Using a Prime-Boost Vaccination Strategy That Proved Effective for High Resolution Epitope Mapping to Characterize the Elusive Immunogenicity of Survivin
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Cell Cultures
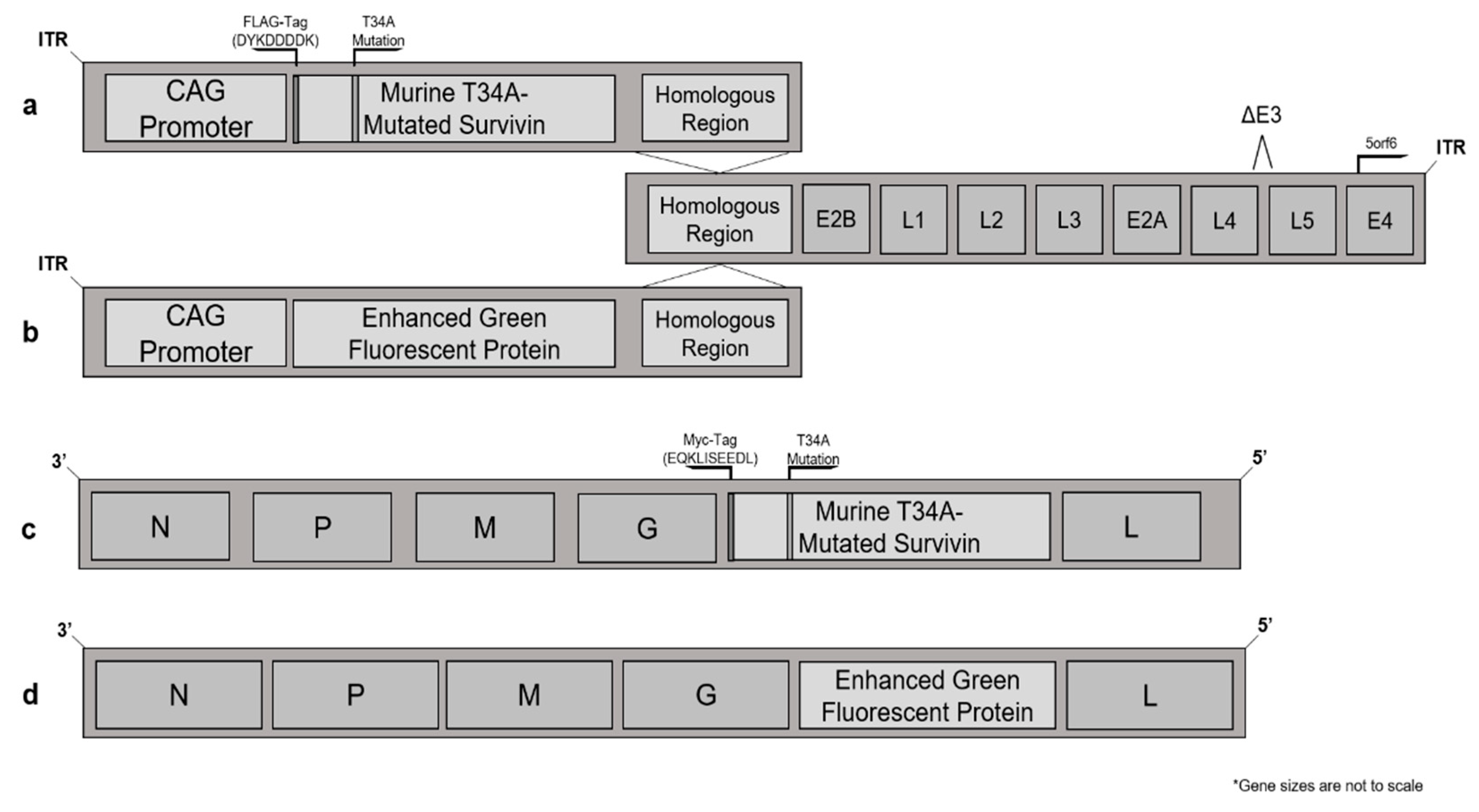
2.3. Plasmids and Vector Construction
2.4. Viruses
2.5. Vaccination Protocol
2.6. Sample Processing
2.7. Quantification of T Cells by Flow Cytometry
2.8. Peptides
2.9. Western Blotting
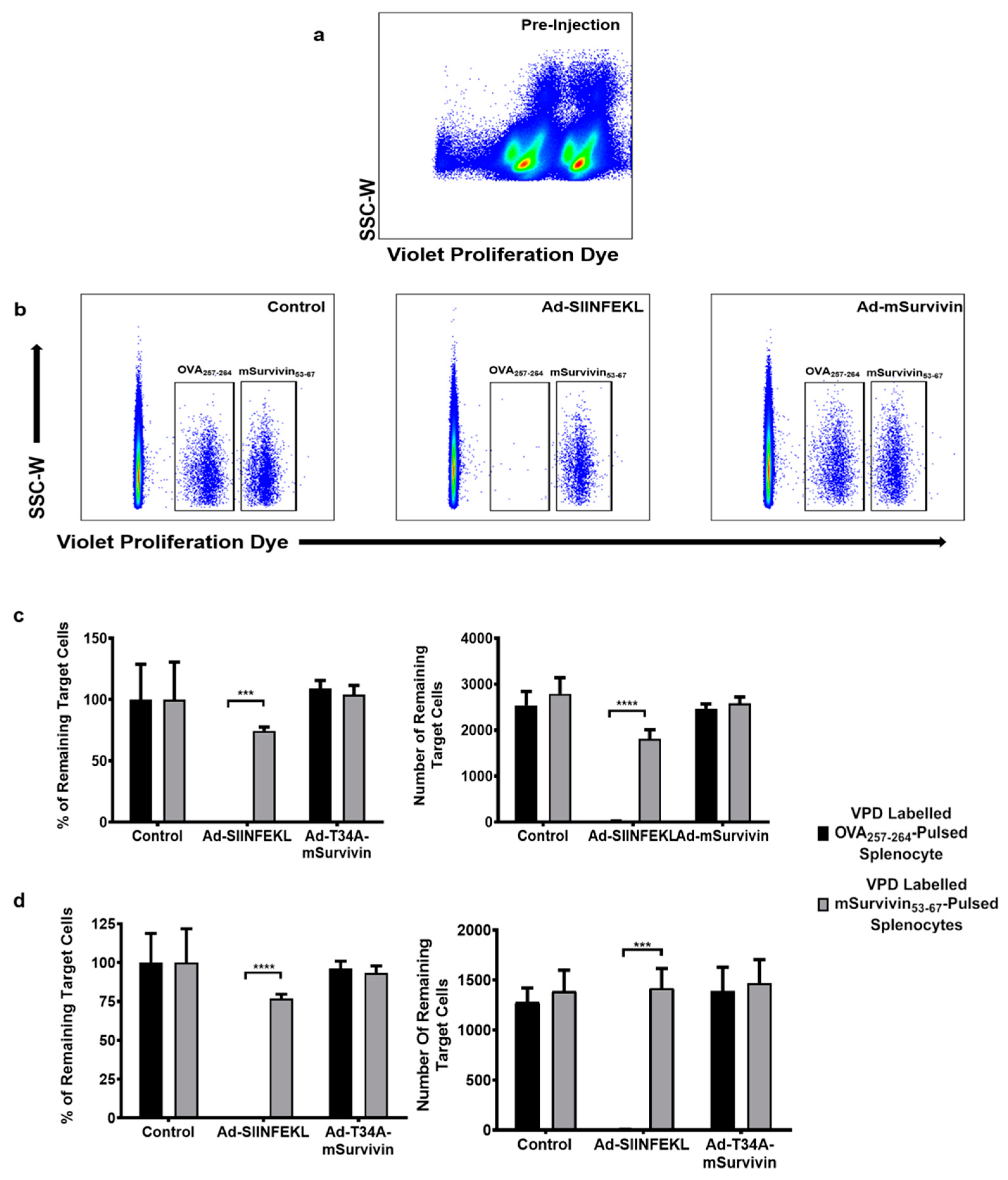
2.10. In Vivo Cytotoxicity Assay
2.11. In Vitro Infection of Antigen-Presenting Cells (APCs) with Adenovirus
2.12. In Vivo Immunological Checkpoint Inhibition with Anti-Cytotoxic T Lymphocyte-Associated Protein 4 (CTLA4)
2.13. Statistics
3. Results
3.1. Murine Survivin Was Successfully Tagged, Mutated, Cloned into, and Then Expressed by Adenovirus and Maraba Virus Vectors
3.2. Vaccination against Murine Survivin Was Unable to Induce a Survivin-Specific Cytotoxic T Cell Response
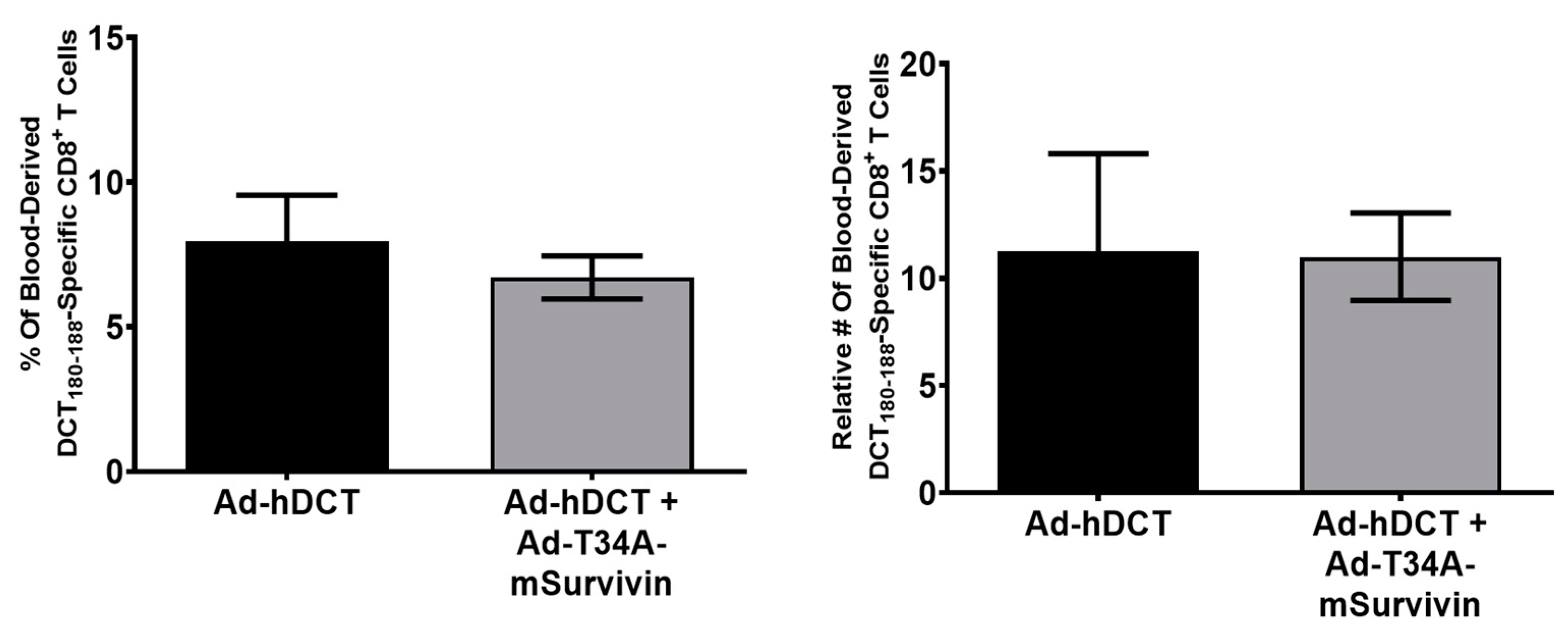
3.3. Co-Vaccination with Ad-T34A-mSurvivin and Ad-DCT Did Not Dampen Nor Eliminate DCT-Specific T Cell Responses
3.4. Ad-T34A-mSurvivin Did Not Significantly Impact the Viability of Antigen-Presenting Cells
3.5. Antibody-Mediated Blockade of CTLA4 Prior to Priming with Ad-T34A-mSurvivin and Boosting with MG1-T34A-mSurvivin Did Not Unveil Survivin-Specific T Cell Responses
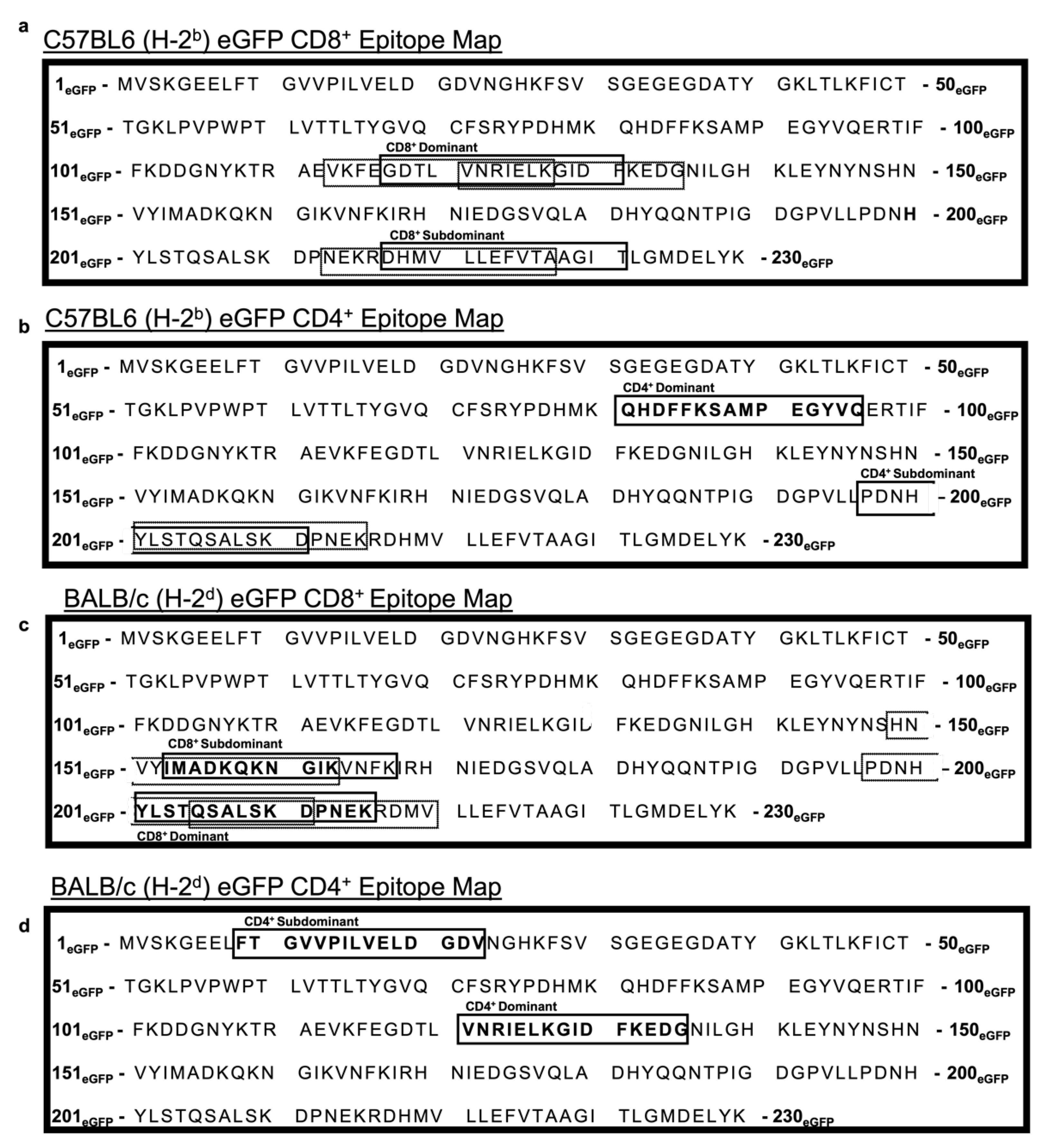
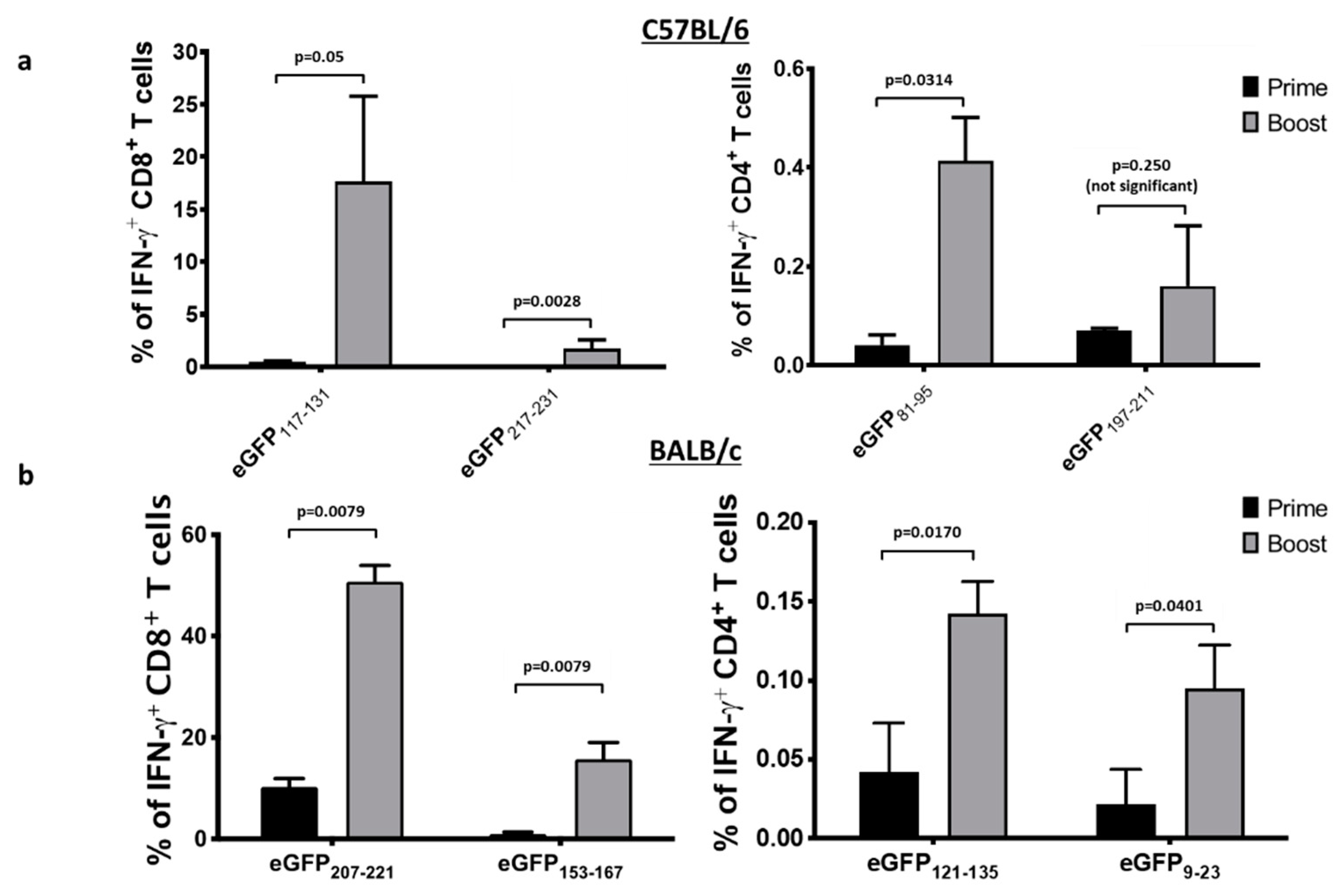
3.6. An Ad Prime Followed by Boosting with VSV Facilitated the Discovery of Novel eGFP Epitopes in C57BL/6 and BALB/c Mice
3.7. An Ad Prime Followed by Boosting with VSV Led to Massive T Cell Responses to eGFP
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- O’Donnell, J.S.; Teng, M.W.L.; Smyth, M.J. Cancer immunoediting and resistance to T cell-based immunotherapy. Nat. Rev. Clin. Oncol. 2019, 16, 151–167. [Google Scholar] [CrossRef] [PubMed]
- Cheever, M.A.; Allison, J.P.; Ferris, A.S.; Finn, O.J.; Hastings, B.M.; Hecht, T.T.; Mellman, I.; Prindiville, S.A.; Viner, J.L.; Weiner, L.M.; et al. The prioritization of cancer antigens: A national cancer institute pilot project for the acceleration of translational research. Clin. Cancer Res. 2009, 15, 5323–5337. [Google Scholar] [CrossRef]
