TGF-Beta1-induced ADAM12 expression in cultured human hepatic stellate cells. J. Hepatol. 2005, 43, 1038–1044. [Google Scholar] [CrossRef]
- Gruel, J.; Leborgne, M.; LeMeur, N.; Théret, N. In silico investigation of ADAM12 effect on TGF-Beta Receptors trafficking. BMC Res. Notes 2009, 2, 193. [Google Scholar] [CrossRef]
- Ruff, M.; Leyme, A.; Le Cann, F.; Bonnier, D.; Le Seyec, J.; Chesnel, F.; Fattet, L.; Rimokh, R.; Baffet, G.; Théret, N. The disintegrin and metalloprotease ADAM12 is associated with TGF-β-Induced epithelial to mesenchymal transition. PLoS ONE 2015, 10, e0139179. [Google Scholar] [CrossRef]
- Dekky, B.; Ruff, M.; Bonnier, D.; Legagneux, V.; Théret, N. Proteomic screening identifies the zonula occludens protein ZO-1 as a new partner for ADAM12 in invadopodia-like structures. Oncotarget 2018, 9, 21366–21382. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Eckert, M.A.; Santiago-Medina, M.; Lwin, T.M.; Kim, J.; Courtneidge, S.A.; Yang, J. ADAM12 Induction by Twist1 promotes tumor invasion and metastasis via regulation of invadopodia and focal adhesions. J. Cell Sci. 2017, 130, 2036–2048. [Google Scholar] [CrossRef] [PubMed]
- Laurent, M.-A.; Bonnier, D.; Théret, N.; Tufféry, P.; Moroy, G. In Silico Characterization of the interaction between LSKL peptide, a LAP-TGF-Beta derived peptide, and ADAMTS. Comput. Biol. Chem. 2016, 61, 155–161. [Google Scholar] [CrossRef]
- Amann, T.; Bataille, F.; Spruss, T.; Mühlbauer, M.; Gäbele, E.; Schölmerich, J.; Kiefer, P.; Bosserhoff, A.-K.; Hellerbrand, C. Activated hepatic stellate cells promote tumorigenicity of hepatocellular carcinoma. Cancer Sci. 2009, 100, 646–653. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).