Expression of Extracellular Matrix-Related Genes and Their Regulatory microRNAs in Problematic Colorectal Polyps
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
2.1. Patient Characteristics
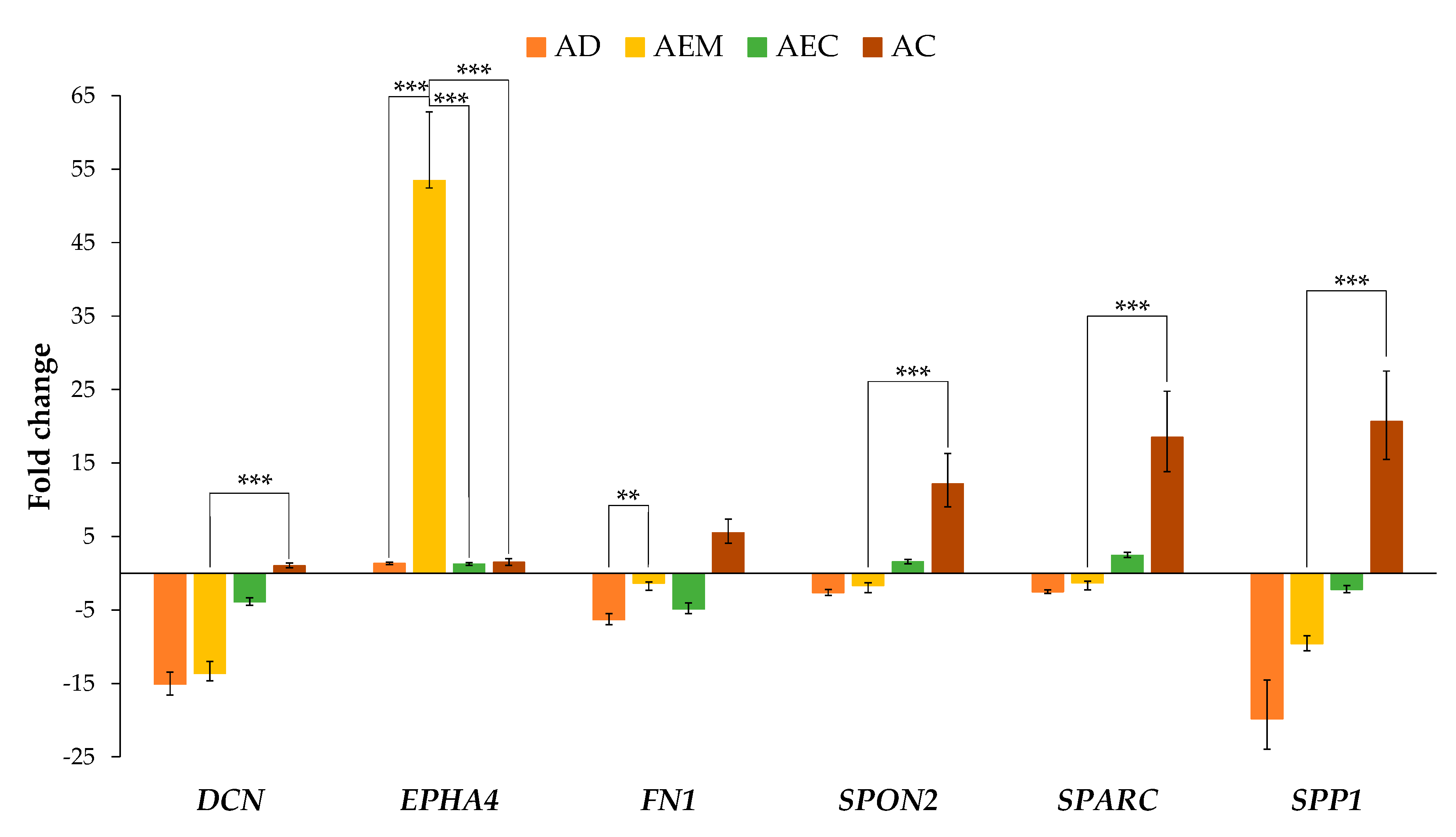
2.2. Expression of ECM-Related Genes in Adenoma with Epithelial Misplacement Compared to Other Groups
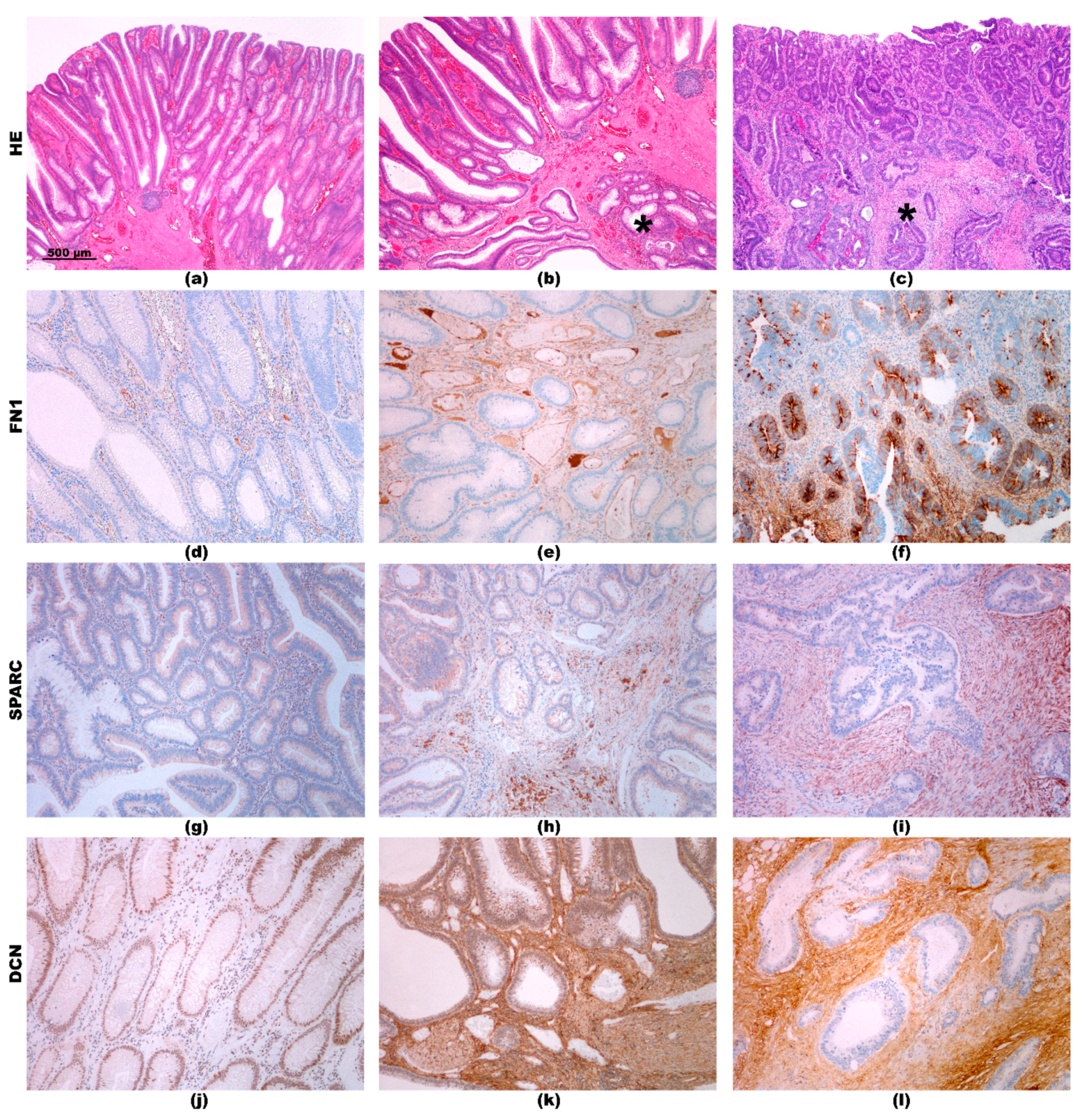
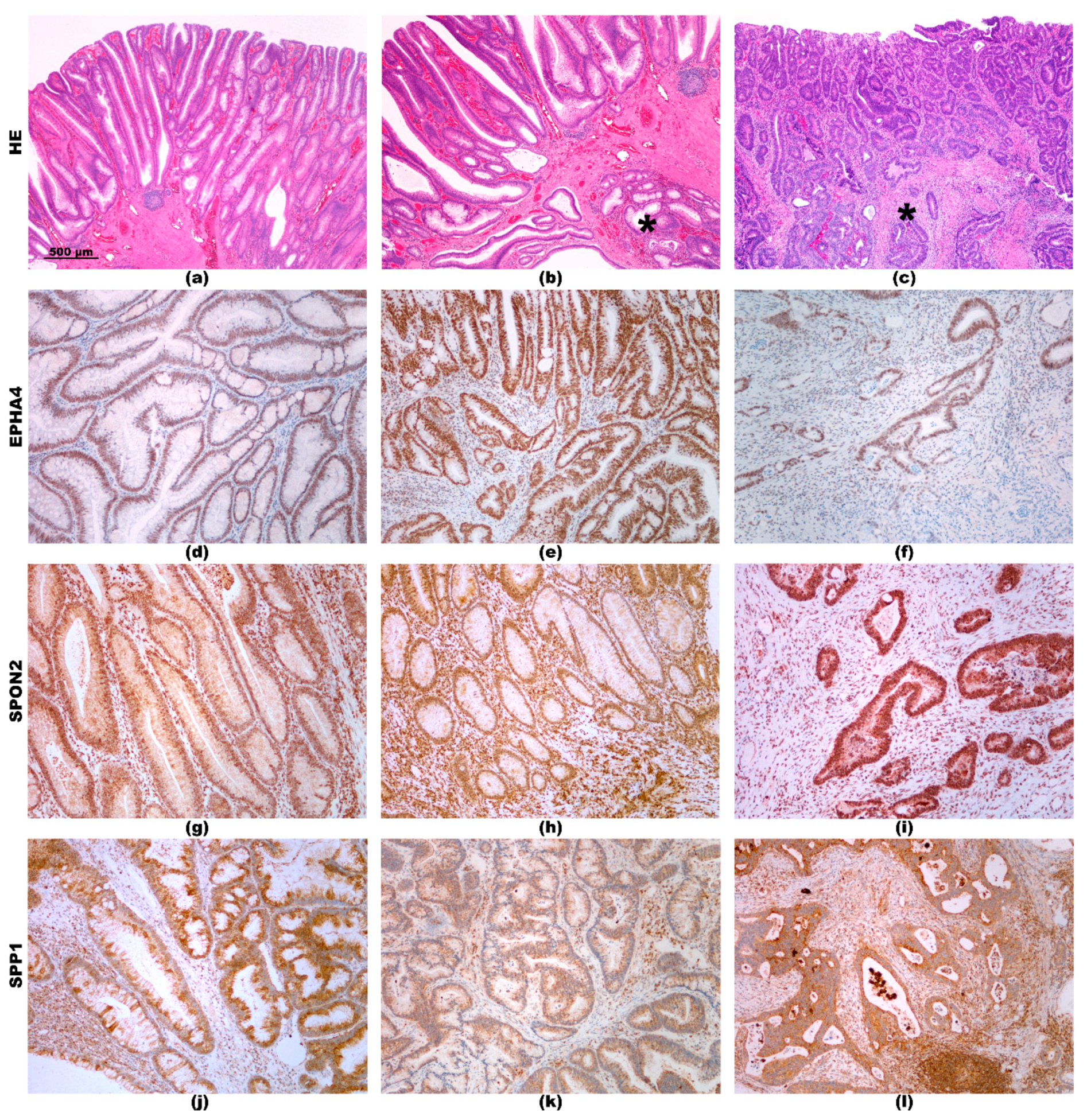
2.3. Immunohistochemistry
2.4. Determining Regulatory microRNAs for the ECM-Related Genes
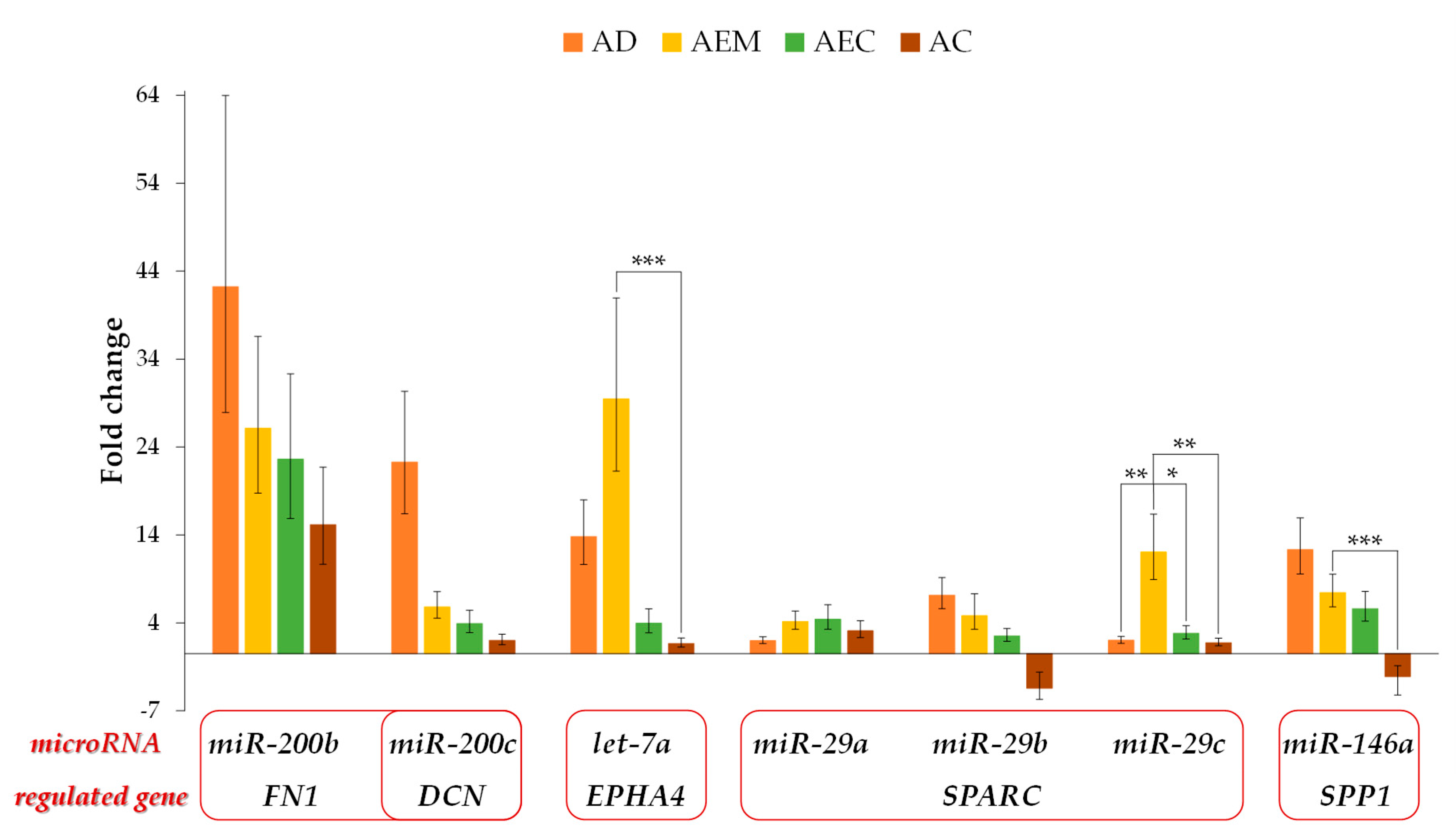
2.5. Expression of the Potential Regulatory microRNAs for the ECM-Related Genes
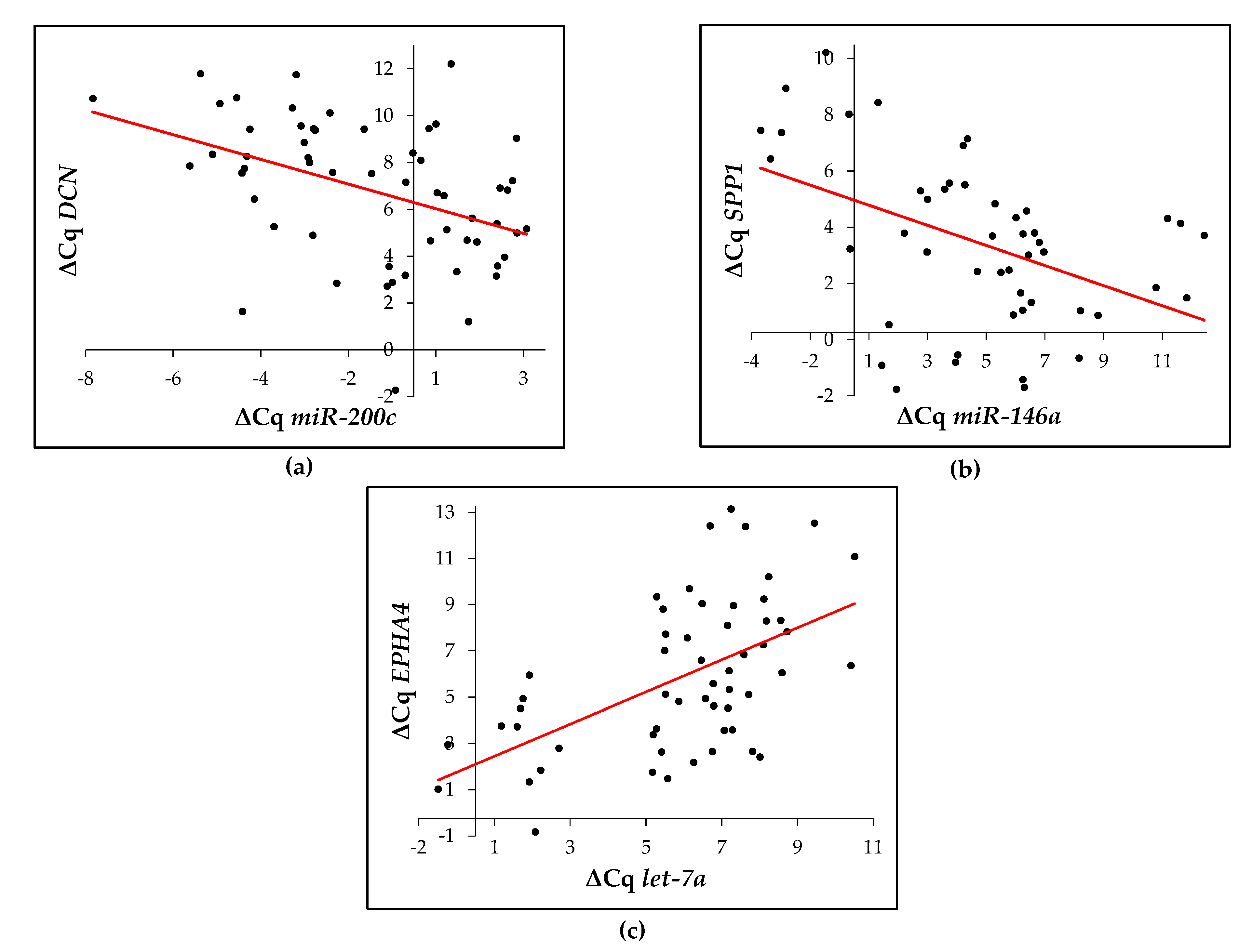
2.6. Correlation between the Expression of Potential Regulating microRNA and Their Target ECM-Related Gene
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Identification of the Potential Regulatory microRNAs
4.3. Isolation of RNA from Formalin-Fixed Paraffin-Embedded Tissue
4.4. Reverse Transcription and Quantitative PCR
4.5. Immunohistochemistry
4.6. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Simon, K. Colorectal cancer development and advances in screening. Clin. Interv. Aging 2016, 11, 967–976. [Google Scholar] [CrossRef]
- Zlajpah, M.; Hauptman, N.; Bostjancic, E.; Zidar, N. Differential expression of extracellular matrixrelated genes DCN, EPHA4, FN1, SPARC, SPON2 and SPP1 in colorectal carcinogenesis. Oncol. Rep. 2019. [Google Scholar] [CrossRef]
- Mathonnet, M.; Perraud, A.; Christou, N.; Akil, H.; Melin, C.; Battu, S.; Jauberteau, M.O.; Denizot, Y. Hallmarks in colorectal cancer: Angiogenesis and cancer stem-like cells. World J. Gastroenterol. 2014, 20, 4189–4196. [Google Scholar] [CrossRef]
- Kuipers, E.J.; Grady, W.M.; Lieberman, D.; Seufferlein, T.; Sung, J.J.; Boelens, P.G.; van de Velde, C.J.; Watanabe, T. Colorectal cancer. Nat. Rev. Dis. Prim. 2015, 1, 15065. [Google Scholar] [CrossRef]
- Hauptman, N.; Bostjancic, E.; Zlajpah, M.; Rankovic, B.; Zidar, N. Bioinformatics analysis reveals most prominent gene candidates to distinguish colorectal adenoma from adenocarcinoma. Biomed. Res. Int. 2018, 2018, 9416515. [Google Scholar] [CrossRef]