- Ambrosini, G.; Adida, C.; Altieri, D.C. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat. Med. 1997, 3, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.-C. The molecular mechanisms of chemoresistance in cancers. Oncotarget 2017, 8, 59950–59964. [Google Scholar] [CrossRef] [PubMed]
- Conway, E.M.; Zwerts, F.; Van Eygen, V.; DeVriese, A.; Nagai, N.; Luo, W.; Collen, D. Survivin-dependent angiogenesis in ischemic brain: Molecular mechanisms of hypoxia-induced up-regulation. Am. J. Pathol. 2003, 163, 935–946. [Google Scholar] [CrossRef]
- Levitsky, A.; Erlandsson, M.C.; van Vollenhoven, R.F.; Bokarewa, M.I. Serum survivin predicts responses to treatment in active rheumatoid arthritis: A post hoc analysis from the SWEFOT trial. BMC Med. 2015, 13, 247. [Google Scholar] [CrossRef] [PubMed]
- Altznauer, F.; Martinelli, S.; Yousefi, S.; Thürig, C.; Schmid, I.; Conway, E.M.; Schöni, M.H.; Vogt, P.; Mueller, C.; Fey, M.F.; et al. Inflammation-associated cell cycle-independent block of apoptosis by survivin in terminally differentiated neutrophils. J. Exp. Med. 2004, 199, 1343–1354. [Google Scholar] [CrossRef] [PubMed]
- Bridle, B.W.; Stephenson, K.B.; Boudreau, J.E.; Koshy, S.; Kazdhan, N.; Pullenayegum, E.; Brunellière, J.; Bramson, J.L.; Lichty, B.D.; Wan, Y. Potentiating Cancer Immunotherapy Using an Oncolytic Virus. Mol. Ther. 2010, 18, 1430–1439. [Google Scholar] [CrossRef]
- Miyazaki, A.; Kobayashi, J.; Torigoe, T.; Hirohashi, Y.; Yamamoto, T.; Yamaguchi, A.; Asanuma, H.; Takahashi, A.; Michifuri, Y.; Nakamori, K.; et al. Phase I clinical trial of survivin-derived peptide vaccine therapy for patients with advanced or recurrent oral cancer. Cancer Sci. 2011, 102, 324–329. [Google Scholar] [CrossRef]
- Becker, J.C.; Andersen, M.H.; Hofmeister-Müller, V.; Wobser, M.; Frey, L.; Sandig, C.; Walter, S.; Singh-Jasuja, H.; Kämpgen, E.; Opitz, A.; et al. Survivin-specific T-cell reactivity correlates with tumor response and patient survival: A phase-II peptide vaccination trial in metastatic melanoma. Cancer Immunol. Immunother. 2012, 61, 2091–2103. [Google Scholar] [CrossRef]
