Deep-Learning Assessed Muscular Hypodensity Independently Predicts Mortality in DLBCL Patients Younger Than 60 Years
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients Selection
2.2. Ground Truth Generation
2.3. DLASA Trainings and Validation
2.4. Evaluation of Body Composition in a Prospective Cohort Using the Algorithm
2.5. Outcomes and Statistics
3. Results
3.1. Segmentation Algorithm Training and Validation
3.2. Anthropometric Evaluation in the Exploratory Set
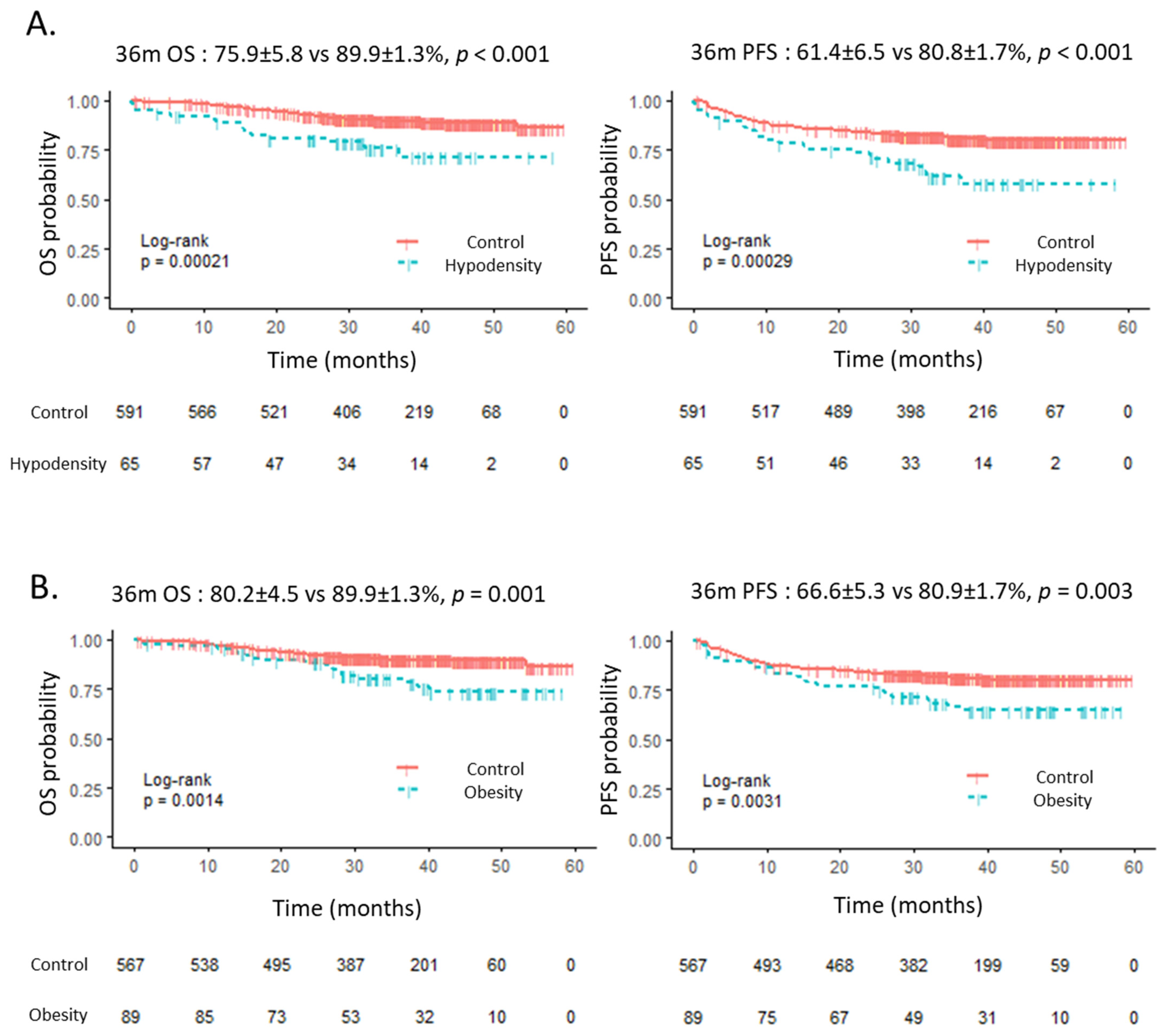
3.3. Outcome According to Anthropometric Evaluation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tisdale, M.J. Cachexia in cancer patients. Nat. Rev. Cancer 2002, 2, 862–871. [Google Scholar] [CrossRef] [PubMed]
- Dewys, W.D.; Begg, C.; Lavin, P.T.; Band, P.R.; Bennett, J.M.; Bertino, J.R.; Cohen, M.H.; Douglass, H.O.; Engstrom, P.F.; Ezdinli, E.Z.; et al. Prognostic effect of weight loss prior to chemotherapy in cancer patients. Eastern Cooperative Oncology Group. Am. J. Med. 1980, 69, 491–497. [Google Scholar] [CrossRef]
- Fearon, K.; Strasser, F.; Anker, S.D.; Bosaeus, I.; Bruera, E.; Fainsinger, R.L.; Jatoi, A.; Loprinzi, C.; MacDonald, N.; Mantovani, G.; et al. Definition and classification of cancer cachexia: An international consensus. Lancet Oncol. 2011, 12, 489–495. [Google Scholar] [CrossRef]
- Prado, C.M.M.; Birdsell, L.A.; Baracos, V.E. The emerging role of computerized tomography in assessing cancer cachexia. Curr. Opin. Support. Palliat. Care 2009, 3, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Mourtzakis, M.; Prado, C.M.M.; Lieffers, J.R.; Reiman, T.; McCargar, L.J.; Baracos, V.E. A practical and precise approach to quantification of body composition in cancer patients using computed tomography images acquired during routine care. Appl. Physiol. Nutr. Metab. 2008, 33, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Aubrey, J.; Esfandiari, N.; Baracos, V.E.; Buteau, F.A.; Frenette, J.; Putman, C.T.; Mazurak, V.C. Measurement of skeletal muscle radiation attenuation and basis of its biological variation. Acta Physiol. Oxf. Engl. 2014, 210, 489–497. [Google Scholar] [CrossRef] [Green Version]