- Loughrey, M.B.; Shepherd, N.A. The pathology of bowel cancer screening. Histopathology 2015, 66, 66–77. [Google Scholar] [CrossRef]
- Shepherd, N.A.; Griggs, R.K. Bowel cancer screening-generated diagnostic conundrum of the century: Pseudoinvasion in sigmoid colonic polyps. Mod. Pathol. 2015, 28 (Suppl. 1), 88–94. [Google Scholar] [CrossRef]
- Griggs, R.K.; Novelli, M.R.; Sanders, D.S.; Warren, B.F.; Williams, G.T.; Quirke, P.; Shepherd, N.A. Challenging diagnostic issues in adenomatous polyps with epithelial misplacement in bowel cancer screening: 5 years’ experience of the Bowel Cancer Screening Programme Expert Board. Histopathology 2017, 70, 466–472. [Google Scholar] [CrossRef]
- Falzone, L.; Scola, L.; Zanghi, A.; Biondi, A.; Di Cataldo, A.; Libra, M.; Candido, S. Integrated analysis of colorectal cancer microRNA datasets: Identification of microRNAs associated with tumor development. Aging (Albany NY) 2018, 10, 1000–1014. [Google Scholar] [CrossRef]
- Nagy, Z.B.; Wichmann, B.; Kalmar, A.; Galamb, O.; Bartak, B.K.; Spisak, S.; Tulassay, Z.; Molnar, B. Colorectal adenoma and carcinoma specific miRNA profiles in biopsy and their expression in plasma specimens. Clin. Epigenet. 2017, 9, 22. [Google Scholar] [CrossRef]
- Slattery, M.L.; Herrick, J.S.; Pellatt, D.F.; Stevens, J.R.; Mullany, L.E.; Wolff, E.; Hoffman, M.D.; Samowitz, W.S.; Wolff, R.K. MicroRNA profiles in colorectal carcinomas, adenomas and normal colonic mucosa: Variations in miRNA expression and disease progression. Carcinogenesis 2016, 37, 245–261. [Google Scholar] [CrossRef]
- Zlajpah, M.; Bostjancic, E.; Zidar, N. (Epi)genetic regulation of osteopontin in colorectal cancerogenesis. Epigenomics 2020, 12, 1389–1403. [Google Scholar] [CrossRef]
- Karagkouni, D.; Paraskevopoulou, M.D.; Chatzopoulos, S.; Vlachos, I.S.; Tastsoglou, S.; Kanellos, I.; Papadimitriou, D.; Kavakiotis, I.; Maniou, S.; Skoufos, G.; et al. DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018, 46, D239–D245. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4. [Google Scholar] [CrossRef]