- Lennerz, V.; Gross, S.; Gallerani, E.; Sessa, C.; Mach, N.; Boehm, S.; Hess, D.; von Boehmer, L.; Knuth, A.; Ochsenbein, A.F.; et al. Immunologic response to the survivin-derived multi-epitope vaccine EMD640744 in patients with advanced solid tumors. Cancer Immunol. Immunother. 2014, 63, 381–394. [Google Scholar] [CrossRef]
- Hofmann, U.B.; Voigt, H.; Andersen, M.H.; Straten, P.T.; Becker, J.C.; Eggert, A.O. Identification and characterization of survivin-derived H-2Kb-restricted CTL epitopes. Eur. J. Immunol. 2009, 39, 1419–1424. [Google Scholar] [CrossRef] [PubMed]
- Ciesielski, M.J.; Kozbor, D.; Castanaro, C.A.; Barone, T.A.; Fenstermaker, R.A. Therapeutic effect of a T helper cell supported CTL response induced by a survivin peptide vaccine against murine cerebral glioma. Cancer Immunol. Immunother. 2008, 57, 1827–1835. [Google Scholar] [CrossRef]
- Muchmore, S.W.; Chen, J.; Jakob, C.; Zakula, D.; Matayoshi, E.D.; Wu, W.; Zhang, H.; Li, F.; Ng, S.C.; Altieri, D.C. Crystal structure and mutagenic analysis of the inhibitor-of-apoptosis protein survivin. Mol. Cell 2000, 6, 173–182. [Google Scholar] [CrossRef]
- Qiu, F.; Zhao, X. In vivo antitumor activity of liposome-plasmid DNA encoding mutant survivin-T34A in cervical cancer. Mol. Med. Rep. 2018, 18, 841–847. [Google Scholar] [CrossRef] [PubMed]
- Mesri, M.; Wall, N.R.; Li, J.; Kim, R.W.; Altieri, D.C. Cancer gene therapy using a survivin mutant adenovirus. J. Clin. Investig. 2001, 108, 981–990. [Google Scholar] [CrossRef]
- Bridle, B.W.; Nguyen, A.; Salem, O.; Zhang, L.; Koshy, S.; Clouthier, D.; Chen, L.; Pol, J.; Swift, S.L.; Bowdish, D.M.E.; et al. Privileged Antigen Presentation in Splenic B Cell Follicles Maximizes T Cell Responses in Prime-Boost Vaccination. J. Immunol. 2016, 196, 4587–4595. [Google Scholar] [CrossRef] [PubMed]