- van Vugt, J.L.A.; Levolger, S.; Gharbharan, A.; Koek, M.; Niessen, W.J.; Burger, J.W.A.; Willemsen, S.P.; de Bruin, R.W.F.; IJzermans, J.N.M. A comparative study of software programmes for cross-sectional skeletal muscle and adipose tissue measurements on abdominal computed tomography scans of rectal cancer patients. J. Cachexia Sarcopenia Muscle 2017, 8, 285–297. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Lect. Notes Comput. Sci. 2015, 9351, 234–241. [Google Scholar]
- Guo, Z.; Guo, N.; Gong, K.; Zhong, S.; Li, Q. Gross tumor volume segmentation for head and neck cancer radiotherapy using deep dense multi-modality network. Phys. Med. Biol. 2019, 64, 205015. [Google Scholar] [CrossRef]
- Li, J.; Sarma, K.V.; Ho, K.C.; Gertych, A.; Knudsen, B.S.; Arnold, C.W. A Multi-scale U-Net for Semantic Segmentation of Histological Images from Radical Prostatectomies. AMIA Annu. Symp. Proc. 2017, 2017, 1140–1148. [Google Scholar]
- Norman, B.; Pedoia, V.; Majumdar, S. Use of 2D U-Net Convolutional Neural Networks for Automated Cartilage and Meniscus Segmentation of Knee MR Imaging Data to Determine Relaxometry and Morphometry. Radiology 2018, 288, 177–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanc-Durand, P.; Jégou, S.; Kanoun, S.; Berriolo-Riedinger, A.; Bodet-Milin, C.; Kraeber-Bodéré, F.; Carlier, T.; Le Gouill, S.; Casasnovas, R.-O.; Meignan, M.; et al. Fully automatic segmentation of diffuse large B cell lymphoma lesions on 3D FDG-PET/CT for total metabolic tumour volume prediction using a convolutional neural network. Eur. J. Nucl. Med. Mol. Imaging 2020, 48, 1362–1370. [Google Scholar] [CrossRef] [PubMed]
- Burns, J.E.; Yao, J.; Chalhoub, D.; Chen, J.J.; Summers, R.M. A Machine Learning Algorithm to Estimate Sarcopenia on Abdominal CT. Acad. Radiol. 2020, 27, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Dabiri, S.; Popuri, K.; Cespedes Feliciano, E.M.; Caan, B.J.; Baracos, V.E.; Beg, M.F. Muscle segmentation in axial computed tomography (CT) images at the lumbar (L3) and thoracic (T4) levels for body composition analysis. Comput. Med. Imaging Graph. 2019, 75, 47–55. [Google Scholar] [CrossRef]