- Hsu, J.B.; Chiu, C.M.; Hsu, S.D.; Huang, W.Y.; Chien, C.H.; Lee, T.Y.; Huang, H.D. miRTar: An integrated system for identifying miRNA-target interactions in human. BMC Bioinform. 2011, 12, 300. [Google Scholar] [CrossRef]
- Jevsinek Skok, D.; Hauptman, N.; Bostjancic, E.; Zidar, N. The integrative knowledge base for miRNA-mRNA expression in colorectal cancer. Sci. Rep. 2019, 9, 18065. [Google Scholar] [CrossRef]
- Tang, R.Y.; Wang, Z.; Chen, H.Q.; Zhu, S.B. Negative correlation between miR-200c and decorin plays an important role in the pathogenesis of colorectal carcinoma. Biomed. Res. Int. 2017, 2017, 1038984. [Google Scholar] [CrossRef]
- Yang, X.; Hu, Q.; Hu, L.X.; Lin, X.R.; Liu, J.Q.; Lin, X.; Dinglin, X.X.; Zeng, J.Y.; Hu, H.; Luo, M.L.; et al. miR-200b regulates epithelial-mesenchymal transition of chemo-resistant breast cancer cells by targeting FN1. Discov. Med. 2017, 24, 75–85. [Google Scholar]
- Kun-Peng, Z.; Chun-Lin, Z.; Xiao-Long, M.; Lei, Z. Fibronectin-1 modulated by the long noncoding RNA OIP5-AS1/miR-200b-3p axis contributes to doxorubicin resistance of osteosarcoma cells. J. Cell. Physiol. 2019, 234, 6927–6939. [Google Scholar] [CrossRef]
- Chen, H.B.; Zheng, H.T. MicroRNA-200c represses migration and invasion of gastric cancer SGC-7901 cells by inhibiting expression of fibronectin 1. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 1753–1758. [Google Scholar]
- Zhang, H.; Sun, Z.; Li, Y.; Fan, D.; Jiang, H. MicroRNA-200c binding to FN1 suppresses the proliferation, migration and invasion of gastric cancer cells. Biomed. Pharmacother. 2017, 88, 285–292. [Google Scholar] [CrossRef]
- Liu, Q.; Guo, X.; Que, S.; Yang, X.; Fan, H.; Liu, M.; Li, X.; Tang, H. LncRNA RSU1P2 contributes to tumorigenesis by acting as a ceRNA against let-7a in cervical cancer cells. Oncotarget 2017, 8, 43768–43781. [Google Scholar] [CrossRef]
- Deng, B.; Qu, L.; Li, J.; Fang, J.; Yang, S.; Cao, Z.; Mei, Z.; Sun, X. MiRNA-211 suppresses cell proliferation, migration and invasion by targeting SPARC in human hepatocellular carcinoma. Sci. Rep. 2016, 6, 26679. [Google Scholar] [CrossRef]
