Simple Summary
NK cells are innate immune cells that form one of the initial responses to infections and cancers. There have been increasing number of studies investigating the anti-tumor effects of NK cells. Immunotherapy targeting NK cell may enhance the therapeutic efficacy of current immunotherapy regimes. Through pro-inflammatory cytokine production, enhancing B cell production of antibodies, facilitate and activate dendritic cells, activate T cells and participating in anti-tumor immunity through the granzyme B pathway and antibody-dependent cellular cytotoxicity, the versatility of NK cells provides an attractive immunotherapy option. This review highlights NK cell biology, NK cell antitumor immunity, NK cell immune evasive mechanisms and novel immunotherapies that aim to target NK cells.
Abstract
Natural killer (NK) cells are a key component of an innate immune system. They are important not only in initiating, but also in augmenting adaptive immune responses. NK cell activation is mediated by a carefully orchestrated balance between the signals from inhibitory and activating NK cell receptors. NK cells are potent producers of proinflammatory cytokines and are also able to elicit strong antitumor responses through secretion of perforin and granzyme B. Tumors can develop many mechanisms to evade NK cell antitumor responses, such as upregulating ligands for inhibitory receptors, secreting anti-inflammatory cytokines and recruiting immunosuppressive cells. Enhancing NK cell responses will likely augment the effectiveness of immunotherapies, and strategies to accomplish this are currently being evaluated in clinical trials. A comprehensive understanding of NK cell biology will likely provide additional opportunities to further leverage the antitumor effects of NK cells. In this review, we therefore sought to highlight NK cell biology, tumor evasion of NK cells and clinical trials that target NK cells.
1. Introduction
Along with innate lymphoid cell 1 (ILC1), natural killer (NK) cells are a part of the group 1 innate lymphoid cells (ILC) family that differentiate from common lymphoid progenitor cells [1]. Unlike ILC1, which display more of a T helper cell-like phenotype and are noncytotoxic, NK cells are cytotoxic [2]. NK cells not only detect and destroy viruses and other pathogens, but also cells undergoing early malignant transformation, without previous priming, hence they are members of the innate immune system. While NK cells constitute only 5–15% of total lymphocytes in peripheral blood [3], their innate ability to detect and target transformed cells (e.g., tumor cells) makes them key cells in antitumor immune surveillance. Mature circulating NK cells carry granules of granzymes and perforins and store transcript for IFN-γ, which they can utilize to destroy tumor cells at first contact in minutes [4]. The innate ability of NK cells to release a potent cytotoxic response has contributed to increasing interest in their utility as a therapeutic target in cancer.
The contribution of NK cells in the immune response against cancer was recorded as early as the 1970s [5,6]. One of the first NK cell studies investigated the effects of NK cell depletion on the growth of melanoma and found that depleted NK cells in melanoma-bearing mice resulted in higher tumor burden compared to non-NK cell depleted mice [7]. Subsequent studies also found that NK cells produce IFN-γ, which is a critical ingredient for the activation and recruitment of cytotoxic T lymphocytes (CTLs) [8]. These cytokines are also essential for an effective adaptive immune response against cancer through macrophage and dendritic cells (DC) activation, M2-macrophage and CD4+ T cell differentiation and suppression of myeloid derived suppressor cells (MDSC) and Tregs (extensively reviewed in [9,10]). This review focuses on the importance of NK cells in mediating an effective anticancer immune response in the context of melanoma and opportunities to overcome tumor resistance to modern immunotherapies.
1.1. NK Cells Subtypes
NK cells are generally characterized into two major subsets based on their expression of CD56 (adhesion molecule) and CD16 (Fc gamma receptor). NK cells are often classified as CD56brightCD16neg or CD56dimCD16bright, which represent functionally distinct NK cell phenotypes [11] (Figure 1). CD56brightCD16neg NK cells represent 10–15% of the total NK cell population and are mainly present in lymph nodes (~75%). They are potent producers of cytokines, especially IFN-γ, and have poor cytolytic capacity [11]. In contrast, CD56dimCD16bright NK cells represent 90% of peripheral blood NK cells and possess strong cytolytic capacity but are relatively poor producers of cytokines [12].
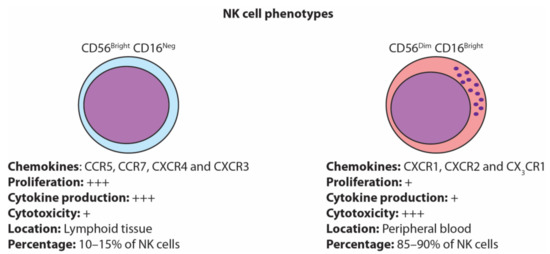
Figure 1.
Natural killer (NK) cell phenotypes. NK cells are generally classified as either the immature CD56bright CD16neg or the mature CD56dim CD16bright.
1.2. NK Cell Activation
NK cell activation is highly dynamic and dependent upon the cumulative effect of activating and inhibitory signals [13]. NK cells have a repertoire of receptors that send activating or inhibitory signals into the NK cell [14]. The ligands for NK cell activating receptors are only upregulated upon cellular stress such as a viral infection or malignant transformation [15]. On the other hand, the ligands for NK cell inhibitory receptors are largely confined to the various heavy chains of the MHC Class I molecule expressed on all healthy nucleated cells [16]. Changes in this balance, for example, the absence of MHC class I expression, are considered abnormal and push NK cells towards an activated state. When the cumulative effect of activating and inhibitory signals surpasses the NK cell activating threshold, NK cell activation occurs [17,18].
Under normal circumstances, healthy nucleated cells, engagement between MHC class I and various killer cell immunoglobulin-like receptors (KIRs) and/or NKG2A/CD94 provide inhibitory signals to NK cells [16]. When this is coupled with the lack of activating receptor stimulation, NK cells are rendered anergic [14]. However, when the target cell becomes virally or malignantly transformed, the cell may downregulate MHC class I and/or upregulate ligands that are stimulated upon cellular stress. The lack of inhibitory receptor activation due to MHC class I downregulation, coupled with large signals being propagated by NK cell-activating receptors, due to the upregulation of stress ligands, cause NK cell activation [14]. Therefore, for NK cells to perform effector functions, stimulation of multiple activating receptors is required to overcome the threshold of activation.
2. NK Cell-Activating Receptors and Their Ligands
2.1. Natural Cytotoxicity Receptors (NCRs)
NCRs are NK cell-activating receptors that that play an important role in NK cell-mediated tumor killing [19]. There are three NCRs expressed on human NK cells: NCR1 (NKp46/CD335), NCR2 (NKp44/CD336) and NCR3 (NKp30/CD337). While NCR1 and NCR3 are constitutively expressed on resting NK cells [20,21,22], NCR2 is only induced upon NK cell activation [23].
The diversity of NCR ligands enables NK cells to effectively detect microbial infections or tumor cells. NCR ligands can be microbial proteins that are presented on their plasma membrane or proteins that are upregulated upon cellular stress. However, due to difficulties in copurifying NCR with NCR ligands, many tumoral NCR ligands remain unknown [24]. Using NCR-Ig fusion proteins that target the extracellular domain of NKp30, NKp44 and NKp46, the density of various NCR ligands was quantified without knowing the exact identity of all the NCR ligands [25]. In metastatic melanoma, NKp30 ligands were minimally expressed [25], while NKp44 and NKp46 ligands had heterogenous expression that seemed to depend on tumor heterogeneity and site of disease [25,26].
There are several NCR ligands that have been discovered in melanoma cells. B7-H6 is a protein that is not expressed on healthy cells and is a ligand for NKp30; however, upon malignant transformation, the activation of the proto-oncogene Myc causes upregulation of B7-H6 [27]. B7-H6 is expressed on the surface of melanoma cells [28], and this protein is shown to be susceptible to membrane shedding by metalloproteases [29]. In fact, B7-H6 is detectable at high levels in the serum of melanoma patients, highlighting that B7-H6 shedding may represent an immune evasive mechanism by melanoma cells to evade potential NK cell mediated killing. Human leukocyte antigen-B-associated transcript 3 (BAT3) is another NKp30 ligand that was shown to upregulate surface expression on melanoma cells upon the activation of the pattern recognition receptor, retinoic acid-inducible gene I (RIG-I), and its upregulation led to potent antitumor activity in melanoma mice models [30]. Galectin-3 is another NKp30 ligand that is expressed on melanoma cells [31]. Despite galectin-3 increasing metastatic potential of tumor cells by increasing its motility via metalloproteinase regulation [32], strong galectin-3 expression in melanoma patients was associated with improved survival [31].
2.2. DNAX Accessory Molecule-1 (DNAM-1/CD226)
The expression of the activating receptor DNAM-1 varies on NK cells, with NK cells that highly express DNAM-1 producing higher levels of proinflammatory cytokines and chemokines (e.g., IFN-g, IL-6, GM-CSF and CCL5), having increased proliferation rates, and being responsive to IL-15 stimulation [33]. DNAM-1 has two known ligands, CD155/PVR and CD112/nectin-2 [34,35,36]. CD155 is expressed in the majority of melanomas, while only 26% of melanomas express CD112 [37]. Interestingly, CD155 expression was seen in the majority of primary and metastatic melanoma, with CD155 showing significant correlation with known negative melanoma prognostic markers such as lymph node involvement and Breslow thickness of the primary tumor [38]. Tumor-expressed CD155 can bind to DNAM-1 on NK cells and enhance the ability of the NK cell to effectively suppress and kill tumor cells in a DNAM-1-dependent manner [26,39]. In fact, CD155 expression on melanomas has also shown to be associated with a poorer response of metastatic melanoma patients to anti-PD-1 monotherapy and combination anti-PD-1 and CTLA-4 [40]. CD155 also exists in a soluble form secreted by tumor cells [41], which impacts NK cell functionality by impairing NK cell activation via DNAM-1 [42]. In fact, patients with advanced gastric cancer had significantly higher levels of soluble CD155 (sCD155), and sCD155 levels directly correlated with tumor burden [43]. The secretion of sCD155 is not restricted to gastric cancer, as other cancers such as breast, prostate and melanoma also secrete sCD155 [44]. Unlike other soluble NK cell ligands, sCD155 is not a result of protease activity [44], but rather due to the CD155 isotype that lacks the transmembrane domain [41].
2.3. Natural-Killer Group 2 Member D (NKG2D)
NKG2D is an activating receptor that belongs to the family of C-type lectin-like receptors and is expressed on all NK cells, along with a subset of activated dendritic and CD8+ T cells [45]. In NK cells, NKG2D expression correlates with degranulation and IFN-g expression in NK cells [46]. NKG2D receptors have eight known ligands that are upregulated upon cellular stress, which include MICA, MICB and ULBP1, 2, 3, 4, 5 and 6. Of all the NKG2D ligands, MICA was shown to be highly expressed in melanoma [47]. However, these ligands can also be shed off the tumor surface through disulphide isomerase (ERp5) and several proteases belonging to disintegrin and metalloproteinase (ADAM) and matrix metalloproteinase (MMP) families [48,49,50,51]. In fact, blocking MICA and MICB shedding from the tumor surface reduced melanoma metastasis and improved the antitumor immunity in an NKG2D- and CD16-dependent manner [52]. The absence of MICB and ULBP-1 in baseline serum, signifying that MICB and ULBP-1 had reduced shedding, is significantly correlated with improved survival in patients with metastatic melanoma treated with immune checkpoint blockade [53].
2.4. CD16a
There are two isotypes of CD16: CD16a (FcγRIIIa) and CD16b (FcγRIIIb). CD16a is expressed by several immune cell types including NK cells, macrophages, monocytes, neutrophils and T cells, while CD16b is exclusively expressed by neutrophils [54]. In NK cells, CD16a binds to the Fc portion of the immunoglobulins IgG1 and IgG3, and, to a minimal extent, with IgG2 and IgG4 [55], and mediates antibody-dependent cell-mediated cytotoxicity (ADCC). Unlike other activating NK cell receptors, the engagement of CD16a to IgG is sufficient for NK cells to release perforin and granzyme B, leading to tumor cell death [56,57,58]. In addition to ADCC, CD16-IgG ligation decreases CD16 surface expression, increases IFN-g production and upregulates CD54 on NK cells [59].
CD16a is a low-affinity Fc receptor; however, a recent study has uncovered the diversity of 234 CD16 polymorphisms which alter the affinity of CD16 to IgG between individuals. Certain polymorphisms also improved the reactivity of NK cells to all four IgG isotypes (IgG1, IgG2, IgG3 and IgG4). For instance, the presence of homozygous valine (V/V) or heterozygous valine/phenylalanine (V/F) at amino acid 158 enhances CD16 binding affinity to IgG1 and IgG3 compared to homozygous phenylalanine (F/F) phenotype [60,61]. As a result, a lower antibody concentration is needed to effectively activate CD16 with a V158V or V158F polymorphism [59]. In this study, 62% of participants had either V158F (55%) or V158V (7%) CD16 polymorphisms that increased CD16 binding affinity, while 38% had F158F polymorphism, which caused CD16 binding affinity to be lower than that of V158F and V158V [60]. This highlights that a significant proportion of society has polymorphisms, at least at amino acid 158, that increase CD16 binding affinity. Further studies are required to delineate whether multiple CD16 polymorphisms are present and whether CD16 binding affinities contribute to a patient’s response to immunotherapies.
2.5. Other NK Cell Activating Molecules
CD69 is a membrane-bound type II C-type lectin and is associated with NK cell activation [62]. Not only is CD69 a marker of NK cell activation, it also plays a functional role in improving NK cell cytotoxicity and various effector functions [63]. CD69 has various ligands, including galectin-1 [62], S100-A8/S100-A9 [62] and myosin light chains 9 and 10 [64]. CD69 induction is mediated by IL-2, IFN-α, CD16 cross-linking and activation of NK cell receptors [65], which stimulated signaling via PTK [65,66] or PKC pathways [65]. While PKC blockade resulted in the abolishment of CD69 induction by unknown NK cell activating ligands, PKC blockade did not affect CD69 induction by IL-2, IFN-α and CD16 cross-linking, PTK blockade abrogated CD69 expression in NK cells stimulated by IL-2, IFN-α, CD16 cross-linking and activation of NK cell-activating ligands [65]. CD69 was shown in many studies to induce NK cell cytotoxicity against tumor cells [67,68,69,70] and that CD69 blockade reduced NK cell cytotoxic capacity to the same level as not activated NK cells [67]. Interestingly, anti-CD69 agonists not only improved NK cell cytotoxicity, but also decreased TGF-β1 expression, increased TGF-α expression, induced NK cell proliferation, upregulated CD25 and ICAM-1 expression [63]. This highlights the multifaceted role of CD69 in regulating NK cell effector functions and may be an attractive future immunotherapy target to improve NK cell responsiveness.
CD44 is a cell surface glycoprotein that not only indicates NK cell activation, but also induces cytokine production and enhances NK cell-mediated cytotoxicity [71,72,73]. CD44 is constitutively expressed on NK cells and when it binds with hyaluronan [72], the main ligand for CD44, coupled with the recruitment of serine-threonine, tyrosine and PI3K signalling, perforin- and granzyme B-dependent NK cell cytotoxicity occurs [73]. In fact, mAb activating CD44 was shown to improve NK cell cytotoxicity [74]. Not only is CD44 implicated in NK cell cytotoxicity, CD44–CD44 ligand interaction upregulates CD69 expression [71], TNF-α production [71], IFN-γ production [72], enhanced CD16-mediated ADCC [75] and the adhesion molecules LFA-3 and LFA-1 [75]. Interestingly, the upregulation of adhesion molecules enhances the binding of NK cells to target cells [75].
3. NK Cell Inhibitory Receptors and Their Ligands
3.1. Killer-Cell Immunoglobulin-Like Receptors (KIRs)
Although the majority of KIRs are inhibitory receptors, some are activating receptors; KIR gene family encode receptors that recognize HLA-A, B, C or G, a major component of MHC class I. The 14 KIR genes and 2 pseudogenes enable great diversity through KIR haplotype and allelic variations [76,77], thus resulting in approximately 1000 allelic variations [78]. Inhibitory KIRs have long cytoplasmic tails (L) accompanied by immunoreceptor tyrosine-based inhibitory motifs (ITIMs), while activating KIRs have a short cytoplasmic tail (S) accompanied by immunoreceptor tyrosine-based activating motifs (ITAMs), with the exception of KIR2DL4, which is an activating KIR that has a long cytoplasmic tail [76,79].
The function of activating KIRs is not well known and activating KIRs are thought to have evolved from inhibitory KIRs [80]. Many NK cells do not possess activating KIRs, suggesting that these receptors are not essential for NK cell functionality [81]. NK cells activated by KIR2DL4 function to stimulate NK cell cytokine production rather than cytotoxicity, highlighting that activating KIRs may shape NK cell responses [79]. In contrast, inhibitory KIRs play an essential role not only in mediating inhibitory signals to suppress NK cell activation, but also in educating NK cells by tuning the threshold for NK cell activation. Without inhibitory KIRs, NK cells become unresponsive [82]. MHC class I downregulation is a tumor evasive mechanism [83] that frequently occurs, and through KIRs, NK cells possess the ability to recognize and eradicate these MHC class I-deficient tumor cells.
Despite several studies, there are no consistent results that highlight an association between KIR receptors with response or prognosis. The expression of KIR2DS5 (activating KIR) was shown to be lower in rapidly progressing patients with advanced melanoma compared to those who progressed slowly [84]. Within the same cohort, stage III or IV melanoma patients had higher expression of KIR2DL2 compared to stage I melanoma patients [84]. In a different study, KIR2DL3 was found to be a protective marker for nodular and ulcerated melanoma [85]. More recently, no NK cell KIR genotypes or KIR/KIR-ligand combinations were shown not to correlate with response to anti-PD-1 therapy [86]. The inconsistencies in attributing certain KIRs with response may be due to great haplotype and allelic diversity within and between ethnicities [87] or may potentially reflect the level of NK cell reactivity via NK cell education, as KIR receptors are known to play an important role in NK cell education [78]. Monoclonal antibody blocking of KIR receptors may improve NK cell functionality and improve patient survival, as demonstrated by the promising effects of KIR-HLA mismatch NK cells in the treatment of leukemia [88].