- Edwards, K.; Chhabra, A.; Dormer, J.; Jones, P.; Boutin, R.D.; Lenchik, L.; Fei, B. Abdominal muscle segmentation from CT using a convolutional neural network. Proc. SPIE Int. Soc. Opt. Eng. 2020, 11317, 113170L. [Google Scholar]
- Lee, H.; Troschel, F.M.; Tajmir, S.; Fuchs, G.; Mario, J.; Fintelmann, F.J.; Do, S. Pixel-Level Deep Segmentation: Artificial Intelligence Quantifies Muscle on Computed Tomography for Body Morphometric Analysis. J. Digit. Imaging 2017, 30, 487–498. [Google Scholar] [CrossRef]
- Lee, J.S.; Kim, Y.S.; Kim, E.Y.; Jin, W. Prognostic significance of CT-determined sarcopenia in patients with advanced gastric cancer. PLoS ONE 2018, 13, e0202700. [Google Scholar] [CrossRef] [Green Version]
- Cho, Y.; Kim, J.W.; Keum, K.C.; Lee, C.G.; Jeung, H.C.; Lee, I.J. Prognostic Significance of Sarcopenia With Inflammation in Patients With Head and Neck Cancer Who Underwent Definitive Chemoradiotherapy. Front. Oncol. 2018, 8, 457. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.Y.; Lee, H.Y.; Kim, Y.S.; Park, I.; Ahn, H.K.; Cho, E.K.; Jeong, Y.M.; Kim, J.H. Prognostic significance of cachexia score assessed by CT in male patients with small cell lung cancer. Eur. J. Cancer Care 2018, 27, e12695. [Google Scholar] [CrossRef]
- Daly, L.E.; Power, D.G.; O’Reilly, Á.; Donnellan, P.; Cushen, S.J.; O’Sullivan, K.; Twomey, M.; Woodlock, D.P.; Redmond, H.P.; Ryan, A.M. The impact of body composition parameters on ipilimumab toxicity and survival in patients with metastatic melanoma. Br. J. Cancer 2017, 116, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.; Birdsell, L.; Macdonald, N.; Reiman, T.; Clandinin, M.T.; McCargar, L.J.; Murphy, R.; Ghosh, S.; Sawyer, M.B.; Baracos, V.E. Cancer cachexia in the age of obesity: Skeletal muscle depletion is a powerful prognostic factor, independent of body mass index. J. Clin. Oncol. 2013, 31, 1539–1547. [Google Scholar] [CrossRef] [PubMed]