- Qiu, F.; Sun, R.; Deng, N.; Guo, T.; Cao, Y.; Yu, Y.; Wang, X.; Zou, B.; Zhang, S.; Jing, T.; et al. miR-29a/b enhances cell migration and invasion in nasopharyngeal carcinoma progression by regulating SPARC and COL3A1 gene expression. PLoS ONE 2015, 10, e0120969. [Google Scholar] [CrossRef]
- Wang, C.; Gao, C.; Zhuang, J.L.; Ding, C.; Wang, Y. A combined approach identifies three mRNAs that are down-regulated by microRNA-29b and promote invasion ability in the breast cancer cell line MCF-7. J. Cancer Res. Clin. Oncol. 2012, 138, 2127–2136. [Google Scholar] [CrossRef]
- Zhang, S.; Jin, J.; Tian, X.; Wu, L. hsa-miR-29c-3p regulates biological function of colorectal cancer by targeting SPARC. Oncotarget 2017, 8, 104508–104524. [Google Scholar] [CrossRef]
- Xu, Y.; Ge, K.; Lu, J.; Huang, J.; Wei, W.; Huang, Q. MicroRNA-493 suppresses hepatocellular carcinoma tumorigenesis through down-regulation of anthrax toxin receptor 1 (ANTXR1) and R-Spondin 2 (RSPO2). Biomed. Pharmacother. 2017, 93, 334–343. [Google Scholar] [CrossRef]
- Tu, M.; Li, Y.; Zeng, C.; Deng, Z.; Gao, S.; Xiao, W.; Luo, W.; Jiang, W.; Li, L.; Lei, G. MicroRNA-127-5p regulates osteopontin expression and osteopontin-mediated proliferation of human chondrocytes. Sci. Rep. 2016, 6, 25032. [Google Scholar] [CrossRef]
- Shevde, L.A.; Metge, B.J.; Mitra, A.; Xi, Y.; Ju, J.; King, J.A.; Samant, R.S. Spheroid-forming subpopulation of breast cancer cells demonstrates vasculogenic mimicry via hsa-miR-299-5p regulated de novo expression of osteopontin. J. Cell. Mol. Med. 2010, 14, 1693–1706. [Google Scholar] [CrossRef]
- Kou, C.J.; Kandpal, R.P. Differential expression patterns of Eph receptors and ephrin ligands in human cancers. Biomed. Res. Int. 2018, 2018, 7390104. [Google Scholar] [CrossRef]
- Giaginis, C.; Tsoukalas, N.; Bournakis, E.; Alexandrou, P.; Kavantzas, N.; Patsouris, E.; Theocharis, S. Ephrin (Eph) receptor A1, A4, A5 and A7 expression in human non-small cell lung carcinoma: Associations with clinicopathological parameters, tumor proliferative capacity and patients’ survival. BMC Clin. Pathol. 2014, 14, 8. [Google Scholar] [CrossRef]