3.2. CD94/NKG2A
NKG2A/CD94 is a heterodimeric receptor that induces inhibitory signals; high expression is associated with an NK cell exhaustion phenotype [89]. NKG2A/CD94 recognizes the nonclassical MHC class I molecule, HLA-E [90]. HLA-E is not recognized by KIR receptors, has limited polymorphism and is expressed in low levels in normal tissue [91,92]. Generally, HLA-E is absent or expressed in low amounts by melanomas. However, IFN-γ significantly upregulates the expression of HLA-E on the tumor cell surface [93]. Given that NK cells are known potent producers of IFN-γ [94], it is likely that the presence of NK cells could cause an upregulation of HLA-E on melanoma cells. This in turn enables the tumor to become resistant to NK cell killing. This resistance to NK cell killing can be effectively reversed through NKG2A blockade [95]. NKG2A blockade has been shown to improve NK cell cytotoxicity against tumor cells, but it also increases IFN-γ production [96].
3.3. TACTILE (CD96)
TACTILE (CD96) has been shown to bind CD155 with higher affinity than DNAM-1 [97]. However, the role of TACTILE is unclear, as some studies have shown it to be associated with NK cell activation [98], while others demonstrated that the presence of ITIM-like motifs compete with DNAM-1, which consequently regulates NK cell-mediated cytotoxicity [97,99].
3.4. Other Immune Checkpoint Receptors
Programmed cell death protein 1 (PD-1), cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), T-cell immunoglobulin and mucin domain containing-3 (TIM-3) and T cell immunoreceptor with Ig and ITIM domains (TIGIT) are all immune checkpoint receptors that are known to be expressed by T cells, and when stimulated, inhibit T cell activation. Like T cells, PD-1, CTLA-4, TIM-3 and TIGIT are also expressed on NK cells, whereby the expression of these receptors suppresses NK cell response. This will be explored later in this review.
4. NK Cell-Mediated Antitumor Immunity
4.1. NK Cell Recruitment to the Tumor Microenvironment
Chemokine receptor profiles contribute to the function of NK cells and govern their location within the body [100]. CD56bright and CD56dim express different chemokine receptor profiles that contribute to their phenotype [100]. CD56bright NK cells constitutively express the receptors CCR7, CCR5, CXCR4 and CXCR3 [100] that attract CD56bright NK cells to secondary lymphoid organs [101]. Conversely, CD56dim NK cells uniquely express CX3CR1, CXCR2 and CXCR1 [100,102,103]. However, CD56dim NK cells can express lower levels of CXCR3, CXCR4 and CCR7 in the presence of IL-18 [104]. CCR7 can also be acquired on CD56dim NK cells via trogocytosis with mature DCs in a KIR2DS1-dependent and IL-18-independent manner [105,106]. In malignancies, several chemokines, including CCL5, CCL27 and CX3CL1 [107], can be produced by tumor cells, which in turn contributes to the recruitment of NK cells. In B16-F10 mouse model of melanoma, genetic silencing of CCL5 in tumor cells completely abrogated NK cell recruitment to the tumor microenvironment (TME) [108]. In particular, CX3CL1, a chemokine that recruits CXCR3-expressing CD56dim NK cells, is a positive prognostic factor for patient survival and NK cell infiltration in five different malignancies [109,110,111,112,113]. This suggests that CD56dim NK cells and their chemoattractants are important for tumor control in patients with malignant disease.
4.2. Antitumor Effects of NK Cells in the TME
NK cells within the TME have many effects on tumor cells (Figure 2); in particular, they have the capacity to directly lyse tumor cells. NK cell-mediated cytotoxicity is initiated by the accumulation of activating signals that overpowers the inhibitory signals. When NK cell cytotoxicity is initiated, NK cells secrete perforin, which forms membrane pores on the target membrane, which would then allow granzyme B to enter the target cell and cause apoptosis through caspase activation [114]. NK cells can also induce apoptosis in its target cell through the interaction between Fas ligand (FasL/CD95L) or TNF-related apoptosis-inducing ligand (TRAIL) with death receptors on the target cell [115].
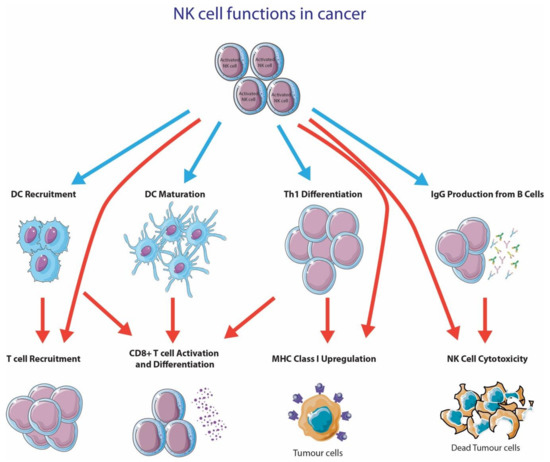
Figure 2.
NK cell functions in cancer. This figure was created using Servier Medical Art templates, which are licensed under a Creative Commons Attribution 3.0 Unported License; https://smart.servier.com (accessed 10 October 2020).
NK cells also play an important role in augmenting the antitumor capacity of other immune cells. In particular, NK cells produce various proinflammatory cytokines like IFN-γ, GM-CSGF, TNF and many others [116]. NK cells are also implicated to produce T cell recruiting chemokines (IL-8, MIP-1a and RANTES) upon ADCC [117]. One critical cytokine produced by NK cells is IFN-γ, as it has profound effects on a broad range of immune cells [118]. IFN-γ has been shown to enhance tumor antigen presentation on MHC class I [119], reduce angiogenesis [120], induce production of IgG from B cells [121,122], activate macrophages and NK cells [123,124], promote Th1 differentiation [125] and have antiproliferative [126] and apoptotic effects [127].
Recent studies have suggested that NK cells play a critical role in DC recruitment to the tumor microenvironment [107]. DCs are essential for CD8+ T cell activation, which is one of the first steps towards alerting the adaptive immune system for proliferation and the execution of T cell effector functions [128]. The recruitment of Batf3+ DCs (cDC1) was shown to be crucial for CD8+ T cell recruitment [128] and the generation of an immunologically inflamed tumor [129]. Böttcher and colleagues have recently discovered that NK cells played an essential role in recruiting CD8+ T cells into the TME via CCL5 and XCL1 secretion [130]. In fact, several studies have found that tumor-infiltrating NK cells are an important source of XCL1 [130,131]. In fact, the XCL1-XCR1 axis is shown to be exclusively connected to cross-presenting DCs [132,133,134]. The recruitment of XCR1+ cross-presenting DCs via XCL1-secreting NK cells is essential for generating a robust T cell response. In addition to DC recruitment via NK cell-dependent XCL1, NK cells also convert infiltrating monocytes into DCs [135]. This conversion is dependent on direct cell-to-cell contact between NK cells and monocytes, and NK cell-secreted cytokines, GM-CSF and CD154 [135].
4.3. NK Cells Are Important for DC Maturation and Activation
DCs are essential innate immune cells that play an important role in bridging the innate and adaptive immune systems. T cells only activate and proliferate when professional antigen presenting cells, like DCs, present antigens on either MHC class I or II to CD4+ or CD8+ T cells, respectively. In the past decade, NK cells have gained significant attention due to their critical role in DC maturation or activation.
Before DC maturation/activation, DCs remain in an immature state, where they are specialized for antigen capture [136,137,138,139]. Immature DCs in cancer are deleterious to effective antitumor immunity [140,141], due to several reasons. Immature DCs lack expression of costimulatory molecules [142] and therefore, when they present antigens, it results in apoptosis [143,144] or anergy [144,145] or development into Treg [146]. However, activated NK cells can lyse immature DCs via NKp30, despite the increased expression of MHC class I [147]. By contrast, mature DCs are protected from NK cell lysis [147], likely due to their expression of NKp30 ligands. Additionally, a hallmark of DC maturation is the upregulation of MHC class I [148]; therefore, the lack of MHC class I expression on immature DCs may also contribute to NK cells selectively lysing immature DCs. Therefore, NK cell-mediated DC editing promotes antigen-specific T cell proliferation, but most importantly, generates a greater protective response during cancer vaccinations [149].
5. NK Cell Evasion Mechanisms Utilized by Melanoma
Many cancers upregulate the ligands for NK cell-activating receptors during malignant transformation. However, many cancers, including melanoma, develop various ways to evade NK cell recognition and lysis. NK cell evasion can be mediated through cell-to-cell contact with melanoma cells, secretion of cytokines/molecules by tumor cells or by immunosuppressive cells and the creation of a hypoxic tumor microenvironment (Figure 3).
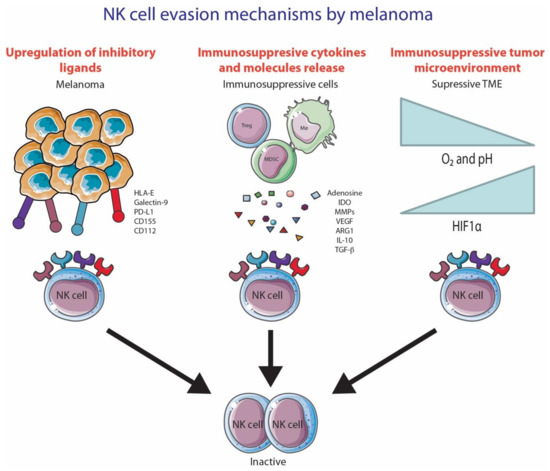
Figure 3.
NK cell evasion mechanisms by melanoma. (1) NK cell inhibition due to the upregulation of inhibitory ligands; (2) Secretion of immunosuppressive cytokines/molecules by immunosuppressive cells; (3) Hypoxic tumor microenvironment. This figure was created using Servier Medical Art templates, which are licensed under a Creative Commons Attribution 3.0 Unported License; https://smart.servier.com (accessed 10 October 2020).
5.1. Cell-to-Cell Contact
NK cells have many inhibitory receptors on their cell surface that constantly engage with their environment. Tumor cells are able to upregulate ligands for these inhibitory receptors to inhibit NK cell responses. Regulatory T cells (Tregs) are inhibitory T cells that are recruited to the tumor environment. Tregs inhibit NK cell responses through membrane-bound TGF-b interaction [150] and CTLA-4 [151]. Antigen presenting cells can also anergize NK cells through CD80 or CD86 interaction with CTLA-4 on NK cells [152]. Cell-to-cell contact between tumor-associated fibroblasts can cause the downregulation of an important NK cell-activating receptor, DNAM-1 [153]. However, the mechanism by which this cell-to-cell contact inhibition occurs is unknown.
Ironically, cell-to-cell contact between NK cells and tumor cells has been shown to not only impact the expression of activating receptors, but also NK cell survival via several mechanisms [154,155,156,157]. In one study, NK cell interaction with tumor cells led to NK cell depletion in a caspase- and Fas-independent manner [157]. This depletion was mediated through CD18 and ICAM [157]. In another study, NK cells incubated in IL-2, followed by coculture with leukemia cells, caused NK cell apoptosis [155]. Interestingly, NK cell apoptosis was mediated through CD16 engagement [155] due to the upregulation of Fas on NK cells [156]. The source of FasL causing NK cell apoptosis may be endogenous [154]. In fact, the engagement of NCRs on NK cells with NCR ligand+ tumor cells caused upregulation and secretion of FasL in NK cells. The autologous FasL interacted with Fas and induced NK cell apoptosis through caspase 3 [154]. The CD16 expression level was also shown to decrease after CD16 and CD16 ligand interaction [155,158,159]. Studies have elucidated that matrix metalloprotein (MMP) 25 [159] and ADAM17 as well were responsible for cleaving CD16 from the NK cell membrane. Although ADAM17 [158] is uniformly expressed on NK cells, MMP25 was upregulated on NK cells by IL-2 [159]. Interestingly, upon CD16 engagement, MMP25 was shown to migrate and accumulate at the immunological synapse between the NK and target cell [159].
The down regulation of MHC class I by melanoma has been shown to be a major mechanism of resistance of melanoma patients to anti-PD-1 based immunotherapies [83]. Ironically, tumor cells also upregulate the expression of MHC class I to evade NK cell activation against the tumor. Balsamo and colleagues found that when NK cells are cultured in low effector/target ratios with melanoma cells, the tumor cells upregulate MHC class I, thereby evading the NK cell response [160]. However, this increase in MHC class I expression may improve T cell-mediated killing of melanoma. In that case, tumors have many well-described immunosuppressive mechanisms, many of which are current targets of immune checkpoint inhibitors, such as PD-1 and PD-L1. Therefore, it is likely that tumors alter their immune evasive phenotype to adapt to their immunological challenges.
Lastly, even though it is well established that NK cells are able to target cells that have low MHC expression and/or overexpress the natural ligands of NK cell-activating receptors, chronic stimulation of activating receptors (by abnormally high levels of ligands or abnormally low levels of MHC) have been found to induce NK cell exhaustion [161].
5.2. Secretion of Cytokines/Molecules from Tumor Cells and Immunosuppressive Immune Cells
Melanoma cells can respond to an IFN-γ mediated NK cell attack through the secretion of potent immunosuppressive enzymes indoleamine 2,3-dioxgenase (IDO) and prostaglandin E2 (PGE2). These enzymes decrease the expression of activating receptors NKp30 and NKG2D on NK cells [162]. IDO is also able to inhibit NK cell functionality, as it catalyzes the production of L-kynurenine, which is known to interfere with IL-2-induced upregulation of NK cell-activating receptors NKp46 and NKG2D [163]. Other immunosuppressive cytokines/molecules secreted by melanoma and known to inhibit NK cell function include adenosine [164], TGF-β [165,166] and IL-10 [167,168].
Through chemokines, tumors are able to recruit immunosuppressive immune cells to the tumor microenvironment, which inhibits NK cell activity. The presence of M2 macrophages, myeloid-derived suppressor cell (MDSC), T regulatory cells (Tregs) and fibroblasts plays a role in suppressing NK cell functions through the secretion of various different molecules and cytokines (extensively reviewed in [169]). Notably, TGF-β is one of many immunosuppressive cytokines/molecules secreted by these suppressive cells, and it has been reported to have an effect in two ways. Firstly, TGF-β can directly decrease the expression of activating receptors NKp30 and NKG2D [34]. Secondly, TGF-β is able to convert NK cells, whether it is CD56dim or CD56bright, into a phenotype resembling decidual NK cells (dNK cells), which are poorly cytotoxic and secrete VEGF [170].
In addition, tumor-secreted enzymes are able to shed the activating ligands B7-H6, MICA, MICB and ULBP2 from NK cells through the activity of metalloproteases [29,50,171,172,173]. These ligands have been shown to induce constant stimulation and subsequent endocytosis and degradation of its activating receptors, thereby decreasing NK cell function [48]. In fact, studies have shown that the shedding of activating ligands, such as MICA and UBLP2, is associated with lower survival in stage IV melanoma patients [171,174].
5.3. Hypoxic Tumor Microenvironment
The hypoxic conditions created by tumor cells cause alterations both in tumor cells and immune cells, as indicated by the upregulation of HIF-1α (extensively reviewed in [169]). In NK cells, hypoxia causes the activation of autophagy, consequently causing the degradation of granzyme B [175]. Additionally, hypoxia reduces the response of NK cells to proinflammatory cytokines, IL-2, IL-12, IL-15 and IL-21, and consequently inhibits their ability to upregulate activating receptors such as NCRs and NKG2D [176]. Hypoxia affects not only the expression of activating receptors, but also the expression of activating ligands on tumor cells. For instance, hypoxia was shown to downregulate surface expression of MICA, a ligand for the NK cell-activating receptor NKG2D, in a HIF-1a-dependent manner [177].
6. NK Cells in the Context of Immunotherapies
There is mounting evidence that NK cells infiltrate various tumors. For instance, NK cells were shown to infiltrate renal cell carcinoma [178,179,180,181,182], melanoma [183,184], breast cancer [185,186,187,188], hepatocellular carcinoma [189,190,191], lung cancer [192,193], prostate cancer [194], bladder cancer [195] and rectal cancer [196]. Although many studies show the presence of intratumoral CD56+ NK cells, studies have also found that intratumoral NK cell infiltration is very low or nonexistent in glioblastoma [197], colorectal cancer (CRC) [198], melanoma [199,200], renal cell carcinoma [157], hepatocellular carcinoma [200] and breast cancer [200]. The reasons behind the discrepancy between tumors with high and low NK cell infiltration are diverse: cohort selection, site of biopsy/sample, the appropriate use of immune markers to identify NK cells and the use of whole slides vs. tissue microarrays (TMAs). In particular, the use of TMA vs. whole slides may significantly influence the scoring of CD56+ NK cells, especially considering that NK cells are not an abundant cell type in the TME [201,202]. In fact, it was shown that TMAs were not an appropriate tool for analyzing less abundant biomarkers and particularly immune cells, and whole slide sections should be used instead [201,202]. Nevertheless, intratumoral NK cell infiltration is shown to be associated with improved overall survival in solid cancers [203,204].
The diverse functions and importance of NK cells to antitumor therapy have put NK cells as an attractive target for immunotherapy, mainly via blocking inhibitory receptors expressed by these innate immune cells. In this section, we will review the role of NK cells for different treatment strategies in melanoma (Figure 4). There are numerous clinical trials that are attempting to understand the significance of augmenting NK cell responses in the treatment of various malignancies (Table 1).