- Lanic, H.; Kraut-Tauzia, J.; Modzelewski, R.; Clatot, F.; Mareschal, S.; Picquenot, J.M.; Stamatoullas, A.; Leprêtre, S.; Tilly, H.; Jardin, F. Sarcopenia is an independent prognostic factor in elderly patients with diffuse large B-cell lymphoma treated with immunochemotherapy. Leuk. Lymphoma 2014, 55, 817–823. [Google Scholar] [CrossRef] [PubMed]
- Jabbour, J.; Manana, B.; Zahreddine, A.; Saade, C.; Charafeddine, M.; Bazarbachi, A.; Blaise, D.; El-Cheikh, J. Sarcopenic obesity derived from PET/CT predicts mortality in lymphoma patients undergoing hematopoietic stem cell transplantation. Curr. Res. Transl. Med. 2019, 67, 93–99. [Google Scholar] [CrossRef]
- Chu, M.P.; Lieffers, J.; Ghosh, S.; Belch, A.; Chua, N.S.; Fontaine, A.; Sangha, R.; Turner, R.A.; Baracos, V.E.; Sawyer, M.B. Skeletal muscle density is an independent predictor of diffuse large B-cell lymphoma outcomes treated with rituximab-based chemoimmunotherapy. J. Cachexia Sarcopenia Muscle 2017, 8, 298–304. [Google Scholar] [CrossRef]
- Le Gouill, S.; Ghesquieres, H.; Obéric, L.; Morschhauser, F.; Tilly, H.; Ribrag, V.; Lamy, T.; Thieblemont, C.; Maisonneuve, H.; Gressin, R.; et al. Obinutuzumab versus Rituximab in young patients with advanced DLBCL, a PET-guided and randomized phase 3 study by LYSA. Blood 2020, 137, 2307–2320. [Google Scholar] [CrossRef]
- Le Gouill, S.; Thieblemont, C.; Oberic, L.; Moreau, A.; Bouabdallah, K.; Dartigeas, C.; Damaj, G.; Gastinne, T.; Ribrag, V.; Feugier, P.; et al. Rituximab after Autologous Stem-Cell Transplantation in Mantle-Cell Lymphoma. N. Engl. J. Med. 2017, 377, 1250–1260. [Google Scholar] [CrossRef]
- Dice, L.R. Measures of the Amount of Ecologic Association Between Species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Cheson, B.D.; Pfistner, B.; Juweid, M.E.; Gascoyne, R.D.; Specht, L.; Horning, S.J.; Coiffier, B.; Fisher, R.I.; Hagenbeek, A.; Zucca, E.; et al. Revised response criteria for malignant lymphoma. J. Clin. Oncol. 2007, 25, 579–586. [Google Scholar] [CrossRef]