- Petrey, A.C.; de la Motte, C.A. The extracellular matrix in IBD: A dynamic mediator of inflammation. Curr. Opin. Gastroenterol. 2017, 33, 234–238. [Google Scholar] [CrossRef]
- Sorokin, L. The impact of the extracellular matrix on inflammation. Nat. Rev. Immunol. 2010, 10, 712–723. [Google Scholar] [CrossRef]
- Denhardt, D.T.; Noda, M.; O’Regan, A.W.; Pavlin, D.; Berman, J.S. Osteopontin as a means to cope with environmental insults: Regulation of inflammation, tissue remodeling, and cell survival. J. Clin. Investig. 2001, 107, 1055–1061. [Google Scholar] [CrossRef]
- Lu, P.; Takai, K.; Weaver, V.M.; Werb, Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef]
- Lu, P.; Weaver, V.M.; Werb, Z. The extracellular matrix: A dynamic niche in cancer progression. J. Cell Biol. 2012, 196, 395–406. [Google Scholar] [CrossRef]
- Pickup, M.W.; Mouw, J.K.; Weaver, V.M. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014, 15, 1243–1253. [Google Scholar] [CrossRef]
- Li, Z.L.; Wang, Z.J.; Wei, G.H.; Yang, Y.; Wang, X.W. Changes in extracellular matrix in different stages of colorectal cancer and their effects on proliferation of cancer cells. World. J. Gastrointest. Oncol. 2020, 12, 267–275. [Google Scholar] [CrossRef]
- Napoli, S.; Scuderi, C.; Gattuso, G.; Bella, V.D.; Candido, S.; Basile, M.S.; Libra, M.; Falzone, L. Functional roles of matrix metalloproteinases and their inhibitors in melanoma. Cells 2020, 9, 1151. [Google Scholar] [CrossRef]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Santi, A.; Kugeratski, F.G.; Zanivan, S. Cancer associated fibroblasts: The architects of stroma remodeling. Proteomics 2018, 18, e1700167. [Google Scholar] [CrossRef]
- Cox, T.R.; Erler, J.T. Remodeling and homeostasis of the extracellular matrix: Implications for fibrotic diseases and cancer. Dis. Model. Mech. 2011, 4, 165–178. [Google Scholar] [CrossRef]
- Caja, L.; Dituri, F.; Mancarella, S.; Caballero-Diaz, D.; Moustakas, A.; Giannelli, G.; Fabregat, I. TGF-beta and the tissue microenvironment: Relevance in fibrosis and cancer. Int. J. Mol. Sci. 2018, 19, 1294. [Google Scholar] [CrossRef]