- Abbink, P.; Lemckert, A.A.C.; Ewald, B.A.; Lynch, D.M.; Denholtz, M.; Smits, S.; Holterman, L.; Damen, I.; Vogels, R.; Thorner, A.R.; et al. Comparative seroprevalence and immunogenicity of six rare serotype recombinant adenovirus vaccine vectors from subgroups B and D. J. Virol. 2007, 81, 4654–4663. [Google Scholar] [CrossRef]
- Pol, J.G.; Zhang, L.; Bridle, B.W.; Stephenson, K.B.; Rességuier, J.; Hanson, S.; Chen, L.; Kazdhan, N.; Bramson, J.L.; Stojdl, D.F.; et al. Maraba virus as a potent oncolytic vaccine vector. Mol. Ther. 2014, 22, 420–429. [Google Scholar] [CrossRef] [PubMed]
- Brun, J.; McManus, D.; Lefebvre, C.; Hu, K.; Falls, T.; Atkins, H.; Bell, J.C.; McCart, J.A.; Mahoney, D.; Stojdl, D.F. Identification of genetically modified Maraba virus as an oncolytic rhabdovirus. Mol. Ther. 2010, 18, 1440–1449. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, J.; Takaki, S.; Araki, K.; Tashiro, F.; Tominaga, A.; Takatsu, K.; Yamamura, K. Expression vector system based on the chicken beta-actin promoter directs efficient production of interleukin-5. Gene 1989, 79, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Bridle, B.W.; Boudreau, J.E.; Lichty, B.D.; Brunellière, J.; Stephenson, K.; Koshy, S.; Bramson, J.L.; Wan, Y. Vesicular stomatitis virus as a novel cancer vaccine vector to prime antitumor immunity amenable to rapid boosting with adenovirus. Mol. Ther. 2009, 17, 1814–1821. [Google Scholar] [CrossRef] [PubMed]
- Bridle, B.W.; Clouthier, D.; Zhang, L.; Pol, J.; Chen, L.; Lichty, B.D.; Bramson, J.L.; Wan, Y. Oncolytic vesicular stomatitis virus quantitatively and qualitatively improves primary CD8+ T-cell responses to anticancer vaccines. Oncoimmunology 2013, 2, e26013. [Google Scholar] [CrossRef] [PubMed]
- Lane, C.; Leitch, J.; Tan, X.; Hadjati, J.; Bramson, J.L.; Wan, Y. Vaccination-induced autoimmune vitiligo is a consequence of secondary trauma to the skin. Cancer Res. 2004, 64, 1509–1514. [Google Scholar] [CrossRef] [PubMed]
- Mould, R.C.; AuYeung, A.W.K.; van Vloten, J.P.; Susta, L.; Mutsaers, A.J.; Petrik, J.J.; Wood, G.A.; Wootton, S.K.; Karimi, K.; Bridle, B.W. Enhancing Immune Responses to Cancer Vaccines Using Multi-Site Injections. Sci. Rep. 2017, 7, 8322. [Google Scholar] [CrossRef]