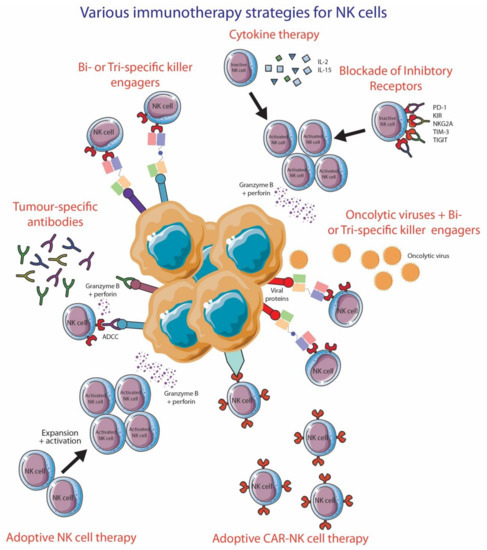
Figure 4.
Various immunotherapy strategies to improve NK cell functionality. This figure was created using Servier Medical Art templates, which are licensed under a Creative Commons Attribution 3.0 Unported License; https://smart.servier.com (accessed 10 October 2020).

Table 1.
Table of clinical trials involving drugs that optimize NK cell functionality.
6.1. Anti-PD-1 and Anti-CTLA-4
Anti-PD-1 and anti-CTLA-4 have revolutionized the treatment of metastatic melanoma, and checkpoint inhibitors have become a standard of care for those with advanced disease. Anti-PD-1 and anti-CTLA-4 play a pivotal role in regulating T cell function, but also have a role in NK cell functionality.
CTLA-4 was shown to be expressed on activated mouse NK cells, and CLTA-4 expression inhibits IFN-γ production by NK cells [152]. In fact, the therapeutic benefit of anti-CTLA-4 was partially dependent on NK cells, as the depletion of both NK and CD8+ T cells significantly decreased the therapeutic response to anti-CTLA-4 [205]. There is little data on the expression and impact of CTLA-4 on human NK cells. One study highlighted that patients treated with anti-CTLA-4 induced NK cell activation and enhanced cytotoxicity [206].
PD-1 is another immune checkpoint that is expressed on NK cells upon activation [94]. In vitro studies show that blocking the PD-1 pathway allows T cell exhaustion to be partly overcome, resulting in enhanced proliferation of CD8+ and CD4+ T cells and increasing their cytokine production [207,208]. NK cells do not express PD-1 as abundantly as T cells, but rather in the range from 1 to 29% [184,209,210], as a proportion of NK cells, with one study showing that 30–40% of NK cells had strong PD-1 upregulation [211], highlighting PD-1 expression heterogeneity on NK cells. However, unlike T cells, PD-1 expression does not induce or imply NK cell exhaustion [211,212]. Given that PD-1 expression is induced on NK cells by IL-2 [213], IL-15 [214] and IL-18 [215], PD-1 expression may represent NK cell activation or maturation status, as demonstrated by a study highlighting that only the mature CD56dim NK cells, and not CD56bright, express PD-1 [216]. NK cells have also shown the ability to acquire PD-1 expression via trogocytosis with leukemia cells, which did not depend on PD-1-PD-L1 ligation, highlighting the dynamic ability of NK cells to regulate its immune response [217].
Nevertheless, PD-1 expression on NK cells is implicated in NK cell dysfunction by repressing PI3K/AKT signalling activation in NK cells [218]. PD-1 ligation with PD-L1 led to decreased ability of NK cells to secrete IFN-γ, express granzyme B + perforin and CD107a, a marker indicating NK cell degranulation [213]. Numerous studies have shown that PD-1 blockade improves NK cell activation and cytotoxicity on PD-1+ NK cells and not PD-1- NK cells [209,218,219,220,221]. PD-1 blockade was also shown to significantly increase not only the expression of NK cell-activating receptors NK2GD, NKp44 and NKp30 [220], but also CXCR4 expression on NK cells and the formation of immune complexes between anti-PD-1-treated NK cells and tumor cells [221].
In mouse models of melanoma, NK or CD8+ T cell depletion caused significant reduction in the survival of melanoma when treated with anti-PD-1 + anti-CTLA-4, supporting the hypothesis that NK and CD8+ T cells cooperate with each other to eradicate the tumor in response to these therapies [222]. In fact, CD16+ NK cells were shown to be significantly more abundant in advanced melanoma patients treated with anti-PD-1 therapy [184]. In the same study, Lee and colleagues found that patients with low MHC class I but high intratumoral NK cell density showed superior survival when compared to patients with low MHC class I and low NK cells. This suggests that NK cells possess the ability improve the therapeutic efficacy of anti-PD-1 therapy by potentially eradicating tumors with MHC class I downregulation. In contrast, CD69+ NK cells were shown to negatively correlate in melanoma patients with progression-free survival (PFS) less than 6 months, while CD69+ NK cells were positively correlated with PFS in melanoma patients with PFS greater than 6 months [223]. The discrepancy may be due to other resistance mechanisms that the tumor may employ to evade NK cell recognition and eradication directly [224] or indirectly through the action of other tumor-mediated immune evasion [225]. Subrahmanyam and colleagues found that MIP1β+ CD69+ NK cells were significantly higher in melanoma patients responding to anti-PD-1 therapy [226]. NK cells represent a promising target for immunotherapy to improve the therapeutic efficacy of anti-PD-1 therapy.
6.2. Anti-KIR
Many studies have consistently reported that KIRs are upregulated in patients with melanoma, compared to healthy control. In one study, KIR3DL1 and KIR2DL3 were expressed at higher levels on the NK cells of melanoma patients [227], while other studies found an increase in KIR2DL1 [228,229] and KIR2DL2/2DL3 [46,230]. Given that KIRs inhibit NK cell activation, there are several clinical trials underway examining drugs that inhibit KIR, including Lirilumab (ClinicalTrials.gov Identifier: NCT01750580, NCT0174739) and ASP98374 (ClinicalTrials.gov Identifier: NCT03260322).
6.3. Anti-NKG2A
NKG2A/CD94 is an inhibitory receptor that is associated with NK cell exhaustion. High NKG2A/CD94 expression on NK cells was associated with poor prognosis of patients with liver cancer [89]. The induction of NKG2A expression may have been stimulated via IL-10 [89]. Nevertheless, in preclinical models, the blockade of NKG2A was shown to improve the antitumor response by NK cells [95,96]. Currently, monalizumab (anti-NKG2A) is in phase 1/2 trials for the treatment of advanced solid tumors (ClinicalTrials.gov Identifier: NCT02671435).
6.4. Anti-TIM-3
TIM-3 is another inhibitory receptor that binds to galectin-9 [231], phosphatidylserine on apoptotic cells [232], high mobility group box 1 [233] and carcinoembryonic antigen-related cell adhesion molecule 1 [234]. TIM-3 is associated with an exhausted NK cell phenotype that has a dysfunctional cytotoxic capacity [227,235]. Blocking TIM-3 improves the cytotoxic ability of NK cells [227,236]. High TIM-3 expression was shown to be correlated with poor prognosis in lung adenocarcinoma and metastatic melanoma [227,236]. Currently, there are several phase 1 trials using anti-TIM-3 for advanced solid tumors (ClinicalTrials.gov Identifier: NCT03708328, NCT03489343). Interestingly, in mouse models, the therapeutic effect of anti-PD-1 + anti-TIM-3 was dependent on NK cells bearing MHC class I-deficient tumors [237].
6.5. Anti-TIGIT
TIGIT is an immunomodulatory receptor expressed by T cells and NK cells. TIGIT shares the same ligands with DNAM-1 and TACTILE; CD112 (Nectin-2) and CD155 (PVR) [99,238]. Not only does this receptor play a role in inhibiting NK cell responses, it was shown to play a role in NK cell education. In the absence of TIGIT, NK cells are rendered unresponsive to stimuli [238]. There has been increasing interest in anti-TIGIT in clinical trials (ClinicalTrials.gov Identifier: NCT02913313, NCT02676869), and several studies have identified that TIGIT negatively regulates NK cell function by suppressing IFN-γ production and cytotoxicity [97,99,239,240]. Blocking TIGIT was shown to improve NK cell cytotoxicity and slow tumor growth in melanoma mouse models [241]. Several studies have associated TIGIT expression with NK cell exhaustion [241,242]. However, given that TIGIT competes for ligands of the activating receptor, DNAM-1, it is likely that high expression of TIGIT sequesters the ligands from DNAM-1.
6.6. Adoptive NK Cell Transfer
Chimeric antigen receptor (CAR)-modified T cells have become an effective immunotherapy option. CAR-T cell therapy has shown remarkable outcomes in B cell lymphoma, with complete response rates between 88 and 93% (sample size ranging from 16 to 30) [243,244,245]. However, severe toxicity (i.e., cytokine release syndrome), graft-versus-host disease, autoimmunity and various T cell resistance mechanisms (i.e., MHC class I loss) are still a concerning issue [246]. CAR-NK cell therapy is an emerging field that has shown great patient safety, limited/no toxicity and is not affected by resistance mechanisms seen in CAR-T cell therapy [247]. In addition to excellent patient safety, CAR-NK cells are not restricted by antigen specificity or strict antigen presentation rules; in fact, CAR-NK cells have numerous activation pathways that ensure effective NK cell activation to tumor targets [224]. CAR-NK cell therapy is a rapidly evolving field, as evidenced by numerous clinical trials using CAR-NK cell therapy in multiple myeloma (BCMA; ClinicalTrials.gov Identifier: NCT03940833), pancreatic cancer (ROBO1; ClinicalTrials.gov Identifier: NCT03941457), glioblastoma (HER2; ClinicalTrials.gov Identifier: NCT03383978), B-cell acute lymphoblastic leukemia (CD19; ClinicalTrials.gov Identifier: NCT00995137) and various solid tumors (NKG2D ligands; ClinicalTrials.gov Identifier: NCT03940820). For instance, a recent phase 1 and 2 clinical trial (CAR-NK cell therapy) highlighted that patients with CD19-positive lymphoid tumors did not develop cytokine release syndrome, neurotoxicity, graft-versus-host disease or increases in inflammatory cytokines, and the maximum tolerated dose was not reached [248]. Most importantly, 73% had a complete response (n = 11) with the infused CAR-NK cells persisting at low levels for a minimum of 12 months [248]. When CAR-NK cells were used in a phase 1 and 2 trial involving lung-, pancreatic-, colon- and ovarian cancer, the treatment was well-tolerated and no adverse reactions were seen [246]; no further results were reported. However, adoptive cell therapy has historically had variable success in solid tumors due to the highly complex and immunosuppressive tumor microenvironment [225,249]. While many unique immunosuppressive mechanisms are largely absent in hematological cancers, the tumor microenvironment in solid tumors has numerous and complicated resistance mechanisms that make it difficult for infused CAR-NK cells to not only reach the target site but, prevent itself from being overwhelmed by immunosuppressive signals [225]. It is likely that effective CAR-NK cell therapy is dependent on multiple immunotherapy treatments that address solid tumor-specific immunosuppressive mechanisms such as PD-1, hypoxia or MHC class I downregulation.
6.7. Bispecific Killer Cell Engagers (BiKE) and Trispecific Killer Cell Engagers (TriKE)
NK cells possess the ability to become activated and efficiently remove tumor cells through ADCC. A major component of this is the CD16 receptor, which is expressed on mature and differentiated NK cells. CD16 engagement to Fc portions of antibodies opsonized on target cells is sufficient for NK cell release of perforin and granzyme [56,57,58]. In addition to CD16-mediated killing of tumor cells, CD16 engagement also leads to NK cell activation, proliferation [250] and the production of proinflammatory cytokines such as IFN-γ [158]. This makes CD16 an attractive target for immunotherapy as the treatment with monoclonal antibodies, particularly monoclonal antibodies that target tumor-specific antigens or antigens overly expressed on tumor cells enable those cells to become tagged for clearance by CD16+ NK cells. In fact, NK cell-mediated ADCC in tumor-antigen targeting monoclonal antibodies, such as rituximab, contributes to the response of treated patients [251,252]. However, a limitation of CD16 is that differences in CD16 allotype affinity to antibodies mean variations in antibodies being able to elicit appropriate ADCC in NK cells. BiKE and TriKE are able to bypass this by being able to bind to CD16 with strong affinity to elicit ADCC.
BiKEs and TriKEs are emerging as promising immunotherapy options to fully utilize the ADCC capacity of NK cells to effectively eradicate tumor cells. BiKEs consist of two single-chain variable fragments (scFv); scFv is only the antigen binding region of an antibody [253]. One scFv is specific for CD16, while the other scFv is specific for an antigen, in this instance, tumor-specific antigen, and these two scFv are connected together by a short peptide that enables great flexibility [253]. When BiKEs bind to their targets, ADCC and proinflammatory cytokine production occurs in NK cells. TriKEs are very similar to BiKEs, but have three scFv, where the additional scFv is specific to another antigen of choice or another NK cell receptor, or contains a cytokine, like IL-15, integrated in the peptide linker between two scFv.
IL-15 has been chosen as a superior candidate over IL-2 due to its superior safety and toxicity profile and benefits to NK cell activation, proliferation and effector functions. Despite being an important proinflammatory cytokine, administering IL-2 to patients is highly toxic and potentially life-threatening. The most common cause of IL-2 cytotoxicity is vascular leak syndrome that causes hypovolemia and fluid accumulation in the interstitial tissue [254]. IL-2 may also cause the activation of the immunosuppressive CD25+ Treg that can inhibit NK cell function [255]. In contrast, IL-15 does not cause vascular leak syndrome or activate Tregs. However, in a phase I clinical trial, patients with various solid tumors had eight serious adverse events (bowel ischemia, pneumonitis, papilledema, uveitis and grade 3 hypotension), and further studies are required to evaluate the safety and toxicity of IL-15 in humans [256]. In saying this, within the same study, IL-15 dramatically induced a 358-fold expansion of circulating CD56brightNK cells [257]. IL-15 was also shown to be important in regulating NK cell survival [258], improving NK cell cytotoxicity through the upregulation of NKG2D, perforin and TRAIL [259] and enhancing NK cell-mediated ADCC [260]. These enhancing effects of IL-15 on NK cells highlight that IL-15 is an attractive candidate to be used as a linker in TriKE.
TriKE containing IL-15 is shown to be a superior therapy option to BiKE [261]. When compared to BiKE, TriKE elicited superior NK cell cytotoxicity, increased IFN-γ and TNFα production, NK cell survival and proliferation [261] when leukemia cell lines were used as targets for NK cells. Within the same study, in vivo studies found that TriKEs had superior antitumor activity and sustained NK cell survival for at least three weeks. The same effects were seen when ovarian, prostate and lung cancers were used, highlighting that TriKEs are effective against solid tumors [262]. Although TriKEs used in these studies contained IL-15, and IL-15 elicited severe toxicity (bowel ischemia, pneumonitis, papilledema, uveitis and grade 3 hypotension) and death in humans [256], IL-15-incorporting TriKEs showed no observable detrimental effects in mice [263].
A caveat with BiKE and TriKE therapy is that they target overexpressing tumor antigens that are also expressed on healthy cells. Additionally, not all tumor cells may express tumor antigens, with some tumors not expressing those antigens at all. In a large number of cancers, a targetable tumor antigen has not yet been identified, making BiKE and TriKE therapy a challenge. A potential option could be to use oncolytic viruses, since a natural aspect of the viral lifecycle is the expression of viral proteins on the cell surface. Here, BiKE and TriKE therapy could be used to target viral protein expression on tumor cells after the delivery of oncolytic viruses [264]. In melanoma, talimogene laherparepvec (T-VEC) has been used in several clinical trials [265]. T-VEC is a herpes simplex virus-1 (HSV-1) that has been genetically engineered to target melanoma cells. Here, HSV-1 proteins could be used as targets to BiKE and TriKE and, given that HSV-1 proteins are not expressed by human cells, this may enable a safer and more effective immunotherapy target compared to targeting tumor antigens.
7. Conclusions
NK cells are multifunctional immune cells that ensure injury and disease are effectively addressed. They play numerous concurrent roles such as efficiently participating in tumor eradication. Recent studies have revealed that NK cells play an essential role in activating both CD4+ and CD8+ T cell responses through intricate NK-DC interactions. In malignancies, NK cells may play a critical role in efficiently coordinating the adaptive immune system. Given the success of CAR-NK cells in hematological malignancies, the use of CAR-NK cells in solid tumors should concurrently consider addressing the diverse resistance mechanisms seen in solid tumors for optimal therapeutic efficacy.
Author Contributions
Conceptualization, H.L., G.V.L., R.A.S. and J.S.W.; writing—original draft preparation, H.L.; writing—review and editing, H.L., J.S.W., G.V.L., R.A.S., I.P.D.S. and U.P. visualization, H.L.; supervision, J.S.W., G.V.L. and R.A.S. All authors have read and agreed to the published version of the manuscript.
Funding
R.A.S. and G.V.L. are supported by a National Health and Medical Research Council of Australia (NHMRC) Program Grant (APP1093017) and R.A.S. is supported by an NHMRC Practitioner Fellowship (APP1141295). G.V.L. is supported by The Medical Foundation, The University of Sydney. Support from Melanoma Institute Australia, The Ainsworth Foundation and Fairfax Foundation is also gratefully acknowledged.
Conflicts of Interest
R.A.S. has received fees for professional services from Qbiotics, Merck Sharp & Dohme, GlaxoSmithKline Australia, Bristol-Myers Squibb, Dermpedia, Novartis., Myriad, NeraCare and Amgen. G.V.L. is consultant advisor for Aduro Biotech Inc, Amgen Inc, Array Biopharma inc, Boehringer Ingelheim International GmbH, Bristol-Myers Squibb, Hexel AG, Highlight Therapeutics S.L., Merck Sharpe & Dohme, Novartis Pharma AG, OncoSec, Pierre Fabre, QBiotics Group Limited, Regeneron Pharmaceuticals Inc, SkylineDX B.V., Specialised Therapeutics Australia Pty Ltd. All remaining authors have declared no conflicts of interests.