- Zhang, W.; Ge, Y.; Cheng, Q.; Zhang, Q.; Fang, L.; Zheng, J. Decorin is a pivotal effector in the extracellular matrix and tumour microenvironment. Oncotarget 2018, 9, 5480–5491. [Google Scholar] [CrossRef]
- Bi, X.L.; Yang, W. Biological functions of decorin in cancer. Chin. J. Cancer 2013, 32, 266–269. [Google Scholar] [CrossRef]
- Drev, D.; Harpain, F.; Beer, A.; Stift, A.; Gruber, E.S.; Klimpfinger, M.; Thalhammer, S.; Reti, A.; Kenner, L.; Bergmann, M.; et al. Impact of fibroblast-derived SPARC on invasiveness of colorectal cancer cells. Cancers 2019, 11, 1421. [Google Scholar] [CrossRef]
- Viana Lde, S.; Affonso, R.J., Jr.; Silva, S.R.; Denadai, M.V.; Matos, D.; Salinas de Souza, C.; Waisberg, J. Relationship between the expression of the extracellular matrix genes SPARC, SPP1, FN1, ITGA5 and ITGAV and clinicopathological parameters of tumor progression and colorectal cancer dissemination. Oncology 2013, 84, 81–91. [Google Scholar] [CrossRef]
- Feng, Y.; Hu, Y.; Mao, Q.; Guo, Y.; Liu, Y.; Xue, W.; Cheng, S. Upregulation of Spondin-2 protein expression correlates with poor prognosis in hepatocellular carcinoma. J. Int. Med. Res. 2019, 47, 569–579. [Google Scholar] [CrossRef]
- Fan, Y.; Zhang, X.; Yang, Z.H.; Sun, X.W.; Li, S.N.; Zhong, L.; Cheng, X.; Wang, Y.; Ma, Y.R. The polymorphisms of osteopontin gene and plasma osteopontin protein levels with susceptibility to colorectal carcinoma. DNA Cell Biol. 2013, 32, 594–600. [Google Scholar] [CrossRef]
- Lamort, A.S.; Giopanou, I.; Psallidas, I.; Stathopoulos, G.T. Osteopontin as a link between inflammation and cancer: The thorax in the spotlight. Cells 2019, 8, 815. [Google Scholar] [CrossRef]
- Valcz, G.; Sipos, F.; Krenacs, T.; Molnar, J.; Patai, A.V.; Leiszter, K.; Toth, K.; Solymosi, N.; Galamb, O.; Molnar, B.; et al. Elevated osteopontin expression and proliferative/apoptotic ratio in the colorectal adenoma-dysplasia-carcinoma sequence. Pathol. Oncol. Res. 2010, 16, 541–545. [Google Scholar] [CrossRef]