- Ciesielski, M.J.; Ahluwalia, M.S.; Munich, S.A.; Orton, M.; Barone, T.; Chanan-Khan, A.; Fenstermaker, R.A. Antitumor cytotoxic T-cell response induced by a survivin peptide mimic. Cancer Immunol. Immunother. 2010, 59, 1211–1221. [Google Scholar] [CrossRef] [PubMed]
- Helft, J.; Böttcher, J.; Chakravarty, P.; Zelenay, S.; Huotari, J.; Schraml, B.U.; Goubau, D.; Reis e Sousa, C. GM-CSF Mouse Bone Marrow Cultures Comprise a Heterogeneous Population of CD11c+MHCII+ Macrophages and Dendritic Cells. Immunity 2015, 42, 1197–1211. [Google Scholar] [CrossRef]
- Qin, J.Y.; Zhang, L.; Clift, K.L.; Hulur, I.; Xiang, A.P.; Ren, B.Z.; Lahn, B.T. Systematic comparison of constitutive promoters and the doxycycline-inducible promoter. PLoS ONE 2010, 5, e10611. [Google Scholar] [CrossRef]
- Aspe, J.R.; Wall, N.R. Survivin-T34A: Molecular mechanism and therapeutic potential. OncoTargets Ther. 2010, 3, 247–254. [Google Scholar] [CrossRef]
- Adida, C.; Crotty, P.L.; McGrath, J.; Berrebi, D.; Diebold, J.; Altieri, D.C. Developmentally regulated expression of the novel cancer anti-apoptosis gene survivin in human and mouse differentiation. Am. J. Pathol. 1998, 152, 43–49. [Google Scholar]
- Leisegang, M.; Wilde, S.; Spranger, S.; Milosevic, S.; Frankenberger, B.; Uckert, W.; Schendel, D.J. MHC-restricted fratricide of human lymphocytes expressing survivin-specific transgenic T cell receptors. J. Clin. Investig. 2010, 120, 3869–3877. [Google Scholar] [CrossRef]
- Kornacker, M.; Verneris, M.R.; Kornacker, B.; Scheffold, C.; Negrin, R.S. Survivin expression correlates with apoptosis resistance after lymphocyte activation and is found preferentially in memory T cells. Immunol. Lett. 2001, 76, 169–173. [Google Scholar] [CrossRef]
- Krummel, M.F.; Allison, J.P. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J. Exp. Med. 1995, 182, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Grosso, J.F.; Jure-Kunkel, M.N. CTLA-4 blockade in tumor models: An overview of preclinical and translational research. Cancer Immun. 2013, 13, 1–14. [Google Scholar]