- Belharbi, S.; Chatelain, C.; Hérault, R.; Adam, S.; Thureau, S.; Chastan, M.; Modzelewski, R. Spotting L3 slice in CT scans using deep convolutional network and transfer learning. Comput. Biol. Med. 2017, 87, 95–103. [Google Scholar] [CrossRef]
- Morsbach, F.; Zhang, Y.-H.; Martin, L.; Lindqvist, C.; Brismar, T. Body composition evaluation with computed tomography: Contrast media and slice thickness cause methodological errors. Nutrition 2019, 59, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, G.; Chretien, Y.R.; Mario, J.; Do, S.; Eikermann, M.; Liu, B.; Yang, K.; Fintelmann, F.J. Quantifying the effect of slice thickness, intravenous contrast and tube current on muscle segmentation: Implications for body composition analysis. Eur. Radiol. 2018, 28, 2455–2463. [Google Scholar] [CrossRef] [PubMed]
- Sabel, M.S.; Lee, J.; Cai, S.; Englesbe, M.J.; Holcombe, S.; Wang, S. Sarcopenia as a prognostic factor among patients with stage III melanoma. Ann. Surg. Oncol. 2011, 18, 3579–3585. [Google Scholar] [CrossRef] [PubMed]
- Antoun, S.; Lanoy, E.; Iacovelli, R.; Albiges-Sauvin, L.; Loriot, Y.; Merad-Taoufik, M.; Fizazi, K.; di Palma, M.; Baracos, V.E.; Escudier, B. Skeletal muscle density predicts prognosis in patients with metastatic renal cell carcinoma treated with targeted therapies. Cancer 2013, 119, 3377–3384. [Google Scholar] [CrossRef]
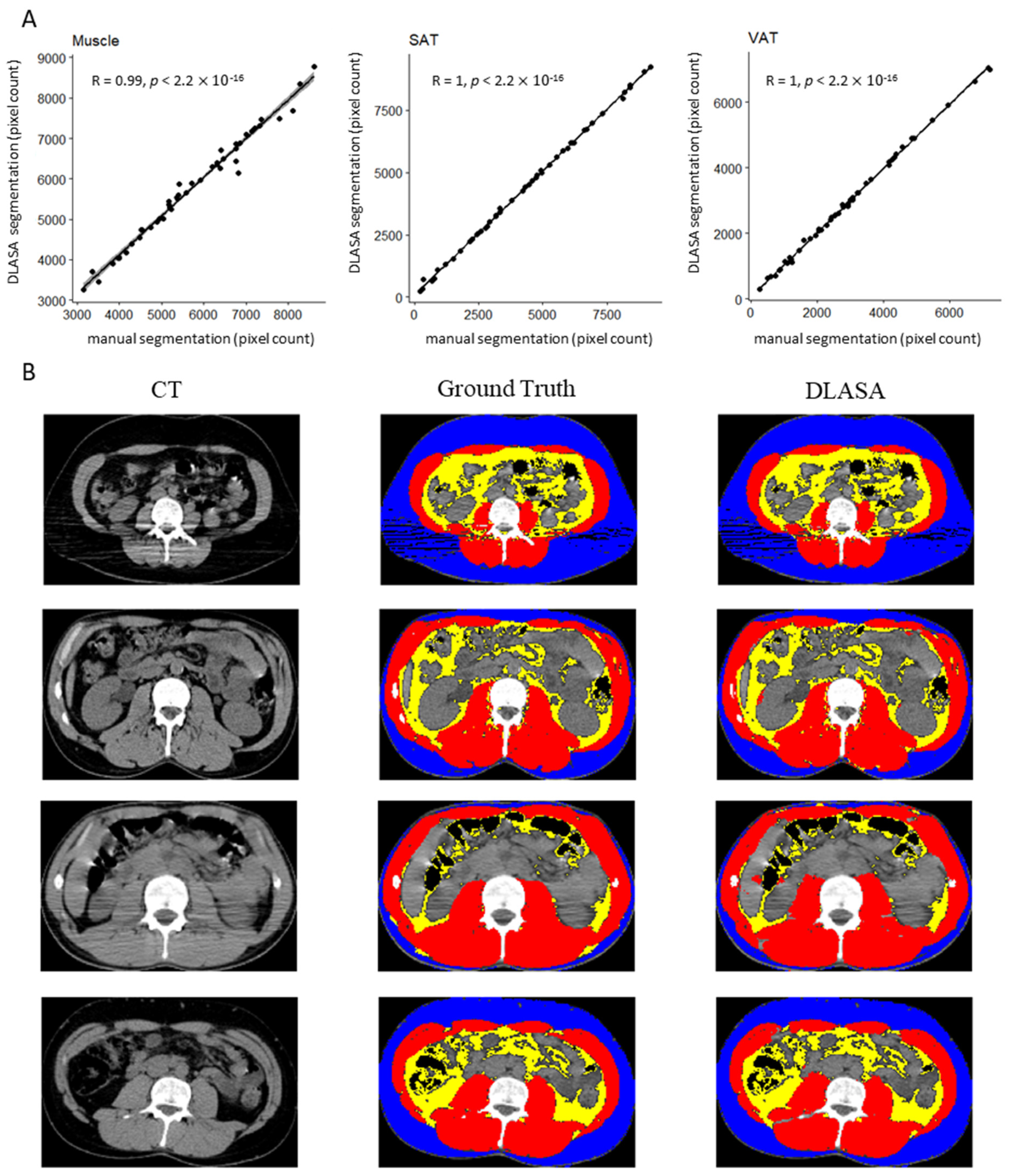
| Variable | Ground Truth | DLASA | p Value |
|---|---|---|---|
| SMI (cm2/m2), median (IQR) | 48.6 (44.5–56.3) | 49.1 (44.8–57.4) | 0.65 |
| Abs difference (mean ± SD) | 1.18 ± 1.2 | ||
| MA (HU), median (IQR) | 38.9 (33.8–43.0) | 38.6 (33.4–42.5) | 0.80 |
| Abs difference, HU (mean ± SD) | 0.55 ± 0.4 | ||
| SAT (cm2/m2), median (IQR) | 35.2 (21.5–61.5) | 35.3 (22.4–62.0) | 0.84 |
| Abs difference (mean ± SD) | 0.71 ± 0.6 | ||
| VAT (cm2/m2), median (IQR) | 22.9 (12.0–40.5) | 22.9 (12.3–40.4) | 0.97 |