References
- Spits, H.; Artis, D.; Colonna, M.; Diefenbach, A.; Di Santo, J.P.; Eberl, G.; Koyasu, S.; Locksley, R.M.; McKenzie, A.N.; Mebius, R.E.; et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nat. Rev. Immunol. 2013, 13, 145–149. [Google Scholar] [CrossRef]
- O’Sullivan, T.E. Dazed and Confused: NK Cells. Front. Immunol. 2019, 10, 2235. [Google Scholar] [CrossRef] [PubMed]
- Guillerey, C.; Huntington, N.D.; Smyth, M.J. Targeting natural killer cells in cancer immunotherapy. Nat. Immunol. 2016, 17, 1025–1036. [Google Scholar] [CrossRef]
- Lanier, L.L. NK cell recognition. Annu. Rev. Immunol. 2005, 23, 225–274. [Google Scholar] [CrossRef]
- Hersey, P.; Edwards, A.; McCarthy, W.H. Tumour-related changes in natural killer cell activity in melanoma patients. Influence of stage of disease, tumour thickness and age of patients. Int. J. Cancer 1980, 25, 187–194. [Google Scholar] [CrossRef]
- Hersey, P.; Edwards, A.; Honeyman, M.; McCarthy, W.H. Low natural-killer-cell activity in familial melanoma patients and their relatives. Br. J. Cancer 1979, 40, 113–122. [Google Scholar] [CrossRef]
- Kurosawa, S.; Harada, M.; Matsuzaki, G.; Shinomiya, Y.; Terao, H.; Kobayashi, N.; Nomoto, K. Early-appearing tumour-infiltrating natural killer cells play a crucial role in the generation of anti-tumour T lymphocytes. Immunology 1995, 85, 338–346. [Google Scholar]
- Tato, C.M.; Martins, G.A.; High, F.A.; DiCioccio, C.B.; Reiner, S.L.; Hunter, C.A. Cutting Edge: Innate production of IFN-gamma by NK cells is independent of epigenetic modification of the IFN-gamma promoter. J. Immunol. 2004, 173, 1514–1517. [Google Scholar] [CrossRef] [PubMed]
- Pallmer, K.; Oxenius, A. Recognition and Regulation of T Cells by NK Cells. Front. Immunol. 2016, 7, 251. [Google Scholar] [CrossRef] [PubMed]
- Crouse, J.; Xu, H.C.; Lang, P.A.; Oxenius, A. NK cells regulating T cell responses: Mechanisms and outcome. Trends Immunol. 2015, 36, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Poli, A.; Michel, T.; Theresine, M.; Andres, E.; Hentges, F.; Zimmer, J. CD56bright natural killer (NK) cells: An important NK cell subset. Immunology 2009, 126, 458–465. [Google Scholar] [CrossRef]
- Montaldo, E.; Del Zotto, G.; Della Chiesa, M.; Mingari, M.C.; Moretta, A.; De Maria, A.; Moretta, L. Human NK cell receptors/markers: A tool to analyze NK cell development, subsets and function. Cytom. A 2013, 83, 702–713. [Google Scholar] [CrossRef]
- Abel, A.M.; Yang, C.; Thakar, M.S.; Malarkannan, S. Natural Killer Cells: Development, Maturation, and Clinical Utilization. Front. Immunol. 2018, 9, 1869. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Lal, G. The Molecular Mechanism of Natural Killer Cells Function and Its Importance in Cancer Immunotherapy. Front. Immunol. 2017, 8, 1124. [Google Scholar] [CrossRef]
- Chan, C.J.; Smyth, M.J.; Martinet, L. Molecular mechanisms of natural killer cell activation in response to cellular stress. Cell Death Differ. 2014, 21, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S. Natural killer cell cytotoxicity and its regulation by inhibitory receptors. Immunology 2018, 154, 383–393. [Google Scholar] [CrossRef]
- Brodin, P.; Lakshmikanth, T.; Johansson, S.; Karre, K.; Hoglund, P. The strength of inhibitory input during education quantitatively tunes the functional responsiveness of individual natural killer cells. Blood 2009, 113, 2434–2441. [Google Scholar] [CrossRef]
- Holmes, T.D.; El-Sherbiny, Y.M.; Davison, A.; Clough, S.L.; Blair, G.E.; Cook, G.P. A human NK cell activation/inhibition threshold allows small changes in the target cell surface phenotype to dramatically alter susceptibility to NK cells. J. Immunol. 2011, 186, 1538–1545. [Google Scholar] [CrossRef] [PubMed]
- Moretta, A.; Bottino, C.; Vitale, M.; Pende, D.; Cantoni, C.; Mingari, M.C.; Biassoni, R.; Moretta, L. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu. Rev. Immunol. 2001, 19, 197–223. [Google Scholar] [CrossRef]
- Sivori, S.; Vitale, M.; Morelli, L.; Sanseverino, L.; Augugliaro, R.; Bottino, C.; Moretta, L.; Moretta, A. p46, a novel natural killer cell-specific surface molecule that mediates cell activation. J. Exp. Med. 1997, 186, 1129–1136. [Google Scholar] [CrossRef]
- Pende, D.; Parolini, S.; Pessino, A.; Sivori, S.; Augugliaro, R.; Morelli, L.; Marcenaro, E.; Accame, L.; Malaspina, A.; Biassoni, R.; et al. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J. Exp. Med. 1999, 190, 1505–1516. [Google Scholar] [CrossRef]
- Samarani, S.; Sagala, P.; Jantchou, P.; Grimard, G.; Faure, C.; Deslandres, C.; Amre, D.K.; Ahmad, A. Phenotypic and Functional Changes in Peripheral Blood Natural Killer Cells in Crohn Disease Patients. Mediat. Inflamm. 2020, 2020, 6401969. [Google Scholar] [CrossRef]
- Vitale, M.; Bottino, C.; Sivori, S.; Sanseverino, L.; Castriconi, R.; Marcenaro, E.; Augugliaro, R.; Moretta, L.; Moretta, A. NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells, is involved in non-major histocompatibility complex-restricted tumor cell lysis. J. Exp. Med. 1998, 187, 2065–2072. [Google Scholar] [CrossRef] [PubMed]
- Koch, J.; Steinle, A.; Watzl, C.; Mandelboim, O. Activating natural cytotoxicity receptors of natural killer cells in cancer and infection. Trends Immunol. 2013, 34, 182–191. [Google Scholar] [CrossRef]
- Markel, G.; Seidman, R.; Besser, M.J.; Zabari, N.; Ortenberg, R.; Shapira, R.; Treves, A.J.; Loewenthal, R.; Orenstein, A.; Nagler, A.; et al. Natural killer lysis receptor (NKLR)/NKLR-ligand matching as a novel approach for enhancing anti-tumor activity of allogeneic NK cells. PLoS ONE 2009, 4, e5597. [Google Scholar] [CrossRef]
- Lakshmikanth, T.; Burke, S.; Ali, T.H.; Kimpfler, S.; Ursini, F.; Ruggeri, L.; Capanni, M.; Umansky, V.; Paschen, A.; Sucker, A.; et al. NCRs and DNAM-1 mediate NK cell recognition and lysis of human and mouse melanoma cell lines in vitro and in vivo. J. Clin. Investig. 2009, 119, 1251–1263. [Google Scholar] [CrossRef]
- Textor, S.; Bossler, F.; Henrich, K.O.; Gartlgruber, M.; Pollmann, J.; Fiegler, N.; Arnold, A.; Westermann, F.; Waldburger, N.; Breuhahn, K.; et al. The proto-oncogene Myc drives expression of the NK cell-activating NKp30 ligand B7-H6 in tumor cells. Oncoimmunology 2016, 5, e1116674. [Google Scholar] [CrossRef] [PubMed]
- Brandt, C.S.; Baratin, M.; Yi, E.C.; Kennedy, J.; Gao, Z.; Fox, B.; Haldeman, B.; Ostrander, C.D.; Kaifu, T.; Chabannon, C.; et al. The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J. Exp. Med. 2009, 206, 1495–1503. [Google Scholar] [CrossRef] [PubMed]
- Schlecker, E.; Fiegler, N.; Arnold, A.; Altevogt, P.; Rose-John, S.; Moldenhauer, G.; Sucker, A.; Paschen, A.; von Strandmann, E.P.; Textor, S.; et al. Metalloprotease-mediated tumor cell shedding of B7-H6, the ligand of the natural killer cell-activating receptor NKp30. Cancer Res. 2014, 74, 3429–3440. [Google Scholar] [CrossRef]
- Dassler-Plenker, J.; Reiners, K.S.; van den Boorn, J.G.; Hansen, H.P.; Putschli, B.; Barnert, S.; Schuberth-Wagner, C.; Schubert, R.; Tuting, T.; Hallek, M.; et al. RIG-I activation induces the release of extracellular vesicles with antitumor activity. Oncoimmunology 2016, 5, e1219827. [Google Scholar] [CrossRef]
- Brown, E.R.; Doig, T.; Anderson, N.; Brenn, T.; Doherty, V.; Xu, Y.; Bartlett, J.M.; Smyth, J.F.; Melton, D.W. Association of galectin-3 expression with melanoma progression and prognosis. Eur. J. Cancer 2012, 48, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.G.; Kim, S.J.; Baek, J.H.; Lee, H.W.; Jeong, S.Y.; Chun, K.H. Galectin-3 increases the motility of mouse melanoma cells by regulating matrix metalloproteinase-1 expression. Exp. Mol. Med. 2012, 44, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Martinet, L.; Smyth, M.J. Balancing natural killer cell activation through paired receptors. Nat. Rev. Immunol. 2015, 15, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Bottino, C.; Castriconi, R.; Pende, D.; Rivera, P.; Nanni, M.; Carnemolla, B.; Cantoni, C.; Grassi, J.; Marcenaro, S.; Reymond, N.; et al. Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J. Exp. Med. 2003, 198, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Tahara-Hanaoka, S.; Shibuya, K.; Onoda, Y.; Zhang, H.; Yamazaki, S.; Miyamoto, A.; Honda, S.; Lanier, L.L.; Shibuya, A. Functional characterization of DNAM-1 (CD226) interaction with its ligands PVR (CD155) and nectin-2 (PRR-2/CD112). Int. Immunol. 2004, 16, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Pende, D.; Bottino, C.; Castriconi, R.; Cantoni, C.; Marcenaro, S.; Rivera, P.; Spaggiari, G.M.; Dondero, A.; Carnemolla, B.; Reymond, N.; et al. PVR (CD155) and Nectin-2 (CD112) as ligands of the human DNAM-1 (CD226) activating receptor: Involvement in tumor cell lysis. Mol. Immunol. 2005, 42, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Casado, J.G.; Pawelec, G.; Morgado, S.; Sanchez-Correa, B.; Delgado, E.; Gayoso, I.; Duran, E.; Solana, R.; Tarazona, R. Expression of adhesion molecules and ligands for activating and costimulatory receptors involved in cell-mediated cytotoxicity in a large panel of human melanoma cell lines. Cancer Immunol. Immunother. 2009, 58, 1517–1526. [Google Scholar] [CrossRef] [PubMed]
- Bevelacqua, V.; Bevelacqua, Y.; Candido, S.; Skarmoutsou, E.; Amoroso, A.; Guarneri, C.; Strazzanti, A.; Gangemi, P.; Mazzarino, M.C.; D’Amico, F.; et al. Nectin like-5 overexpression correlates with the malignant phenotype in cutaneous melanoma. Oncotarget 2012, 3, 882–892. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.J.; Andrews, D.M.; McLaughlin, N.M.; Yagita, H.; Gilfillan, S.; Colonna, M.; Smyth, M.J. DNAM-1/CD155 interactions promote cytokine and NK cell-mediated suppression of poorly immunogenic melanoma metastases. J. Immunol. 2010, 184, 902–911. [Google Scholar] [CrossRef]
- Lepletier, A.; Madore, J.; O’Donnell, J.S.; Johnston, R.L.; Li, X.Y.; McDonald, E.; Ahern, E.; Kuchel, A.; Eastgate, M.; Pearson, S.A.; et al. Tumor CD155 Expression Is Associated with Resistance to Anti-PD1 Immunotherapy in Metastatic Melanoma. Clin. Cancer Res. 2020, 26, 3671–3681. [Google Scholar] [CrossRef]
- Koike, S.; Horie, H.; Ise, I.; Okitsu, A.; Yoshida, M.; Iizuka, N.; Takeuchi, K.; Takegami, T.; Nomoto, A. The poliovirus receptor protein is produced both as membrane-bound and secreted forms. Embo J. 1990, 9, 3217–3224. [Google Scholar] [CrossRef] [PubMed]
- Okumura, G.; Iguchi-Manaka, A.; Murata, R.; Yamashita-Kanemaru, Y.; Shibuya, A.; Shibuya, K. Tumor-derived soluble CD155 inhibits DNAM-1-mediated antitumor activity of natural killer cells. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef] [PubMed]
- Iguchi-Manaka, A.; Okumura, G.; Kojima, H.; Cho, Y.; Hirochika, R.; Bando, H.; Sato, T.; Yoshikawa, H.; Hara, H.; Shibuya, A.; et al. Increased Soluble CD155 in the Serum of Cancer Patients. PLoS ONE 2016, 11, e0152982. [Google Scholar] [CrossRef]
- Baury, B.; Masson, D.; McDermott, B.M., Jr.; Jarry, A.; Blottiere, H.M.; Blanchardie, P.; Laboisse, C.L.; Lustenberger, P.; Racaniello, V.R.; Denis, M.G. Identification of secreted CD155 isoforms. Biochem. Biophys. Res. Commun. 2003, 309, 175–182. [Google Scholar] [CrossRef]
- Nausch, N.; Cerwenka, A. NKG2D ligands in tumor immunity. Oncogene 2008, 27, 5944–5958. [Google Scholar] [CrossRef] [PubMed]
- Mirjacic Martinovic, K.M.; Babovic, N.; Dzodic, R.R.; Jurisic, V.B.; Tanic, N.T.; Konjevic, G.M. Decreased expression of NKG2D, NKp46, DNAM-1 receptors, and intracellular perforin and STAT-1 effector molecules in NK cells and their dim and bright subsets in metastatic melanoma patients. Melanoma Res. 2014, 24, 295–304. [Google Scholar] [CrossRef]
- Cluxton, C.D.; Spillane, C.; O’Toole, S.A.; Sheils, O.; Gardiner, C.M.; O’Leary, J.J. Suppression of Natural Killer cell NKG2D and CD226 anti-tumour cascades by platelet cloaked cancer cells: Implications for the metastatic cascade. PLoS ONE 2019, 14, e0211538. [Google Scholar] [CrossRef] [PubMed]
- Groh, V.; Wu, J.; Yee, C.; Spies, T. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature 2002, 419, 734–738. [Google Scholar] [CrossRef]
- Boutet, P.; Aguera-Gonzalez, S.; Atkinson, S.; Pennington, C.J.; Edwards, D.R.; Murphy, G.; Reyburn, H.T.; Vales-Gomez, M. Cutting edge: The metalloproteinase ADAM17/TNF-alpha-converting enzyme regulates proteolytic shedding of the MHC class I-related chain B protein. J. Immunol. 2009, 182, 49–53. [Google Scholar] [CrossRef]
- Kaiser, B.K.; Yim, D.; Chow, I.T.; Gonzalez, S.; Dai, Z.; Mann, H.H.; Strong, R.K.; Groh, V.; Spies, T. Disulphide-isomerase-enabled shedding of tumour-associated NKG2D ligands. Nature 2007, 447, 482–486. [Google Scholar] [CrossRef]
- Waldhauer, I.; Steinle, A. NK cells and cancer immunosurveillance. Oncogene 2008, 27, 5932–5943. [Google Scholar] [CrossRef]
- Ferrari de Andrade, L.; Tay, R.E.; Pan, D.; Luoma, A.M.; Ito, Y.; Badrinath, S.; Tsoucas, D.; Franz, B.; May, K.F., Jr.; Harvey, C.J.; et al. Antibody-mediated inhibition of MICA and MICB shedding promotes NK cell-driven tumor immunity. Science 2018, 359, 1537–1542. [Google Scholar] [CrossRef]
- Maccalli, C.; Giannarelli, D.; Chiarucci, C.; Cutaia, O.; Giacobini, G.; Hendrickx, W.; Amato, G.; Annesi, D.; Bedognetti, D.; Altomonte, M.; et al. Soluble NKG2D ligands are biomarkers associated with the clinical outcome to immune checkpoint blockade therapy of metastatic melanoma patients. Oncoimmunology 2017, 6, e1323618. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, J.; Newton, R.; Bahaie, N.S.; Long, C.; Walcheck, B. ADAM17 cleaves CD16b (FcgammaRIIIb) in human neutrophils. Biochim. Biophys. Acta 2013, 1833, 680–685. [Google Scholar] [CrossRef] [PubMed]
- De Taeye, S.W.; Bentlage, A.E.H.; Mebius, M.M.; Meesters, J.I.; Lissenberg-Thunnissen, S.; Falck, D.; Senard, T.; Salehi, N.; Wuhrer, M.; Schuurman, J.; et al. FcgammaR Binding and ADCC Activity of Human IgG Allotypes. Front. Immunol. 2020, 11, 740. [Google Scholar] [CrossRef]
- Bryceson, Y.T.; March, M.E.; Ljunggren, H.G.; Long, E.O. Activation, coactivation, and costimulation of resting human natural killer cells. Immunol. Rev. 2006, 214, 73–91. [Google Scholar] [CrossRef] [PubMed]