- Cobleigh, M.A.; Bradfield, C.; Liu, Y.; Mehta, A.; Robek, M.D. The Immune Response to a Vesicular Stomatitis Virus Vaccine Vector Is Independent of Particulate Antigen Secretion and Protein Turnover Rate. J. Virol. 2012, 86, 4253–4261. [Google Scholar] [CrossRef]
- Fukuda, S.; Pelus, L.M. Survivin, a cancer target with an emerging role in normal adult tissues. Mol. Cancer Ther. 2006, 5, 1087–1098. [Google Scholar] [CrossRef]
- Gerdemann, U.; Katari, U.; Christin, A.S.; Cruz, C.R.; Tripic, T.; Rousseau, A.; Gottschalk, S.M.; Savoldo, B.; Vera, J.F.; Heslop, H.E.; et al. Cytotoxic T lymphocytes simultaneously targeting multiple tumor-associated antigens to treat EBV negative lymphoma. Mol. Ther. 2011, 19, 2258–2268. [Google Scholar] [CrossRef]
- Fenstermaker, R.A.; Ciesielski, M.J.; Qiu, J.; Yang, N.; Frank, C.L.; Lee, K.P.; Mechtler, L.R.; Belal, A.; Ahluwalia, M.S.; Hutson, A.D. Clinical study of a survivin long peptide vaccine (SurVaxM) in patients with recurrent malignant glioma. Cancer Immunol. Immunother. 2016, 65, 1339–1352. [Google Scholar] [CrossRef]
- Berinstein, N.L.; Karkada, M.; Oza, A.M.; Odunsi, K.; Villella, J.A.; Nemunaitis, J.J.; Morse, M.A.; Pejovic, T.; Bentley, J.; Buyse, M.; et al. Survivin-targeted immunotherapy drives robust polyfunctional T cell generation and differentiation in advanced ovarian cancer patients. Oncoimmunology 2015, 4, e1026529. [Google Scholar] [CrossRef] [PubMed]
- Skelton, D.; Satake, N.; Kohn, D. The enhanced green fluorescent protein (eGFP) is minimally immunogenic in C57BL/6 mice. Gene Ther. 2001, 8, 1813–1814. [Google Scholar] [CrossRef] [PubMed]
- Ansari, A.M.; Ahmed, A.K.; Matsangos, A.E.; Lay, F.; Born, L.J.; Marti, G.; Harmon, J.W.; Sun, Z. Cellular GFP Toxicity and Immunogenicity: Potential Confounders in in Vivo Cell Tracking Experiments. Stem Cell Rev. 2016, 12, 553–559. [Google Scholar] [CrossRef]
- Pol, J.G.; Atherton, M.J.; Bridle, B.W.; Stephenson, K.B.; Le Boeuf, F.; Hummel, J.L.; Martin, C.G.; Pomoransky, J.; Breitbach, C.J.; Diallo, J.S.; et al. Development and applications of oncolytic Maraba virus vaccines. Oncolytic Virother. 2018, 7, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Han, W.G.H.; Unger, W.W.J.; Wauben, M.H.M. Identification of the immunodominant CTL epitope of EGFP in C57BL/6 mice. Gene Ther. 2008, 15, 700–701. [Google Scholar] [CrossRef] [PubMed]