| Abs difference (mean ± SD) | 0.47 ± 0.6 | ||
| Dice (mean ± SD) | |||
| muscle | 0.97 ± 0.03 | ||
| SAT | 0.97 ± 0.06 | ||
| VAT | 0.97 ± 0.03 | ||
| n = 656 | |||
|---|---|---|---|
| Male | Female | p | |
| n (%) | 367 (55.9) | 289 (44.1) | |
| Age (years) | 49 (40–56) | 47 (36–54) | 0.01 |
| BMI (kg/m2) | 24.6 (22.2–27.8) | 23.4 (20.8–27.3) | 0.02 |
| <18.5, n (%) | 11 (3.0) | 23 (8.0) | 0.01 |
| >30, n (%) | 45 (12.3) | 44 (15.2) | 0.41 |
| SMI (cm2/m2) | 55.5 (50.6–61.4) | 44.9 (40.9–48.1) | <10−15 |
| sarcopenia, n (%) | 179 (48.8) | 46 (15.9) | <10−15 |
| MA (HU) | 37.8 (32.6–43.6) | 34.2 (28.5–40.0) | <10−6 |
| muscular hypodensity, n (%) | 38 (10.3) | 27 (9.3) | 0.77 |
| aaIPI | |||
| low, n(%) | 1 (0.3) | 1 (0.3) | 1 |
| low-int, n(%) | 147 (40.1) | 130 (45.0) | 0.23 |
| high-int, n(%) | 173 (47.1) | 130 (45.0) | 0.64 |
| high, n(%) | 46 (12.5) | 28 (9.7) | 0.31 |
| Obese | Control | p | Sarcopenia | Control | p | Muscular Hypodensity | Control | p | ||
|---|---|---|---|---|---|---|---|---|---|---|
| PET 2 | Neg | 61 (70.1%) | 391 (70.8%) | 0.99 | 149 (69.3%) | 303 (71.5%) | 0.63 | 45 (75.0%) | 407 (70.3%) | 0.54 |
| Pos | 26 (29.9%) | 161 (29.2%) | 66 (30.7%) | 121 (28.5%) | 15 (25.0%) | 172 (29.7%) | ||||
| PET 4 | Neg | 66 (77.6%) | 446 (84.2%) | 0.18 | 171 (82.2%) | 341 (83.8%) | 0.70 | 48 (87.3%) | 464 (82.9%) | 0.52 |
| Pos | 19 (22.4%) | 84 (15.8%) | 37 (17.8%) | 66 (16.2%) | 7 (12.7%) | 96 (17.1%) |
| OS | PFS | |||||
|---|---|---|---|---|---|---|
| Variable | HR | 95% CI | p | HR | 95% CI | p |
| Obesity (BMI > 30 kg/m2) | 2.30 | 1.36–3.87 | 0.002 | 1.84 | 1.22–2.77 | 0.004 |
| Underweightness (BMI < 18.5 kg/m2) | 1.76 | 0.76–4.06 | 0.19 | 1.75 | 0.94–3.24 | 0.08 |
| Muscle hypodensity | 2.80 | 1.58–4.95 | <0.001 | 2.22 | 1.43–3.45 | <0.001 |
| Sarcopenia | 1.20 | 0.74–1.93 | 0.46 | 1.27 | 0.90–1.80 | 0.18 |
| aaIPI | 1.88 | 1.35–2.63 | <0.001 | 1.48 | 1.16–1.90 | 0.002 |
| age | 1.03 | 1.01–1.05 | 0.02 | 1.01 | 0.99–1.03 | 0.19 |
| Rituximab vs. Obinutuzumab | 1.08 | 0.68–1.73 | 0.74 | 0.99 | 0.71–1.40 | 0.96 |