- Bryceson, Y.T.; March, M.E.; Ljunggren, H.G.; Long, E.O. Synergy among receptors on resting NK cells for the activation of natural cytotoxicity and cytokine secretion. Blood 2006, 107, 159–166. [Google Scholar] [CrossRef]
- Bryceson, Y.T.; March, M.E.; Barber, D.F.; Ljunggren, H.G.; Long, E.O. Cytolytic granule polarization and degranulation controlled by different receptors in resting NK cells. J. Exp. Med. 2005, 202, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Bowles, J.A.; Weiner, G.J. CD16 polymorphisms and NK activation induced by monoclonal antibody-coated target cells. J. Immunol. Methods 2005, 304, 88–99. [Google Scholar] [CrossRef]
- Mahaweni, N.M.; Olieslagers, T.I.; Rivas, I.O.; Molenbroeck, S.J.J.; Groeneweg, M.; Bos, G.M.J.; Tilanus, M.G.J.; Voorter, C.E.M.; Wieten, L. A comprehensive overview of FCGR3A gene variability by full-length gene sequencing including the identification of V158F polymorphism. Sci. Rep. 2018, 8, 15983. [Google Scholar] [CrossRef]
- Hatjiharissi, E.; Xu, L.; Santos, D.D.; Hunter, Z.R.; Ciccarelli, B.T.; Verselis, S.; Modica, M.; Cao, Y.; Manning, R.J.; Leleu, X.; et al. Increased natural killer cell expression of CD16, augmented binding and ADCC activity to rituximab among individuals expressing the Fc{gamma}RIIIa-158 V/V and V/F polymorphism. Blood 2007, 110, 2561–2564. [Google Scholar] [CrossRef]
- Cibrian, D.; Sanchez-Madrid, F. CD69: From activation marker to metabolic gatekeeper. Eur. J. Immunol. 2017, 47, 946–953. [Google Scholar] [CrossRef]
- Borrego, F.; Robertson, M.J.; Ritz, J.; Pena, J.; Solana, R. CD69 is a stimulatory receptor for natural killer cell and its cytotoxic effect is blocked by CD94 inhibitory receptor. Immunology 1999, 97, 159–165. [Google Scholar] [CrossRef]
- Hayashizaki, K.; Kimura, M.Y.; Tokoyoda, K.; Hosokawa, H.; Shinoda, K.; Hirahara, K.; Ichikawa, T.; Onodera, A.; Hanazawa, A.; Iwamura, C.; et al. Myosin light chains 9 and 12 are functional ligands for CD69 that regulate airway inflammation. Sci. Immunol. 2016, 1, eaaf9154. [Google Scholar] [CrossRef] [PubMed]
- Borrego, F.; Pena, J.; Solana, R. Regulation of CD69 expression on human natural killer cells: Differential involvement of protein kinase C and protein tyrosine kinases. Eur. J. Immunol. 1993, 23, 1039–1043. [Google Scholar] [CrossRef] [PubMed]
- Pisegna, S.; Zingoni, A.; Pirozzi, G.; Cinque, B.; Cifone, M.G.; Morrone, S.; Piccoli, M.; Frati, L.; Palmieri, G.; Santoni, A. Src-dependent Syk activation controls CD69-mediated signaling and function on human NK cells. J. Immunol. 2002, 169, 68–74. [Google Scholar] [CrossRef]
- Marden, C.M.; North, J.; Anderson, R.; Bakhsh, I.A.; Addison, E.; Pittman, H.; Mackinnon, S.; Lowdell, M.W. CD69 Is Required for Activated NK Cell-Mediated Killing of Resistant Targets. Blood 2005, 106, 3322. [Google Scholar] [CrossRef]
- Clausen, J.; Vergeiner, B.; Enk, M.; Petzer, A.L.; Gastl, G.; Gunsilius, E. Functional significance of the activation-associated receptors CD25 and CD69 on human NK-cells and NK-like T-cells. Immunobiology 2003, 207, 85–93. [Google Scholar] [CrossRef]
- Esplugues, E.; Vega-Ramos, J.; Cartoixa, D.; Vazquez, B.N.; Salaet, I.; Engel, P.; Lauzurica, P. Induction of tumor NK-cell immunity by anti-CD69 antibody therapy. Blood 2005, 105, 4399–4406. [Google Scholar] [CrossRef]
- North, J.; Bakhsh, I.; Marden, C.; Pittman, H.; Addison, E.; Navarrete, C.; Anderson, R.; Lowdell, M.W. Tumor-primed human natural killer cells lyse NK-resistant tumor targets: Evidence of a two-stage process in resting NK cell activation. J. Immunol. 2007, 178, 85–94. [Google Scholar] [CrossRef]
- Galandrini, R.; Piccoli, M.; Frati, L.; Santoni, A. Tyrosine kinase-dependent activation of human NK cell functions upon triggering through CD44 receptor. Eur. J. Immunol. 1996, 26, 2807–2811. [Google Scholar] [CrossRef] [PubMed]
- Sague, S.L.; Tato, C.; Pure, E.; Hunter, C.A. The regulation and activation of CD44 by natural killer (NK) cells and its role in the production of IFN-gamma. J. Interferon Cytokine Res. 2004, 24, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Sconocchia, G.; Titus, J.A.; Segal, D.M. Signaling pathways regulating CD44-dependent cytolysis in natural killer cells. Blood 1997, 90, 716–725. [Google Scholar] [CrossRef]
- Tan, P.H.; Santos, E.B.; Rossbach, H.C.; Sandmaier, B.M. Enhancement of natural killer activity by an antibody to CD44. J. Immunol. 1993, 150, 812–820. [Google Scholar]
- Galandrini, R.; De Maria, R.; Piccoli, M.; Frati, L.; Santoni, A. CD44 triggering enhances human NK cell cytotoxic functions. J. Immunol. 1994, 153, 4399–4407. [Google Scholar]
- Campbell, K.S.; Purdy, A.K. Structure/function of human killer cell immunoglobulin-like receptors: Lessons from polymorphisms, evolution, crystal structures and mutations. Immunology 2011, 132, 315–325. [Google Scholar] [CrossRef]
- Hsu, K.C.; Liu, X.R.; Selvakumar, A.; Mickelson, E.; O’Reilly, R.J.; Dupont, B. Killer Ig-like receptor haplotype analysis by gene content: Evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J. Immunol. 2002, 169, 5118–5129. [Google Scholar] [CrossRef]
- Pende, D.; Falco, M.; Vitale, M.; Cantoni, C.; Vitale, C.; Munari, E.; Bertaina, A.; Moretta, F.; Del Zotto, G.; Pietra, G.; et al. Killer Ig-Like Receptors (KIRs): Their Role in NK Cell Modulation and Developments Leading to Their Clinical Exploitation. Front. Immunol. 2019, 10, 1179. [Google Scholar] [CrossRef]
- Rajagopalan, S.; Fu, J.; Long, E.O. Cutting edge: Induction of IFN-gamma production but not cytotoxicity by the killer cell Ig-like receptor KIR2DL4 (CD158d) in resting NK cells. J. Immunol. 2001, 167, 1877–1881. [Google Scholar] [CrossRef]
- Abi-Rached, L.; Parham, P. Natural selection drives recurrent formation of activating killer cell immunoglobulin-like receptor and Ly49 from inhibitory homologues. J. Exp. Med. 2005, 201, 1319–1332. [Google Scholar] [CrossRef]
- Middleton, D.; Gonzelez, F. The extensive polymorphism of KIR genes. Immunology 2010, 129, 8–19. [Google Scholar] [CrossRef]
- Anfossi, N.; Andre, P.; Guia, S.; Falk, C.S.; Roetynck, S.; Stewart, C.A.; Breso, V.; Frassati, C.; Reviron, D.; Middleton, D.; et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity 2006, 25, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Shklovskaya, E.; Lim, S.Y.; Carlino, M.S.; Menzies, A.M.; Stewart, A.; Pedersen, B.; Irvine, M.; Alavi, S.; Yang, J.Y.H.; et al. Transcriptional downregulation of MHC class I and melanoma de- differentiation in resistance to PD-1 inhibition. Nat. Commun. 2020, 11, 1897. [Google Scholar] [CrossRef] [PubMed]
- Kandilarova, S.M.; Paschen, A.; Mihaylova, A.; Ivanova, M.; Schadendorf, D.; Naumova, E. The Influence of HLA and KIR Genes on Malignant Melanoma Development and Progression. Arch. Immunol. Ther. Exp. 2016, 64, 73–81. [Google Scholar] [CrossRef]
- Campillo, J.A.; Legaz, I.; Lopez-Alvarez, M.R.; Bolarin, J.M.; Las Heras, B.; Muro, M.; Minguela, A.; Moya-Quiles, M.R.; Blanco-Garcia, R.; Martinez-Banaclocha, H.; et al. KIR gene variability in cutaneous malignant melanoma: Influence of KIR2D/HLA-C pairings on disease susceptibility and prognosis. Immunogenetics 2013, 65, 333–343. [Google Scholar] [CrossRef]
- Ishida, Y.; Nakashima, C.; Kojima, H.; Tanaka, H.; Fujimura, T.; Matsushita, S.; Yamamoto, Y.; Yoshino, K.; Fujisawa, Y.; Otsuka, A.; et al. Killer immunoglobulin-like receptor genotype did not correlate with response to anti-PD-1 antibody treatment in a Japanese cohort. Sci. Rep. 2018, 8, 15962. [Google Scholar] [CrossRef] [PubMed]
- Toneva, M.; Lepage, V.; Lafay, G.; Dulphy, N.; Busson, M.; Lester, S.; Vu-Trien, A.; Michaylova, A.; Naumova, E.; McCluskey, J.; et al. Genomic diversity of natural killer cell receptor genes in three populations. Tissue Antigens 2001, 57, 358–362. [Google Scholar] [CrossRef] [PubMed]
- Ruggeri, L.; Capanni, M.; Urbani, E.; Perruccio, K.; Shlomchik, W.D.; Tosti, A.; Posati, S.; Rogaia, D.; Frassoni, F.; Aversa, F.; et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science 2002, 295, 2097–2100. [Google Scholar] [CrossRef]
- Sun, C.; Xu, J.; Huang, Q.; Huang, M.; Wen, H.; Zhang, C.; Wang, J.; Song, J.; Zheng, M.; Sun, H.; et al. High NKG2A expression contributes to NK cell exhaustion and predicts a poor prognosis of patients with liver cancer. Oncoimmunology 2017, 6, e1264562. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Goodlett, D.R.; Ishitani, A.; Marquardt, H.; Geraghty, D.E. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J. Immunol. 1998, 160, 4951–4960. [Google Scholar]
- Joyce, M.G.; Sun, P.D. The structural basis of ligand recognition by natural killer cell receptors. J. Biomed. Biotechnol. 2011, 2011, 203628. [Google Scholar] [CrossRef]
- Seliger, B. Molecular mechanisms of HLA class I-mediated immune evasion of human tumors and their role in resistance to immunotherapies. HLA 2016, 88, 213–220. [Google Scholar] [CrossRef]
- Derre, L.; Corvaisier, M.; Charreau, B.; Moreau, A.; Godefroy, E.; Moreau-Aubry, A.; Jotereau, F.; Gervois, N. Expression and release of HLA-E by melanoma cells and melanocytes: Potential impact on the response of cytotoxic effector cells. J. Immunol. 2006, 177, 3100–3107. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Gu, Y.; Zhang, Q.; Han, Y.; Hou, J.; Lin, L.; Wu, C.; Bao, Y.; Su, X.; Jiang, M.; et al. Identification of resting and type I IFN-activated human NK cell miRNomes reveals microRNA-378 and microRNA-30e as negative regulators of NK cell cytotoxicity. J. Immunol. 2012, 189, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, T.; Seow, S.V.; Wong, D.; Robinson, M.; Campana, D. Blocking expression of inhibitory receptor NKG2A overcomes tumor resistance to NK cells. J. Clin. Investig. 2019, 129, 2094–2106. [Google Scholar] [CrossRef]
- Andre, P.; Denis, C.; Soulas, C.; Bourbon-Caillet, C.; Lopez, J.; Arnoux, T.; Blery, M.; Bonnafous, C.; Gauthier, L.; Morel, A.; et al. Anti-NKG2A mAb Is a Checkpoint Inhibitor that Promotes Anti-tumor Immunity by Unleashing Both T and NK Cells. Cell 2018, 175, 1731–1743.e13. [Google Scholar] [CrossRef]
- Chan, C.J.; Martinet, L.; Gilfillan, S.; Souza-Fonseca-Guimaraes, F.; Chow, M.T.; Town, L.; Ritchie, D.S.; Colonna, M.; Andrews, D.M.; Smyth, M.J. The receptors CD96 and CD226 oppose each other in the regulation of natural killer cell functions. Nat. Immunol. 2014, 15, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, A.; Cella, M.; Giurisato, E.; Shaw, A.S.; Colonna, M. Cutting edge: CD96 (tactile) promotes NK cell-target cell adhesion by interacting with the poliovirus receptor (CD155). J. Immunol. 2004, 172, 3994–3998. [Google Scholar] [CrossRef] [PubMed]
- Stanietsky, N.; Simic, H.; Arapovic, J.; Toporik, A.; Levy, O.; Novik, A.; Levine, Z.; Beiman, M.; Dassa, L.; Achdout, H.; et al. The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc. Natl. Acad. Sci. USA 2009, 106, 17858–17863. [Google Scholar] [CrossRef] [PubMed]
- Carrega, P.; Bonaccorsi, I.; Di Carlo, E.; Morandi, B.; Paul, P.; Rizzello, V.; Cipollone, G.; Navarra, G.; Mingari, M.C.; Moretta, L.; et al. CD56(bright)perforin(low) noncytotoxic human NK cells are abundant in both healthy and neoplastic solid tissues and recirculate to secondary lymphoid organs via afferent lymph. J. Immunol. 2014, 192, 3805–3815. [Google Scholar] [CrossRef]
- Ferlazzo, G.; Thomas, D.; Lin, S.L.; Goodman, K.; Morandi, B.; Muller, W.A.; Moretta, A.; Munz, C. The abundant NK cells in human secondary lymphoid tissues require activation to express killer cell Ig-like receptors and become cytolytic. J. Immunol. 2004, 172, 1455–1462. [Google Scholar] [CrossRef]
- Gregoire, C.; Chasson, L.; Luci, C.; Tomasello, E.; Geissmann, F.; Vivier, E.; Walzer, T. The trafficking of natural killer cells. Immunol. Rev. 2007, 220, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.J.; Qin, S.; Unutmaz, D.; Soler, D.; Murphy, K.E.; Hodge, M.R.; Wu, L.; Butcher, E.C. Unique subpopulations of CD56+ NK and NK-T peripheral blood lymphocytes identified by chemokine receptor expression repertoire. J. Immunol. 2001, 166, 6477–6482. [Google Scholar] [CrossRef] [PubMed]
- Mailliard, R.B.; Alber, S.M.; Shen, H.; Watkins, S.C.; Kirkwood, J.M.; Herberman, R.B.; Kalinski, P. IL-18-induced CD83+CCR7+ NK helper cells. J. Exp. Med. 2005, 202, 941–953. [Google Scholar] [CrossRef] [PubMed]
- Marcenaro, E.; Cantoni, C.; Pesce, S.; Prato, C.; Pende, D.; Agaugue, S.; Moretta, L.; Moretta, A. Uptake of CCR7 and acquisition of migratory properties by human KIR+ NK cells interacting with monocyte-derived DC or EBV cell lines: Regulation by KIR/HLA-class I interaction. Blood 2009, 114, 4108–4116. [Google Scholar] [CrossRef]
- Marcenaro, E.; Pesce, S.; Sivori, S.; Carlomagno, S.; Moretta, L.; Moretta, A. KIR2DS1-dependent acquisition of CCR7 and migratory properties by human NK cells interacting with allogeneic HLA-C2+ DCs or T-cell blasts. Blood 2013, 121, 3396–3401. [Google Scholar] [CrossRef][Green Version]
- Bald, T.; Krummel, M.F.; Smyth, M.J.; Barry, K.C. The NK cell-cancer cycle: Advances and new challenges in NK cell-based immunotherapies. Nat. Immunol. 2020, 21, 835–847. [Google Scholar] [CrossRef]
- Noman, M.Z.; Paggetti, J.; Moussay, E.; Berchem, G.; Janji, B. Driving Natural Killer cells toward the melanoma tumor battlefield: Autophagy as a valuable therapeutic target. Oncoimmunology 2018, 7, e1452583. [Google Scholar] [CrossRef]
- Liu, J.; Li, Y.; Zhu, X.; Li, Q.; Liang, X.; Xie, J.; Hu, S.; Peng, W.; Li, C. Increased CX3CL1 mRNA expression level is a positive prognostic factor in patients with lung adenocarcinoma. Oncol. Lett. 2019, 17, 4877–4890. [Google Scholar] [CrossRef]
- Chen, E.B.; Zhou, Z.J.; Xiao, K.; Zhu, G.Q.; Yang, Y.; Wang, B.; Zhou, S.L.; Chen, Q.; Yin, D.; Wang, Z.; et al. The miR-561-5p/CX3CL1 Signaling Axis Regulates Pulmonary Metastasis in Hepatocellular Carcinoma Involving CX3CR1(+) Natural Killer Cells Infiltration. Theranostics 2019, 9, 4779–4794. [Google Scholar] [CrossRef] [PubMed]