- King, C.; Garza, E.N.; Mazor, R.; Linehan, J.L.; Pastan, I.; Pepper, M.; Baker, D. Removing T-cell epitopes with computational protein design. Proc. Natl. Acad. Sci. USA 2014, 111, 8577–8582. [Google Scholar] [CrossRef]
- Gambotto, A.; Dworacki, G.; Cicinnati, V.; Kenniston, T.; Steitz, J.; Tüting, T.; Robbins, P.D.; DeLeo, A.B. Immunogenicity of enhanced green fluorescent protein (EGFP) in BALB/c mice: Identification of an H2-Kd-restricted CTL epitope. Gene Ther. 2000, 7, 2036–2040. [Google Scholar] [CrossRef] [PubMed]
- Altieri, D.C. Validating survivin as a cancer therapeutic target. Nat. Rev. Cancer 2003, 3, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Alexander-Miller, M.A.; Leggatt, G.R.; Sarin, A.; Berzofsky, J.A. Role of antigen, CD8, and cytotoxic T lymphocyte (CTL) avidity in high dose antigen induction of apoptosis of effector CTL. J. Exp. Med. 1996, 184, 485–492. [Google Scholar] [CrossRef]
- Greenwald, R.J.; Boussiotis, V.A.; Lorsbach, R.B.; Abbas, A.K.; Sharpe, A.H. CTLA-4 regulates induction of anergy in vivo. Immunity 2001, 14, 145–155. [Google Scholar] [CrossRef]
- Onodi, F.; Maherzi-Mechalikh, C.; Mougel, A.; Hamouda, N.B.; Taboas, C.; Gueugnon, F.; Tran, T.; Nozach, H.; Marcon, E.; Gey, A.; et al. High therapeutic efficacy of a new survivin LSP-cancer vaccine containing CD4+ and CD8+ T-cell epitopes. Front. Oncol. 2018, 8, 517. [Google Scholar] [CrossRef]
- Lenardo, M.; Chan, K.M.; Hornung, F.; McFarland, H.; Siegel, R.; Wang, J.; Zheng, L. Mature T lymphocyte apoptosis--immune regulation in a dynamic and unpredictable antigenic environment. Annu. Rev. Immunol. 1999, 17, 221–253. [Google Scholar] [CrossRef] [PubMed]
- Paris, S.; Sesboüé, R. Metastasis models: The green flourescent revolution? Carcinogenesis 2004, 25, 2285–2292. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Makki, A.; Weidt, G.; Blachere, N.E.; Lefrançois, L.; Srivastava, P.K. Immunization against a dominant tumor antigen abrogates immunogenicity of the tumor. Cancer Immun. Arch. 2002, 2, 4. [Google Scholar]