| CHOP vs. ACVBP | 1.11 | 0.70–1.78 | 0.65 | 1.16 | 0.82–1.63 | 0.41 |
| OS | PFS | |||||
|---|---|---|---|---|---|---|
| Variable | HR | 95% CI | p | HR | 95% CI | p |
| Obesity | 2.18 | 1.18–4.06 | 0.01 | 1.81 | 1.11–2.94 | 0.02 |
| Muscle hypodensity | 2.22 | 1.04–4.70 | 0.04 | 2.06 | 1.16–3.67 | 0.01 |
| aaIPI | 1.73 | 1.23–2.44 | 0.002 | 1.40 | 1.09–1.80 | 0.009 |
| age | 1.02 | 0.99–1.04 | 0.14 | 1 | 0.99–1.02 | 0.72 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jullien, M.; Tessoulin, B.; Ghesquières, H.; Oberic, L.; Morschhauser, F.; Tilly, H.; Ribrag, V.; Lamy, T.; Thieblemont, C.; Villemagne, B.; et al. Deep-Learning Assessed Muscular Hypodensity Independently Predicts Mortality in DLBCL Patients Younger Than 60 Years. Cancers 2021, 13, 4503. https://doi.org/10.3390/cancers13184503
Jullien M, Tessoulin B, Ghesquières H, Oberic L, Morschhauser F, Tilly H, Ribrag V, Lamy T, Thieblemont C, Villemagne B, et al. Deep-Learning Assessed Muscular Hypodensity Independently Predicts Mortality in DLBCL Patients Younger Than 60 Years. Cancers. 2021; 13(18):4503. https://doi.org/10.3390/cancers13184503
Chicago/Turabian StyleJullien, Maxime, Benoit Tessoulin, Hervé Ghesquières, Lucie Oberic, Franck Morschhauser, Hervé Tilly, Vincent Ribrag, Thierry Lamy, Catherine Thieblemont, Bruno Villemagne, and et al. 2021. "Deep-Learning Assessed Muscular Hypodensity Independently Predicts Mortality in DLBCL Patients Younger Than 60 Years" Cancers 13, no. 18: 4503. https://doi.org/10.3390/cancers13184503
APA StyleJullien, M., Tessoulin, B., Ghesquières, H., Oberic, L., Morschhauser, F., Tilly, H., Ribrag, V., Lamy, T., Thieblemont, C., Villemagne, B., Gressin, R., Bouabdallah, K., Haioun, C., Damaj, G., Fornecker, L.-M., Schiano De Colella, J.-M., Feugier, P., Hermine, O., Cartron, G., ... Le Gouill, S. (2021). Deep-Learning Assessed Muscular Hypodensity Independently Predicts Mortality in DLBCL Patients Younger Than 60 Years. Cancers, 13(18), 4503. https://doi.org/10.3390/cancers13184503