- Ohta, M.; Tanaka, F.; Yamaguchi, H.; Sadanaga, N.; Inoue, H.; Mori, M. The high expression of Fractalkine results in a better prognosis for colorectal cancer patients. Int. J. Oncol. 2005, 26, 41–47. [Google Scholar] [CrossRef]
- Hyakudomi, M.; Matsubara, T.; Hyakudomi, R.; Yamamoto, T.; Kinugasa, S.; Yamanoi, A.; Maruyama, R.; Tanaka, T. Increased expression of fractalkine is correlated with a better prognosis and an increased number of both CD8+ T cells and natural killer cells in gastric adenocarcinoma. Ann. Surg. Oncol. 2008, 15, 1775–1782. [Google Scholar] [CrossRef]
- Park, M.H.; Lee, J.S.; Yoon, J.H. High expression of CX3CL1 by tumor cells correlates with a good prognosis and increased tumor-infiltrating CD8+ T cells, natural killer cells, and dendritic cells in breast carcinoma. J. Surg. Oncol. 2012, 106, 386–392. [Google Scholar] [CrossRef] [PubMed]
- Voskoboinik, I.; Whisstock, J.C.; Trapani, J.A. Perforin and granzymes: Function, dysfunction and human pathology. Nat. Rev. Immunol. 2015, 15, 388–400. [Google Scholar] [CrossRef]
- Smyth, M.J.; Street, S.E.; Trapani, J.A. Cutting edge: Granzymes A and B are not essential for perforin-mediated tumor rejection. J. Immunol. 2003, 171, 515–518. [Google Scholar] [CrossRef]
- Fauriat, C.; Long, E.O.; Ljunggren, H.G.; Bryceson, Y.T. Regulation of human NK-cell cytokine and chemokine production by target cell recognition. Blood 2010, 115, 2167–2176. [Google Scholar] [CrossRef] [PubMed]
- Roda, J.M.; Parihar, R.; Magro, C.; Nuovo, G.J.; Tridandapani, S.; Carson, W.E., 3rd. Natural killer cells produce T cell-recruiting chemokines in response to antibody-coated tumor cells. Cancer Res. 2006, 66, 517–526. [Google Scholar] [CrossRef]
- Schroder, K.; Hertzog, P.J.; Ravasi, T.; Hume, D.A. Interferon-gamma: An overview of signals, mechanisms and functions. J. Leukoc. Biol. 2004, 75, 163–189. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F. Molecular mechanisms of IFN-gamma to up-regulate MHC class I antigen processing and presentation. Int. Rev. Immunol. 2009, 28, 239–260. [Google Scholar] [CrossRef] [PubMed]
- Beatty, G.; Paterson, Y. IFN-gamma-dependent inhibition of tumor angiogenesis by tumor-infiltrating CD4+ T cells requires tumor responsiveness to IFN-gamma. J. Immunol. 2001, 166, 2276–2282. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, M.I.; Catalan-Dibene, J.; Zlotnik, A. B cells responses and cytokine production are regulated by their immune microenvironment. Cytokine 2015, 74, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Mohr, E.; Cunningham, A.F.; Toellner, K.M.; Bobat, S.; Coughlan, R.E.; Bird, R.A.; MacLennan, I.C.; Serre, K. IFN-{gamma} produced by CD8 T cells induces T-bet-dependent and -independent class switching in B cells in responses to alum-precipitated protein vaccine. Proc. Natl. Acad. Sci. USA 2010, 107, 17292–17297. [Google Scholar] [CrossRef] [PubMed]
- Dallagi, A.; Girouard, J.; Hamelin-Morrissette, J.; Dadzie, R.; Laurent, L.; Vaillancourt, C.; Lafond, J.; Carrier, C.; Reyes-Moreno, C. The activating effect of IFN-gamma on monocytes/macrophages is regulated by the LIF-trophoblast-IL-10 axis via Stat1 inhibition and Stat3 activation. Cell Mol. Immunol. 2015, 12, 326–341. [Google Scholar] [CrossRef]
- Aquino-Lopez, A.; Senyukov, V.V.; Vlasic, Z.; Kleinerman, E.S.; Lee, D.A. Interferon Gamma Induces Changes in Natural Killer (NK) Cell Ligand Expression and Alters NK Cell-Mediated Lysis of Pediatric Cancer Cell Lines. Front. Immunol. 2017, 8, 391. [Google Scholar] [CrossRef]
- Smeltz, R.B.; Chen, J.; Ehrhardt, R.; Shevach, E.M. Role of IFN-gamma in Th1 differentiation: IFN-gamma regulates IL-18R alpha expression by preventing the negative effects of IL-4 and by inducing/maintaining IL-12 receptor beta 2 expression. J. Immunol. 2002, 168, 6165–6172. [Google Scholar] [CrossRef]
- Castro, F.; Cardoso, A.P.; Goncalves, R.M.; Serre, K.; Oliveira, M.J. Interferon-Gamma at the Crossroads of Tumor Immune Surveillance or Evasion. Front. Immunol. 2018, 9, 847. [Google Scholar] [CrossRef]
- Kotredes, K.P.; Gamero, A.M. Interferons as inducers of apoptosis in malignant cells. J. Interferon Cytokine Res. 2013, 33, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Spranger, S.; Bao, R.; Gajewski, T.F. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature 2015, 523, 231–235. [Google Scholar] [CrossRef]
- Spranger, S.; Luke, J.J.; Bao, R.; Zha, Y.; Hernandez, K.M.; Li, Y.; Gajewski, A.P.; Andrade, J.; Gajewski, T.F. Density of immunogenic antigens does not explain the presence or absence of the T-cell-inflamed tumor microenvironment in melanoma. Proc. Natl. Acad. Sci. USA 2016, 113, E7759–E7768. [Google Scholar] [CrossRef] [PubMed]
- Bottcher, J.P.; Bonavita, E.; Chakravarty, P.; Blees, H.; Cabeza-Cabrerizo, M.; Sammicheli, S.; Rogers, N.C.; Sahai, E.; Zelenay, S.; Reis, E.S.C. NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell 2018, 172, 1022–1037 e1014. [Google Scholar] [CrossRef]
- De Andrade, L.F.; Lu, Y.; Luoma, A.; Ito, Y.; Pan, D.; Pyrdol, J.W.; Yoon, C.H.; Yuan, G.C.; Wucherpfennig, K.W. Discovery of specialized NK cell populations infiltrating human melanoma metastases. JCI Insight 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Vu Manh, T.P.; Elhmouzi-Younes, J.; Urien, C.; Ruscanu, S.; Jouneau, L.; Bourge, M.; Moroldo, M.; Foucras, G.; Salmon, H.; Marty, H.; et al. Defining Mononuclear Phagocyte Subset Homology Across Several Distant Warm-Blooded Vertebrates Through Comparative Transcriptomics. Front. Immunol. 2015, 6, 299. [Google Scholar] [CrossRef] [PubMed]
- Crozat, K.; Guiton, R.; Contreras, V.; Feuillet, V.; Dutertre, C.A.; Ventre, E.; Vu Manh, T.P.; Baranek, T.; Storset, A.K.; Marvel, J.; et al. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8alpha+ dendritic cells. J. Exp. Med. 2010, 207, 1283–1292. [Google Scholar] [CrossRef]
- Dorner, B.G.; Dorner, M.B.; Zhou, X.; Opitz, C.; Mora, A.; Guttler, S.; Hutloff, A.; Mages, H.W.; Ranke, K.; Schaefer, M.; et al. Selective expression of the chemokine receptor XCR1 on cross-presenting dendritic cells determines cooperation with CD8+ T cells. Immunity 2009, 31, 823–833. [Google Scholar] [CrossRef]
- Zhang, A.L.; Colmenero, P.; Purath, U.; Teixeira de Matos, C.; Hueber, W.; Klareskog, L.; Tarner, I.H.; Engleman, E.G.; Soderstrom, K. Natural killer cells trigger differentiation of monocytes into dendritic cells. Blood 2007, 110, 2484–2493. [Google Scholar] [CrossRef] [PubMed]
- Steinman, R.M.; Hawiger, D.; Liu, K.; Bonifaz, L.; Bonnyay, D.; Mahnke, K.; Iyoda, T.; Ravetch, J.; Dhodapkar, M.; Inaba, K.; et al. Dendritic cell function in vivo during the steady state: A role in peripheral tolerance. Ann. N. Y. Acad. Sci. 2003, 987, 15–25. [Google Scholar] [CrossRef]
- Valladeau, J.; Ravel, O.; Dezutter-Dambuyant, C.; Moore, K.; Kleijmeer, M.; Liu, Y.; Duvert-Frances, V.; Vincent, C.; Schmitt, D.; Davoust, J.; et al. Langerin, a novel C-type lectin specific to Langerhans cells, is an endocytic receptor that induces the formation of Birbeck granules. Immunity 2000, 12, 71–81. [Google Scholar] [CrossRef]
- Geijtenbeek, T.B.; Torensma, R.; van Vliet, S.J.; van Duijnhoven, G.C.; Adema, G.J.; van Kooyk, Y.; Figdor, C.G. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell 2000, 100, 575–585. [Google Scholar] [CrossRef]
- Sallusto, F.; Cella, M.; Danieli, C.; Lanzavecchia, A. Dendritic cells use macropinocytosis and the mannose receptor to concentrate macromolecules in the major histocompatibility complex class II compartment: Downregulation by cytokines and bacterial products. J. Exp. Med. 1995, 182, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Baleeiro, R.B.; Anselmo, L.B.; Soares, F.A.; Pinto, C.A.; Ramos, O.; Gross, J.L.; Haddad, F.; Younes, R.N.; Tomiyoshi, M.Y.; Bergami-Santos, P.C.; et al. High frequency of immature dendritic cells and altered in situ production of interleukin-4 and tumor necrosis factor-alpha in lung cancer. Cancer Immunol. Immunother. 2008, 57, 1335–1345. [Google Scholar] [CrossRef]
- Johnson, D.J.; Ohashi, P.S. Molecular programming of steady-state dendritic cells: Impact on autoimmunity and tumor immune surveillance. Ann. N. Y. Acad. Sci. 2013, 1284, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Patente, T.A.; Pinho, M.P.; Oliveira, A.A.; Evangelista, G.C.M.; Bergami-Santos, P.C.; Barbuto, J.A.M. Human Dendritic Cells: Their Heterogeneity and Clinical Application Potential in Cancer Immunotherapy. Front. Immunol. 2018, 9, 3176. [Google Scholar] [CrossRef] [PubMed]
- Kurts, C.; Kosaka, H.; Carbone, F.R.; Miller, J.F.; Heath, W.R. Class I-restricted cross-presentation of exogenous self-antigens leads to deletion of autoreactive CD8(+) T cells. J. Exp. Med. 1997, 186, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Manicassamy, S.; Pulendran, B. Dendritic cell control of tolerogenic responses. Immunol. Rev. 2011, 241, 206–227. [Google Scholar] [CrossRef]
- Zhu, H.C.; Qiu, T.; Liu, X.H.; Dong, W.C.; Weng, X.D.; Hu, C.H.; Kuang, Y.L.; Gao, R.H.; Dan, C.; Tao, T. Tolerogenic dendritic cells generated by RelB silencing using shRNA prevent acute rejection. Cell Immunol. 2012, 274, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Sela, U.; Park, C.G.; Park, A.; Olds, P.; Wang, S.; Steinman, R.M.; Fischetti, V.A. Dendritic Cells Induce a Subpopulation of IL-12Rbeta2-Expressing Treg that Specifically Consumes IL-12 to Control Th1 Responses. PLoS ONE 2016, 11, e0146412. [Google Scholar] [CrossRef] [PubMed]
- Ferlazzo, G.; Tsang, M.L.; Moretta, L.; Melioli, G.; Steinman, R.M.; Munz, C. Human dendritic cells activate resting natural killer (NK) cells and are recognized via the NKp30 receptor by activated NK cells. J. Exp. Med. 2002, 195, 343–351. [Google Scholar] [CrossRef]
- Li, J.; Schuler-Thurner, B.; Schuler, G.; Huber, C.; Seliger, B. Bipartite regulation of different components of the MHC class I antigen-processing machinery during dendritic cell maturation. Int. Immunol. 2001, 13, 1515–1523. [Google Scholar] [CrossRef]
- Morandi, B.; Mortara, L.; Chiossone, L.; Accolla, R.S.; Mingari, M.C.; Moretta, L.; Moretta, A.; Ferlazzo, G. Dendritic cell editing by activated natural killer cells results in a more protective cancer-specific immune response. PLoS ONE 2012, 7, e39170. [Google Scholar] [CrossRef]
- Ghiringhelli, F.; Menard, C.; Terme, M.; Flament, C.; Taieb, J.; Chaput, N.; Puig, P.E.; Novault, S.; Escudier, B.; Vivier, E.; et al. CD4+CD25+ regulatory T cells inhibit natural killer cell functions in a transforming growth factor-beta-dependent manner. J. Exp. Med. 2005, 202, 1075–1085. [Google Scholar] [CrossRef] [PubMed]
- Langhans, B.; Alwan, A.W.; Kramer, B.; Glassner, A.; Lutz, P.; Strassburg, C.P.; Nattermann, J.; Spengler, U. Regulatory CD4+ T cells modulate the interaction between NK cells and hepatic stellate cells by acting on either cell type. J. Hepatol. 2015, 62, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Stojanovic, A.; Fiegler, N.; Brunner-Weinzierl, M.; Cerwenka, A. CTLA-4 is expressed by activated mouse NK cells and inhibits NK Cell IFN-gamma production in response to mature dendritic cells. J. Immunol. 2014, 192, 4184–4191. [Google Scholar] [CrossRef]
- Balsamo, M.; Scordamaglia, F.; Pietra, G.; Manzini, C.; Cantoni, C.; Boitano, M.; Queirolo, P.; Vermi, W.; Facchetti, F.; Moretta, A.; et al. Melanoma-associated fibroblasts modulate NK cell phenotype and antitumor cytotoxicity. Proc. Natl. Acad. Sci. USA 2009, 106, 20847–20852. [Google Scholar] [CrossRef] [PubMed]
- Poggi, A.; Massaro, A.M.; Negrini, S.; Contini, P.; Zocchi, M.R. Tumor-induced apoptosis of human IL-2-activated NK cells: Role of natural cytotoxicity receptors. J. Immunol. 2005, 174, 2653–2660. [Google Scholar] [CrossRef]
- Arriga, R.; Caratelli, S.; Coppola, A.; Spagnoli, G.C.; Venditti, A.; Amadori, S.; Lanzilli, G.; Lauro, D.; Palomba, P.; Sconocchia, T.; et al. Enhancement of anti-leukemia activity of NK cells in vitro and in vivo by inhibition of leukemia cell-induced NK cell damage. Oncotarget 2016, 7, 2070–2079. [Google Scholar] [CrossRef] [PubMed]
- Jewett, A.; Cavalcanti, M.; Bonavida, B. Pivotal role of endogenous TNF-alpha in the induction of functional inactivation and apoptosis in NK cells. J. Immunol. 1997, 159, 4815–4822. [Google Scholar]
- Sconocchia, G.; Spagnoli, G.C.; Del Principe, D.; Ferrone, S.; Anselmi, M.; Wongsena, W.; Cervelli, V.; Schultz-Thater, E.; Wyler, S.; Carafa, V.; et al. Defective infiltration of natural killer cells in MICA/B-positive renal cell carcinoma involves beta(2)-integrin-mediated interaction. Neoplasia 2009, 11, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Romee, R.; Foley, B.; Lenvik, T.; Wang, Y.; Zhang, B.; Ankarlo, D.; Luo, X.; Cooley, S.; Verneris, M.; Walcheck, B.; et al. NK cell CD16 surface expression and function is regulated by a disintegrin and metalloprotease-17 (ADAM17). Blood 2013, 121, 3599–3608. [Google Scholar] [CrossRef]
- Peruzzi, G.; Femnou, L.; Gil-Krzewska, A.; Borrego, F.; Weck, J.; Krzewski, K.; Coligan, J.E. Membrane-type 6 matrix metalloproteinase regulates the activation-induced downmodulation of CD16 in human primary NK cells. J. Immunol. 2013, 191, 1883–1894. [Google Scholar] [CrossRef]
- Balsamo, M.; Vermi, W.; Parodi, M.; Pietra, G.; Manzini, C.; Queirolo, P.; Lonardi, S.; Augugliaro, R.; Moretta, A.; Facchetti, F.; et al. Melanoma cells become resistant to NK-cell-mediated killing when exposed to NK-cell numbers compatible with NK-cell infiltration in the tumor. Eur. J. Immunol. 2012, 42, 1833–1842. [Google Scholar] [CrossRef]
- Pradeu, T.; Jaeger, S.; Vivier, E. The speed of change: Towards a discontinuity theory of immunity? Nat. Rev. Immunol. 2013, 13, 764–769. [Google Scholar] [CrossRef]
- Pietra, G.; Manzini, C.; Rivara, S.; Vitale, M.; Cantoni, C.; Petretto, A.; Balsamo, M.; Conte, R.; Benelli, R.; Minghelli, S.; et al. Melanoma cells inhibit natural killer cell function by modulating the expression of activating receptors and cytolytic activity. Cancer Res. 2012, 72, 1407–1415. [Google Scholar] [CrossRef] [PubMed]