- Jackaman, C.; Gardner, J.K.; Tomay, F.; Spowart, J.; Crabb, H.; Dye, D.E.; Fox, S.; Proksch, S.; Metharom, P.; Dhaliwal, S.S.; et al. CD8+ cytotoxic T cell responses to dominant tumor-associated antigens are profoundly weakened by aging yet subdominant responses retain functionality and expand in response to chemotherapy. Oncoimmunology 2019, 8, e1564452. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mould, R.C.; van Vloten, J.P.; AuYeung, A.W.K.; Walsh, S.R.; de Jong, J.; Susta, L.; Mutsaers, A.J.; Petrik, J.J.; Wood, G.A.; Wootton, S.K.; et al. Using a Prime-Boost Vaccination Strategy That Proved Effective for High Resolution Epitope Mapping to Characterize the Elusive Immunogenicity of Survivin. Cancers 2021, 13, 6270. https://doi.org/10.3390/cancers13246270
Mould RC, van Vloten JP, AuYeung AWK, Walsh SR, de Jong J, Susta L, Mutsaers AJ, Petrik JJ, Wood GA, Wootton SK, et al. Using a Prime-Boost Vaccination Strategy That Proved Effective for High Resolution Epitope Mapping to Characterize the Elusive Immunogenicity of Survivin. Cancers. 2021; 13(24):6270. https://doi.org/10.3390/cancers13246270
Chicago/Turabian StyleMould, Robert C., Jacob P. van Vloten, Amanda W. K. AuYeung, Scott R. Walsh, Jondavid de Jong, Leonardo Susta, Anthony J. Mutsaers, James J. Petrik, Geoffrey A. Wood, Sarah K. Wootton, and et al. 2021. "Using a Prime-Boost Vaccination Strategy That Proved Effective for High Resolution Epitope Mapping to Characterize the Elusive Immunogenicity of Survivin" Cancers 13, no. 24: 6270. https://doi.org/10.3390/cancers13246270
APA StyleMould, R. C., van Vloten, J. P., AuYeung, A. W. K., Walsh, S. R., de Jong, J., Susta, L., Mutsaers, A. J., Petrik, J. J., Wood, G. A., Wootton, S. K., Karimi, K., & Bridle, B. W. (2021). Using a Prime-Boost Vaccination Strategy That Proved Effective for High Resolution Epitope Mapping to Characterize the Elusive Immunogenicity of Survivin. Cancers, 13(24), 6270. https://doi.org/10.3390/cancers13246270