- Coulthard, M.G.; Morgan, M.; Woodruff, T.M.; Arumugam, T.V.; Taylor, S.M.; Carpenter, T.C.; Lackmann, M.; Boyd, A.W. Eph/Ephrin signaling in injury and inflammation. Am. J. Pathol. 2012, 181, 1493–1503. [Google Scholar] [CrossRef]
- Pasquale, E.B. Eph-ephrin bidirectional signaling in physiology and disease. Cell 2008, 133, 38–52. [Google Scholar] [CrossRef]
- Ivanov, A.I.; Romanovsky, A.A. Putative dual role of ephrin-Eph receptor interactions in inflammation. IUBMB Life 2006, 58, 389–394. [Google Scholar] [CrossRef]
- Shiuan, E.; Chen, J. Eph receptor tyrosine kinases in tumor immunity. Cancer Res. 2016, 76, 6452–6457. [Google Scholar] [CrossRef]
- Latham, G.J. Normalization of microRNA quantitative RT-PCR data in reduced scale experimental designs. Methods Mol. Biol. 2010, 667, 19–31. [Google Scholar] [CrossRef]
| Demographic Characteristics | Healthy Colon Mucosa | Adenoma | Adenoma with Epithelial Misplacement | Adenoma with Early Carcinoma | Advanced Carcinoma |
|---|---|---|---|---|---|
| male:female | 10:11 | 10:0 | 10:3 | 6:4 | 7:4 |
| Age (mean ± SD) | 48–90 (75 ± 11) | 50–83 (65 ± 11) | 47–78 (58 ± 8) | 56–72 (65 ± 5) | 58–90 (76 ± 11) |
| microRNA | Gene | TarBase [13] | TargetScan [14] | miRTar [15] | miRNA & CRC [16] | Literature |
|---|---|---|---|---|---|---|
| hsa-miR-200c | DCN | - | - | - | + | [17] |
| hsa-miR-200b | FN1 | + | - | - | + | [18,19] |
| hsa-miR-200c | FN1 | + | - | - | + | [20,21] |
| hsa-let-7a | EPHA4 | - | + | - | + | [22] |
| hsa-miR-211 | SPARC | - | - | + | + | [23] |
| hsa-miR-29a | SPARC | + | + | + | + | - |
| hsa-miR-29b | SPARC | + | + | + | + | [24,25] |
| hsa-miR-29c | SPARC | + | + | + | + | [26] |
| hsa-miR-493 | SPON2 | - | - | - | - | [27] |
| hsa-miR-146a [12] | SPP1 | - | - | + | + | - |
| hsa-miR-127 [12] | SPP1 | - | - | + | + | [28] |
| hsa-miR-299 [12] | SPP1 | - | - | + | + | [29] |
| Gene or microRNA | Assay ID |
|---|---|
| B2M | Hs 99999907_m1 |
| IPO8 | Hs 00183533_m1 |
| DCN | Hs 00266491_m1 |
| EPHA4 | Hs 00953178_m1 |
| FN1 | Hs 01549976_m1 |
| SPARC | Hs 00234160_m1 |
| SPP1 | Hs 00959010_m1 |
| SPON2 | Hs 01557678_g1 |