- Della Chiesa, M.; Carlomagno, S.; Frumento, G.; Balsamo, M.; Cantoni, C.; Conte, R.; Moretta, L.; Moretta, A.; Vitale, M. The tryptophan catabolite L-kynurenine inhibits the surface expression of NKp46- and NKG2D-activating receptors and regulates NK-cell function. Blood 2006, 108, 4118–4125. [Google Scholar] [CrossRef] [PubMed]
- Morandi, F.; Morandi, B.; Horenstein, A.L.; Chillemi, A.; Quarona, V.; Zaccarello, G.; Carrega, P.; Ferlazzo, G.; Mingari, M.C.; Moretta, L.; et al. A non-canonical adenosinergic pathway led by CD38 in human melanoma cells induces suppression of T cell proliferation. Oncotarget 2015, 6, 25602–25618. [Google Scholar] [CrossRef]
- Penafuerte, C.; Galipeau, J. TGF beta secreted by B16 melanoma antagonizes cancer gene immunotherapy bystander effect. Cancer Immunol. Immunother. 2008, 57, 1197–1206. [Google Scholar] [CrossRef]
- Busse, A.; Keilholz, U. Role of TGF-beta in melanoma. Curr. Pharm. Biotechnol. 2011, 12, 2165–2175. [Google Scholar] [CrossRef] [PubMed]
- Itakura, E.; Huang, R.R.; Wen, D.R.; Paul, E.; Wunsch, P.H.; Cochran, A.J. IL-10 expression by primary tumor cells correlates with melanoma progression from radial to vertical growth phase and development of metastatic competence. Mod. Pathol. 2011, 24, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Gerlini, G.; Tun-Kyi, A.; Dudli, C.; Burg, G.; Pimpinelli, N.; Nestle, F.O. Metastatic melanoma secreted IL-10 down-regulates CD1 molecules on dendritic cells in metastatic tumor lesions. Am. J. Pathol. 2004, 165, 1853–1863. [Google Scholar] [CrossRef]
- Hasmim, M.; Messai, Y.; Ziani, L.; Thiery, J.; Bouhris, J.H.; Noman, M.Z.; Chouaib, S. Critical Role of Tumor Microenvironment in Shaping NK Cell Functions: Implication of Hypoxic Stress. Front. Immunol. 2015, 6, 482. [Google Scholar] [CrossRef] [PubMed]
- Cerdeira, A.S.; Rajakumar, A.; Royle, C.M.; Lo, A.; Husain, Z.; Thadhani, R.I.; Sukhatme, V.P.; Karumanchi, S.A.; Kopcow, H.D. Conversion of peripheral blood NK cells to a decidual NK-like phenotype by a cocktail of defined factors. J. Immunol. 2013, 190, 3939–3948. [Google Scholar] [CrossRef]
- Holdenrieder, S.; Stieber, P.; Peterfi, A.; Nagel, D.; Steinle, A.; Salih, H.R. Soluble MICA in malignant diseases. Int. J. Cancer 2006, 118, 684–687. [Google Scholar] [CrossRef]
- Fernandez-Messina, L.; Ashiru, O.; Boutet, P.; Aguera-Gonzalez, S.; Skepper, J.N.; Reyburn, H.T.; Vales-Gomez, M. Differential mechanisms of shedding of the glycosylphosphatidylinositol (GPI)-anchored NKG2D ligands. J. Biol. Chem. 2010, 285, 8543–8551. [Google Scholar] [CrossRef] [PubMed]
- Salih, H.R.; Rammensee, H.G.; Steinle, A. Cutting edge: Down-regulation of MICA on human tumors by proteolytic shedding. J. Immunol. 2002, 169, 4098–4102. [Google Scholar] [CrossRef] [PubMed]
- Holdenrieder, S.; Stieber, P.; Peterfi, A.; Nagel, D.; Steinle, A.; Salih, H.R. Soluble MICB in malignant diseases: Analysis of diagnostic significance and correlation with soluble MICA. Cancer Immunol. Immunother. 2006, 55, 1584–1589. [Google Scholar] [CrossRef] [PubMed]
- Baginska, J.; Viry, E.; Berchem, G.; Poli, A.; Noman, M.Z.; van Moer, K.; Medves, S.; Zimmer, J.; Oudin, A.; Niclou, S.P.; et al. Granzyme B degradation by autophagy decreases tumor cell susceptibility to natural killer-mediated lysis under hypoxia. Proc. Natl. Acad. Sci. USA 2013, 110, 17450–17455. [Google Scholar] [CrossRef] [PubMed]
- Balsamo, M.; Manzini, C.; Pietra, G.; Raggi, F.; Blengio, F.; Mingari, M.C.; Varesio, L.; Moretta, L.; Bosco, M.C.; Vitale, M. Hypoxia downregulates the expression of activating receptors involved in NK-cell-mediated target cell killing without affecting ADCC. Eur. J. Immunol. 2013, 43, 2756–2764. [Google Scholar] [CrossRef]
- Yamada, N.; Yamanegi, K.; Ohyama, H.; Hata, M.; Nakasho, K.; Futani, H.; Okamura, H.; Terada, N. Hypoxia downregulates the expression of cell surface MICA without increasing soluble MICA in osteosarcoma cells in a HIF-1alpha-dependent manner. Int. J. Oncol. 2012, 41, 2005–2012. [Google Scholar] [CrossRef]
- Cozar, J.M.; Canton, J.; Tallada, M.; Concha, A.; Cabrera, T.; Garrido, F.; Ruiz-Cabello Osuna, F. Analysis of NK cells and chemokine receptors in tumor infiltrating CD4 T lymphocytes in human renal carcinomas. Cancer Immunol. Immunother. 2005, 54, 858–866. [Google Scholar] [CrossRef] [PubMed]
- Schleypen, J.S.; Baur, N.; Kammerer, R.; Nelson, P.J.; Rohrmann, K.; Grone, E.F.; Hohenfellner, M.; Haferkamp, A.; Pohla, H.; Schendel, D.J.; et al. Cytotoxic markers and frequency predict functional capacity of natural killer cells infiltrating renal cell carcinoma. Clin. Cancer Res. 2006, 12, 718–725. [Google Scholar] [CrossRef]
- Guan, Y.; Chambers, C.B.; Tabatabai, T.; Hatley, H.; Delfino, K.R.; Robinson, K.; Alanee, S.R.; Ran, S.; Torry, D.S.; Wilber, A. Renal cell tumors convert natural killer cells to a proangiogenic phenotype. Oncotarget 2020, 11, 2571–2585. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Zhang, Q.; Zhen, Q.; Zhao, Y.; Liu, N.; Li, T.; Hao, Y.; Zhang, Y.; Luo, C.; Wu, X. Negative regulation of tumor-infiltrating NK cell in clear cell renal cell carcinoma patients through the exosomal pathway. Oncotarget 2017, 8, 37783–37795. [Google Scholar] [CrossRef]
- Geissler, K.; Fornara, P.; Lautenschlager, C.; Holzhausen, H.J.; Seliger, B.; Riemann, D. Immune signature of tumor infiltrating immune cells in renal cancer. Oncoimmunology 2015, 4, e985082. [Google Scholar] [CrossRef]
- Ali, T.H.; Pisanti, S.; Ciaglia, E.; Mortarini, R.; Anichini, A.; Garofalo, C.; Tallerico, R.; Santinami, M.; Gulletta, E.; Ietto, C.; et al. Enrichment of CD56(dim)KIR + CD57 + highly cytotoxic NK cells in tumour-infiltrated lymph nodes of melanoma patients. Nat. Commun. 2014, 5, 5639. [Google Scholar] [CrossRef]
- Lee, H.; Quek, C.; Silva, I.; Tasker, A.; Batten, M.; Rizos, H.; Lim, S.Y.; Nur Gide, T.; Shang, P.; Attrill, G.H.; et al. Integrated molecular and immunophenotypic analysis of NK cells in anti-PD-1 treated metastatic melanoma patients. Oncoimmunology 2019, 8, e1537581. [Google Scholar] [CrossRef]
- Triki, H.; Charfi, S.; Bouzidi, L.; Ben Kridis, W.; Daoud, J.; Chaabane, K.; Sellami-Boudawara, T.; Rebai, A.; Cherif, B. CD155 expression in human breast cancer: Clinical significance and relevance to natural killer cell infiltration. Life Sci. 2019, 231, 116543. [Google Scholar] [CrossRef] [PubMed]
- Honkanen, T.J.; Moilanen, T.; Karihtala, P.; Tiainen, S.; Auvinen, P.; Vayrynen, J.P.; Makinen, M.; Koivunen, J.P. Prognostic and predictive role of spatially positioned tumour infiltrating lymphocytes in metastatic HER2 positive breast cancer treated with trastuzumab. Sci. Rep. 2017, 7, 18027. [Google Scholar] [CrossRef]
- Muntasell, A.; Rojo, F.; Servitja, S.; Rubio-Perez, C.; Cabo, M.; Tamborero, D.; Costa-Garcia, M.; Martinez-Garcia, M.; Menendez, S.; Vazquez, I.; et al. NK Cell Infiltrates and HLA Class I Expression in Primary HER2(+) Breast Cancer Predict and Uncouple Pathological Response and Disease-free Survival. Clin. Cancer Res. 2019, 25, 1535–1545. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wang, Q.; Wang, Z.; Jiang, J.; Yu, S.C.; Ping, Y.F.; Yang, J.; Xu, S.L.; Ye, X.Z.; Xu, C.; et al. Metastatic consequences of immune escape from NK cell cytotoxicity by human breast cancer stem cells. Cancer Res. 2014, 74, 5746–5757. [Google Scholar] [CrossRef] [PubMed]
- Easom, N.J.W.; Stegmann, K.A.; Swadling, L.; Pallett, L.J.; Burton, A.R.; Odera, D.; Schmidt, N.; Huang, W.C.; Fusai, G.; Davidson, B.; et al. IL-15 Overcomes Hepatocellular Carcinoma-Induced NK Cell Dysfunction. Front. Immunol. 2018, 9, 1009. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Zhang, Z.; Zhou, L.; Wang, H.; Fu, J.; Zhang, S.; Shi, M.; Zhang, H.; Yang, Y.; Wu, H.; et al. Functional impairment in circulating and intrahepatic NK cells and relative mechanism in hepatocellular carcinoma patients. Clin. Immunol. 2008, 129, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Liu, L.; Huang, Q.; Liu, H.; Huang, M.; Wang, J.; Wen, H.; Lin, R.; Qu, K.; Li, K.; et al. Accumulation of Tumor-Infiltrating CD49a(+) NK Cells Correlates with Poor Prognosis for Human Hepatocellular Carcinoma. Cancer Immunol. Res. 2019, 7, 1535–1546. [Google Scholar] [CrossRef]
- Carrega, P.; Morandi, B.; Costa, R.; Frumento, G.; Forte, G.; Altavilla, G.; Ratto, G.B.; Mingari, M.C.; Moretta, L.; Ferlazzo, G. Natural killer cells infiltrating human nonsmall-cell lung cancer are enriched in CD56 bright CD16(-) cells and display an impaired capability to kill tumor cells. Cancer 2008, 112, 863–875. [Google Scholar] [CrossRef] [PubMed]
- Lavin, Y.; Kobayashi, S.; Leader, A.; Amir, E.D.; Elefant, N.; Bigenwald, C.; Remark, R.; Sweeney, R.; Becker, C.D.; Levine, J.H.; et al. Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses. Cell 2017, 169, 750–765 e717. [Google Scholar] [CrossRef]
- Pasero, C.; Gravis, G.; Granjeaud, S.; Guerin, M.; Thomassin-Piana, J.; Rocchi, P.; Salem, N.; Walz, J.; Moretta, A.; Olive, D. Highly effective NK cells are associated with good prognosis in patients with metastatic prostate cancer. Oncotarget 2015, 6, 14360–14373. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, N.; Ji, N.; Hurez, V.; Curiel, T.J.; Montgomery, M.O.; Braun, A.J.; Nicolas, M.; Aguilera, M.; Kaushik, D.; Liu, Q.; et al. Intratumoral CD56(bright) natural killer cells are associated with improved survival in bladder cancer. Oncotarget 2018, 9, 36492–36502. [Google Scholar] [CrossRef] [PubMed]
- Alderdice, M.; Dunne, P.D.; Cole, A.J.; O’Reilly, P.G.; McArt, D.G.; Bingham, V.; Fuchs, M.A.; McQuaid, S.; Loughrey, M.B.; Murray, G.I.; et al. Natural killer-like signature observed post therapy in locally advanced rectal cancer is a determinant of pathological response and improved survival. Mod. Pathol. 2017, 30, 1287–1298. [Google Scholar] [CrossRef] [PubMed]
- Kmiecik, J.; Poli, A.; Brons, N.H.; Waha, A.; Eide, G.E.; Enger, P.O.; Zimmer, J.; Chekenya, M. Elevated CD3+ and CD8+ tumor-infiltrating immune cells correlate with prolonged survival in glioblastoma patients despite integrated immunosuppressive mechanisms in the tumor microenvironment and at the systemic level. J. Neuroimmunol. 2013, 264, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Halama, N.; Braun, M.; Kahlert, C.; Spille, A.; Quack, C.; Rahbari, N.; Koch, M.; Weitz, J.; Kloor, M.; Zoernig, I.; et al. Natural killer cells are scarce in colorectal carcinoma tissue despite high levels of chemokines and cytokines. Clin. Cancer Res. 2011, 17, 678–689. [Google Scholar] [CrossRef]
- Erdag, G.; Schaefer, J.T.; Smolkin, M.E.; Deacon, D.H.; Shea, S.M.; Dengel, L.T.; Patterson, J.W.; Slingluff, C.L., Jr. Immunotype and immunohistologic characteristics of tumor-infiltrating immune cells are associated with clinical outcome in metastatic melanoma. Cancer Res. 2012, 72, 1070–1080. [Google Scholar] [CrossRef] [PubMed]
- Sconocchia, G.; Arriga, R.; Tornillo, L.; Terracciano, L.; Ferrone, S.; Spagnoli, G.C. Melanoma cells inhibit NK cell functions. Cancer Res. 2012, 72, 5428–5429, author reply 5430. [Google Scholar] [CrossRef]
- Behling, F.; Schittenhelm, J. Tissue microarrays—Translational biomarker research in the fast lane. Expert Rev. Mol. Diagn. 2018, 18, 833–835. [Google Scholar] [CrossRef] [PubMed]
- Eckel-Passow, J.E.; Lohse, C.M.; Sheinin, Y.; Crispen, P.L.; Krco, C.J.; Kwon, E.D. Tissue microarrays: One size does not fit all. Diagn. Pathol. 2010, 5, 48. [Google Scholar] [CrossRef]
- Nersesian, S.; Schwartz, S.L.; Grantham, S.R.; MacLean, L.K.; Lee, S.N.; Pugh-Toole, M.; Boudreau, J.E. NK cell infiltration is associated with improved overall survival in solid cancers: A systematic review and meta-analysis. Transl. Oncol. 2021, 14, 100930. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, W.; Hu, B.; Wang, P.; Lv, X.; Chen, S.; Shao, Z. Prognostic Significance of Tumor-Infiltrating Natural Killer Cells in Solid Tumors: A Systematic Review and Meta-Analysis. Front. Immunol. 2020, 11, 1242. [Google Scholar] [CrossRef]
- Kohlhapp, F.J.; Broucek, J.R.; Hughes, T.; Huelsmann, E.J.; Lusciks, J.; Zayas, J.P.; Dolubizno, H.; Fleetwood, V.A.; Grin, A.; Hill, G.E.; et al. NK cells and CD8+ T cells cooperate to improve therapeutic responses in melanoma treated with interleukin-2 (IL-2) and CTLA-4 blockade. J. Immunother. Cancer 2015, 3, 18. [Google Scholar] [CrossRef]
- Sanseviero, E.; O’Brien, E.M.; Karras, J.R.; Shabaneh, T.B.; Aksoy, B.A.; Xu, W.; Zheng, C.; Yin, X.; Xu, X.; Karakousis, G.C.; et al. Anti-CTLA-4 Activates Intratumoral NK Cells and Combined with IL15/IL15Ralpha Complexes Enhances Tumor Control. Cancer Immunol. Res. 2019, 7, 1371–1380. [Google Scholar] [CrossRef]
- Radziewicz, H.; Ibegbu, C.C.; Fernandez, M.L.; Workowski, K.A.; Obideen, K.; Wehbi, M.; Hanson, H.L.; Steinberg, J.P.; Masopust, D.; Wherry, E.J.; et al. Liver-infiltrating lymphocytes in chronic human hepatitis C virus infection display an exhausted phenotype with high levels of PD-1 and low levels of CD127 expression. J. Virol. 2007, 81, 2545–2553. [Google Scholar] [CrossRef] [PubMed]
- Golden-Mason, L.; Palmer, B.; Klarquist, J.; Mengshol, J.A.; Castelblanco, N.; Rosen, H.R. Upregulation of PD-1 expression on circulating and intrahepatic hepatitis C virus-specific CD8+ T cells associated with reversible immune dysfunction. J. Virol. 2007, 81, 9249–9258. [Google Scholar] [CrossRef] [PubMed]
- Concha-Benavente, F.; Kansy, B.; Moskovitz, J.; Moy, J.; Chandran, U.; Ferris, R.L. PD-L1 Mediates Dysfunction in Activated PD-1(+) NK Cells in Head and Neck Cancer Patients. Cancer Immunol. Res. 2018, 6, 1548–1560. [Google Scholar] [CrossRef]
- Mariotti, F.R.; Petrini, S.; Ingegnere, T.; Tumino, N.; Besi, F.; Scordamaglia, F.; Munari, E.; Pesce, S.; Marcenaro, E.; Moretta, A.; et al. PD-1 in human NK cells: Evidence of cytoplasmic mRNA and protein expression. Oncoimmunology 2019, 8, 1557030. [Google Scholar] [CrossRef]
- Hsu, J.; Hodgins, J.J.; Marathe, M.; Nicolai, C.J.; Bourgeois-Daigneault, M.C.; Trevino, T.N.; Azimi, C.S.; Scheer, A.K.; Randolph, H.E.; Thompson, T.W.; et al. Contribution of NK cells to immunotherapy mediated by PD-1/PD-L1 blockade. J. Clin. Investig. 2018, 128, 4654–4668. [Google Scholar] [CrossRef]