| RNU6B | 001093 |
| hsa-miR-1274b | 002884 |
| hsa-miR-29a-5p | 002112 |
| hsa-miR-29b-5p | 00041 |
| hsa-miR-29c-5p | 000587 |
| hsa-miR-127-5p | 002229 |
| hsa-miR-146a-5p | 000468 |
| hsa-miR-200b | 002251 |
| hsa-miR-200c | 002300 |
| hsa-miR-211 | 000514 |
| hsa-miR-299-5p | 000600 |
| hsa-miR-493 | 001040 |
| hsa-let-7a | 000377 |
| Antibody | Manufacturer | Catalogue Number | Type | Host Species | Dilution | Antigen Retrieval | Duration of Primary Antibody Incubation (T = 37 °C) |
|---|---|---|---|---|---|---|---|
| DCN | LifeSpan Biosciences | LS-B4312-50 | Monoclonal | Mouse | 1:1000 | No pretreatment | 12 min |
| EPHA4 | SantaCruz Biotechnology | SC-365503 | Monoclonal | Mouse | 1:500 | Cell conditioning solution 1 (Ventana BenchMark) | 12 min |
| FN1 | Cell Signaling | 26836S | Monoclonal | Rabbit | 1:300 | 28 min | |
| SPARC | Invitrogen, Thermo Fisher Scientific, Inc. | 33-5500 | Monoclonal | Mouse | 1:400 | 24 min | |
| SPON2 | PA5-59087 | Polyclonal | Rabbit | 1:20 | 32 min | ||
| SPP1 | MA5-17180 | Monoclonal | Mouse | 1:300 | 24 min |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Žlajpah, M.; Boštjančič, E.; Tepeš, B.; Zidar, N. Expression of Extracellular Matrix-Related Genes and Their Regulatory microRNAs in Problematic Colorectal Polyps. Cancers 2020, 12, 3715. https://doi.org/10.3390/cancers12123715
Žlajpah M, Boštjančič E, Tepeš B, Zidar N. Expression of Extracellular Matrix-Related Genes and Their Regulatory microRNAs in Problematic Colorectal Polyps. Cancers. 2020; 12(12):3715. https://doi.org/10.3390/cancers12123715
Chicago/Turabian StyleŽlajpah, Margareta, Emanuela Boštjančič, Bojan Tepeš, and Nina Zidar. 2020. "Expression of Extracellular Matrix-Related Genes and Their Regulatory microRNAs in Problematic Colorectal Polyps" Cancers 12, no. 12: 3715. https://doi.org/10.3390/cancers12123715
APA StyleŽlajpah, M., Boštjančič, E., Tepeš, B., & Zidar, N. (2020). Expression of Extracellular Matrix-Related Genes and Their Regulatory microRNAs in Problematic Colorectal Polyps. Cancers, 12(12), 3715. https://doi.org/10.3390/cancers12123715