- Alvarez, M.; Simonetta, F.; Baker, J.; Morrison, A.R.; Wenokur, A.S.; Pierini, A.; Berraondo, P.; Negrin, R.S. Indirect Impact of PD-1/PD-L1 Blockade on a Murine Model of NK Cell Exhaustion. Front. Immunol. 2020, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Niu, C.; Li, M.; Zhu, S.; Chen, Y.; Zhou, L.; Xu, D.; Xu, J.; Li, Z.; Li, W.; Cui, J. PD-1-positive Natural Killer Cells have a weaker antitumor function than that of PD-1-negative Natural Killer Cells in Lung Cancer. Int. J. Med. Sci. 2020, 17, 1964–1973. [Google Scholar] [CrossRef]
- Norris, S.; Coleman, A.; Kuri-Cervantes, L.; Bower, M.; Nelson, M.; Goodier, M.R. PD-1 expression on natural killer cells and CD8(+) T cells during chronic HIV-1 infection. Viral Immunol. 2012, 25, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Park, I.H.; Yang, H.N.; Lee, K.J.; Kim, T.S.; Lee, E.S.; Jung, S.Y.; Kwon, Y.; Kong, S.Y. Tumor-derived IL-18 induces PD-1 expression on immunosuppressive NK cells in triple-negative breast cancer. Oncotarget 2017, 8, 32722–32730. [Google Scholar] [CrossRef]
- Pesce, S.; Greppi, M.; Grossi, F.; Del Zotto, G.; Moretta, L.; Sivori, S.; Genova, C.; Marcenaro, E. PD/1-PD-Ls Checkpoint: Insight on the Potential Role of NK Cells. Front. Immunol. 2019, 10, 1242. [Google Scholar] [CrossRef] [PubMed]
- Hasim, M.S.; Vulpis, E.; Sciumè, G.; Shih, H.Y.; Scheer, A.; MacMillan, O.; Petrucci, M.T.; Santoni, A.; Zingoni, A.; Ardolino, M. NK cells acquire PD-1 from the membrane of tumor cells. bioRxiv 2020. [Google Scholar] [CrossRef]
- Liu, Y.; Cheng, Y.; Xu, Y.; Wang, Z.; Du, X.; Li, C.; Peng, J.; Gao, L.; Liang, X.; Ma, C. Increased expression of programmed cell death protein 1 on NK cells inhibits NK-cell-mediated anti-tumor function and indicates poor prognosis in digestive cancers. Oncogene 2017, 36, 6143–6153. [Google Scholar] [CrossRef]
- Oyer, J.L.; Gitto, S.B.; Altomare, D.A.; Copik, A.J. PD-L1 blockade enhances anti-tumor efficacy of NK cells. Oncoimmunology 2018, 7, e1509819. [Google Scholar] [CrossRef]
- Guo, Y.; Feng, X.; Jiang, Y.; Shi, X.; Xing, X.; Liu, X.; Li, N.; Fadeel, B.; Zheng, C. PD1 blockade enhances cytotoxicity of in vitro expanded natural killer cells towards myeloma cells. Oncotarget 2016, 7, 48360–48374. [Google Scholar] [CrossRef]
- Benson, D.M.; Bakan, C.E.; Mishra, A.; Hofmeister, C.C.; Efebera, Y.; Becknell, B.; Baiocchi, R.A.; Zhang, J.; Yu, J.; Smith, M.K.; et al. The PD-1/PD-L1 axis modulates the natural killer cell versus multiple myeloma effect: A therapeutic target for CT-011, a novel monoclonal anti–PD-1 antibody. Blood 2010, 116, 2286–2294. [Google Scholar] [CrossRef]
- Taggart, D.; Andreou, T.; Scott, K.J.; Williams, J.; Rippaus, N.; Brownlie, R.J.; Ilett, E.J.; Salmond, R.J.; Melcher, A.; Lorger, M. Anti-PD-1/anti-CTLA-4 efficacy in melanoma brain metastases depends on extracranial disease and augmentation of CD8(+) T cell trafficking. Proc. Natl. Acad. Sci. USA 2018, 115, E1540–E1549. [Google Scholar] [CrossRef]
- Pico de Coana, Y.; Wolodarski, M.; van der Haar Avila, I.; Nakajima, T.; Rentouli, S.; Lundqvist, A.; Masucci, G.; Hansson, J.; Kiessling, R. PD-1 checkpoint blockade in advanced melanoma patients: NK cells, monocytic subsets and host PD-L1 expression as predictive biomarker candidates. Oncoimmunology 2020, 9, 1786888. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Hu, Y.; Shi, C. Targeting Natural Killer Cells for Tumor Immunotherapy. Front. Immunol. 2020, 11, 60. [Google Scholar] [CrossRef]
- Gide, T.N.; Wilmott, J.S.; Scolyer, R.A.; Long, G.V. Primary and Acquired Resistance to Immune Checkpoint Inhibitors in Metastatic Melanoma. Clin. Cancer Res. 2018, 24, 1260–1270. [Google Scholar] [CrossRef]
- Subrahmanyam, P.B.; Dong, Z.; Gusenleitner, D.; Giobbie-Hurder, A.; Severgnini, M.; Zhou, J.; Manos, M.; Eastman, L.M.; Maecker, H.T.; Hodi, F.S. Distinct predictive biomarker candidates for response to anti-CTLA-4 and anti-PD-1 immunotherapy in melanoma patients. J. Immunother. Cancer 2018, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, I.P.; Gallois, A.; Jimenez-Baranda, S.; Khan, S.; Anderson, A.C.; Kuchroo, V.K.; Osman, I.; Bhardwaj, N. Reversal of NK-cell exhaustion in advanced melanoma by Tim-3 blockade. Cancer Immunol. Res. 2014, 2, 410–422. [Google Scholar] [CrossRef] [PubMed]
- Fregni, G.; Perier, A.; Pittari, G.; Jacobelli, S.; Sastre, X.; Gervois, N.; Allard, M.; Bercovici, N.; Avril, M.F.; Caignard, A. Unique functional status of natural killer cells in metastatic stage IV melanoma patients and its modulation by chemotherapy. Clin. Cancer Res. 2011, 17, 2628–2637. [Google Scholar] [CrossRef]
- Konjevic, G.; Mirjacic Martinovic, K.; Jurisic, V.; Babovic, N.; Spuzic, I. Biomarkers of suppressed natural killer (NK) cell function in metastatic melanoma: Decreased NKG2D and increased CD158a receptors on CD3-CD16+ NK cells. Biomarkers 2009, 14, 258–270. [Google Scholar] [CrossRef] [PubMed]
- Mirjacic Martinovic, K.; Konjevic, G.; Babovic, N.; Inic, M. The stage dependent changes in NK cell activity and the expression of activating and inhibitory NK cell receptors in melanoma patients. J. Surg. Res. 2011, 171, 637–649. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Anderson, A.C.; Schubart, A.; Xiong, H.; Imitola, J.; Khoury, S.J.; Zheng, X.X.; Strom, T.B.; Kuchroo, V.K. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat. Immunol. 2005, 6, 1245–1252. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, M.; Akiba, H.; Takeda, K.; Kojima, Y.; Hashiguchi, M.; Azuma, M.; Yagita, H.; Okumura, K. Tim-3 mediates phagocytosis of apoptotic cells and cross-presentation. Blood 2009, 113, 3821–3830. [Google Scholar] [CrossRef]
- Chiba, S.; Baghdadi, M.; Akiba, H.; Yoshiyama, H.; Kinoshita, I.; Dosaka-Akita, H.; Fujioka, Y.; Ohba, Y.; Gorman, J.V.; Colgan, J.D.; et al. Tumor-infiltrating DCs suppress nucleic acid-mediated innate immune responses through interactions between the receptor TIM-3 and the alarmin HMGB1. Nat. Immunol. 2012, 13, 832–842. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Zhu, C.; Kondo, Y.; Anderson, A.C.; Gandhi, A.; Russell, A.; Dougan, S.K.; Petersen, B.S.; Melum, E.; Pertel, T.; et al. CEACAM1 regulates TIM-3-mediated tolerance and exhaustion. Nature 2015, 517, 386–390. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, Y.; Lian, J.; Yang, H.; Li, F.; Zhao, S.; Qi, Y.; Zhang, Y.; Huang, L. TNF-alpha-induced Tim-3 expression marks the dysfunction of infiltrating natural killer cells in human esophageal cancer. J. Transl. Med. 2019, 17, 165. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Yuan, L.; Gao, Q.; Yuan, P.; Zhao, P.; Yuan, H.; Fan, H.; Li, T.; Qin, P.; Han, L.; et al. Circulating and tumor-infiltrating Tim-3 in patients with colorectal cancer. Oncotarget 2015, 6, 20592–20603. [Google Scholar] [CrossRef]
- Seo, H.; Kim, B.S.; Bae, E.A.; Min, B.S.; Han, Y.D.; Shin, S.J.; Kang, C.Y. IL21 Therapy Combined with PD-1 and Tim-3 Blockade Provides Enhanced NK Cell Antitumor Activity against MHC Class I-Deficient Tumors. Cancer Immunol. Res. 2018, 6, 685–695. [Google Scholar] [CrossRef]
- Deuss, F.A.; Gully, B.S.; Rossjohn, J.; Berry, R. Recognition of nectin-2 by the natural killer cell receptor TIGIT. J. Biol. Chem. 2017. [Google Scholar] [CrossRef]
- Wang, F.; Hou, H.; Wu, S.; Tang, Q.; Liu, W.; Huang, M.; Yin, B.; Huang, J.; Mao, L.; Lu, Y.; et al. TIGIT expression levels on human NK cells correlate with functional heterogeneity among healthy individuals. Eur. J. Immunol. 2015, 45, 2886–2897. [Google Scholar] [CrossRef]
- Sarhan, D.; Cichocki, F.; Zhang, B.; Yingst, A.; Spellman, S.R.; Cooley, S.; Verneris, M.R.; Blazar, B.R.; Miller, J.S. Adaptive NK Cells with Low TIGIT Expression Are Inherently Resistant to Myeloid-Derived Suppressor Cells. Cancer Res. 2016, 76, 5696–5706. [Google Scholar] [CrossRef]
- Zhang, Q.; Bi, J.; Zheng, X.; Chen, Y.; Wang, H.; Wu, W.; Wang, Z.; Wu, Q.; Peng, H.; Wei, H.; et al. Blockade of the checkpoint receptor TIGIT prevents NK cell exhaustion and elicits potent anti-tumor immunity. Nat. Immunol. 2018, 19, 723–732. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Sun, C. The Rise of NK Cell Checkpoints as Promising Therapeutic Targets in Cancer Immunotherapy. Front. Immunol. 2019, 10, 2354. [Google Scholar] [CrossRef] [PubMed]
- Maude, S.L.; Frey, N.; Shaw, P.A.; Aplenc, R.; Barrett, D.M.; Bunin, N.J.; Chew, A.; Gonzalez, V.E.; Zheng, Z.; Lacey, S.F.; et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 2014, 371, 1507–1517. [Google Scholar] [CrossRef] [PubMed]
- Davila, M.L.; Riviere, I.; Wang, X.; Bartido, S.; Park, J.; Curran, K.; Chung, S.S.; Stefanski, J.; Borquez-Ojeda, O.; Olszewska, M.; et al. Efficacy and toxicity management of 19-28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci. Transl. Med. 2014, 6, 224ra225. [Google Scholar] [CrossRef] [PubMed]
- Turtle, C.J.; Hanafi, L.A.; Berger, C.; Gooley, T.A.; Cherian, S.; Hudecek, M.; Sommermeyer, D.; Melville, K.; Pender, B.; Budiarto, T.M.; et al. CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J. Clin. Investig. 2016, 126, 2123–2138. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Dou, M.; Ma, Q.; Yao, R.; Liu, J. Chimeric antigen receptor (CAR)-modified NK cells against cancer: Opportunities and challenges. Int. Immunopharmacol. 2019, 74, 105695. [Google Scholar] [CrossRef]
- Abbasi, J. CAR Natural Killer Cell Therapy Safe and Effective in First Trial. JAMA 2020, 323, 916. [Google Scholar] [CrossRef]
- Liu, E.; Marin, D.; Banerjee, P.; Macapinlac, H.A.; Thompson, P.; Basar, R.; Nassif Kerbauy, L.; Overman, B.; Thall, P.; Kaplan, M.; et al. Use of CAR-Transduced Natural Killer Cells in CD19-Positive Lymphoid Tumors. N. Engl. J. Med. 2020, 382, 545–553. [Google Scholar] [CrossRef]
- Strizova, Z.; Bartunkova, J.; Smrz, D. The challenges of adoptive cell transfer in the treatment of human renal cell carcinoma. Cancer Immunol. Immunother. 2019, 68, 1831–1838. [Google Scholar] [CrossRef] [PubMed]
- Pahl, J.H.W.; Koch, J.; Gotz, J.J.; Arnold, A.; Reusch, U.; Gantke, T.; Rajkovic, E.; Treder, M.; Cerwenka, A. CD16A Activation of NK Cells Promotes NK Cell Proliferation and Memory-Like Cytotoxicity against Cancer Cells. Cancer Immunol. Res. 2018, 6, 517–527. [Google Scholar] [CrossRef]
- Weiner, G.J. Rituximab: Mechanism of action. Semin. Hematol. 2010, 47, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Merkt, W.; Lorenz, H.M.; Watzl, C. Rituximab induces phenotypical and functional changes of NK cells in a non-malignant experimental setting. Arthritis Res. 2016, 18, 206. [Google Scholar] [CrossRef] [PubMed]
- Felices, M.; Lenvik, T.R.; Davis, Z.B.; Miller, J.S.; Vallera, D.A. Generation of BiKEs and TriKEs to Improve NK Cell-Mediated Targeting of Tumor Cells. Methods Mol. Biol. 2016, 1441, 333–346. [Google Scholar] [CrossRef]
- Baluna, R.; Vitetta, E.S. Vascular leak syndrome: A side effect of immunotherapy. Immunopharmacology 1997, 37, 117–132. [Google Scholar] [CrossRef]
- Bachanova, V.; Cooley, S.; Defor, T.E.; Verneris, M.R.; Zhang, B.; McKenna, D.H.; Curtsinger, J.; Panoskaltsis-Mortari, A.; Lewis, D.; Hippen, K.; et al. Clearance of acute myeloid leukemia by haploidentical natural killer cells is improved using IL-2 diphtheria toxin fusion protein. Blood 2014, 123, 3855–3863. [Google Scholar] [CrossRef]
- Conlon, K.C.; Potter, E.L.; Pittaluga, S.; Lee, C.R.; Miljkovic, M.D.; Fleisher, T.A.; Dubois, S.; Bryant, B.R.; Petrus, M.; Perera, L.P.; et al. IL15 by Continuous Intravenous Infusion to Adult Patients with Solid Tumors in a Phase I Trial Induced Dramatic NK-Cell Subset Expansion. Clin. Cancer Res. 2019, 25, 4945–4954. [Google Scholar] [CrossRef]
- Waldmann, T.A.; Dubois, S.; Miljkovic, M.D.; Conlon, K.C. IL-15 in the Combination Immunotherapy of Cancer. Front. Immunol. 2020, 11, 868. [Google Scholar] [CrossRef]
- Carson, W.E.; Fehniger, T.A.; Haldar, S.; Eckhert, K.; Lindemann, M.J.; Lai, C.F.; Croce, C.M.; Baumann, H.; Caligiuri, M.A. A potential role for interleukin-15 in the regulation of human natural killer cell survival. J. Clin. Investig. 1997, 99, 937–943. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, J.; Niu, J.; Zhang, J.; Tian, Z. Interleukin-15 improves cytotoxicity of natural killer cells via up-regulating NKG2D and cytotoxic effector molecule expression as well as STAT1 and ERK1/2 phosphorylation. Cytokine 2008, 42, 128–136. [Google Scholar] [CrossRef]
- Zhang, M.; Wen, B.; Anton, O.M.; Yao, Z.; Dubois, S.; Ju, W.; Sato, N.; DiLillo, D.J.; Bamford, R.N.; Ravetch, J.V.; et al. IL-15 enhanced antibody-dependent cellular cytotoxicity mediated by NK cells and macrophages. Proc. Natl. Acad. Sci. USA 2018, 115, E10915–E10924. [Google Scholar] [CrossRef]
- Vallera, D.A.; Felices, M.; McElmurry, R.; McCullar, V.; Zhou, X.; Schmohl, J.U.; Zhang, B.; Lenvik, A.J.; Panoskaltsis-Mortari, A.; Verneris, M.R.; et al. IL15 Trispecific Killer Engagers (TriKE) Make Natural Killer Cells Specific to CD33+ Targets While Also Inducing Persistence, In Vivo Expansion, and Enhanced Function. Clin. Cancer Res. 2016, 22, 3440–3450. [Google Scholar] [CrossRef]
- Vallera, D.A.; Ferrone, S.; Kodal, B.; Hinderlie, P.; Bendzick, L.; Ettestad, B.; Hallstrom, C.; Zorko, N.A.; Rao, A.; Fujioka, N.; et al. NK-Cell-Mediated Targeting of Various Solid Tumors Using a B7-H3 Tri-Specific Killer Engager In Vitro and In Vivo. Cancers 2020, 12, 2659. [Google Scholar] [CrossRef] [PubMed]
- Tay, S.S.; Carol, H.; Biro, M. TriKEs and BiKEs join CARs on the cancer immunotherapy highway. Hum. Vaccin Immunother. 2016, 12, 2790–2796. [Google Scholar] [CrossRef] [PubMed]
- Stoermer, K.A.; Morrison, T.E. Complement and viral pathogenesis. Virology 2011, 411, 362–373. [Google Scholar] [CrossRef]
- Raman, S.S.; Hecht, J.R.; Chan, E. Talimogene laherparepvec: Review of its mechanism of action and clinical efficacy and safety. Immunotherapy 2019, 11, 705–723. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).