Lipid Droplet-Associated Factors, PNPLA3, TM6SF2, and HSD17B Proteins in Hepatopancreatobiliary Cancer
Abstract
:Simple Summary
Abstract
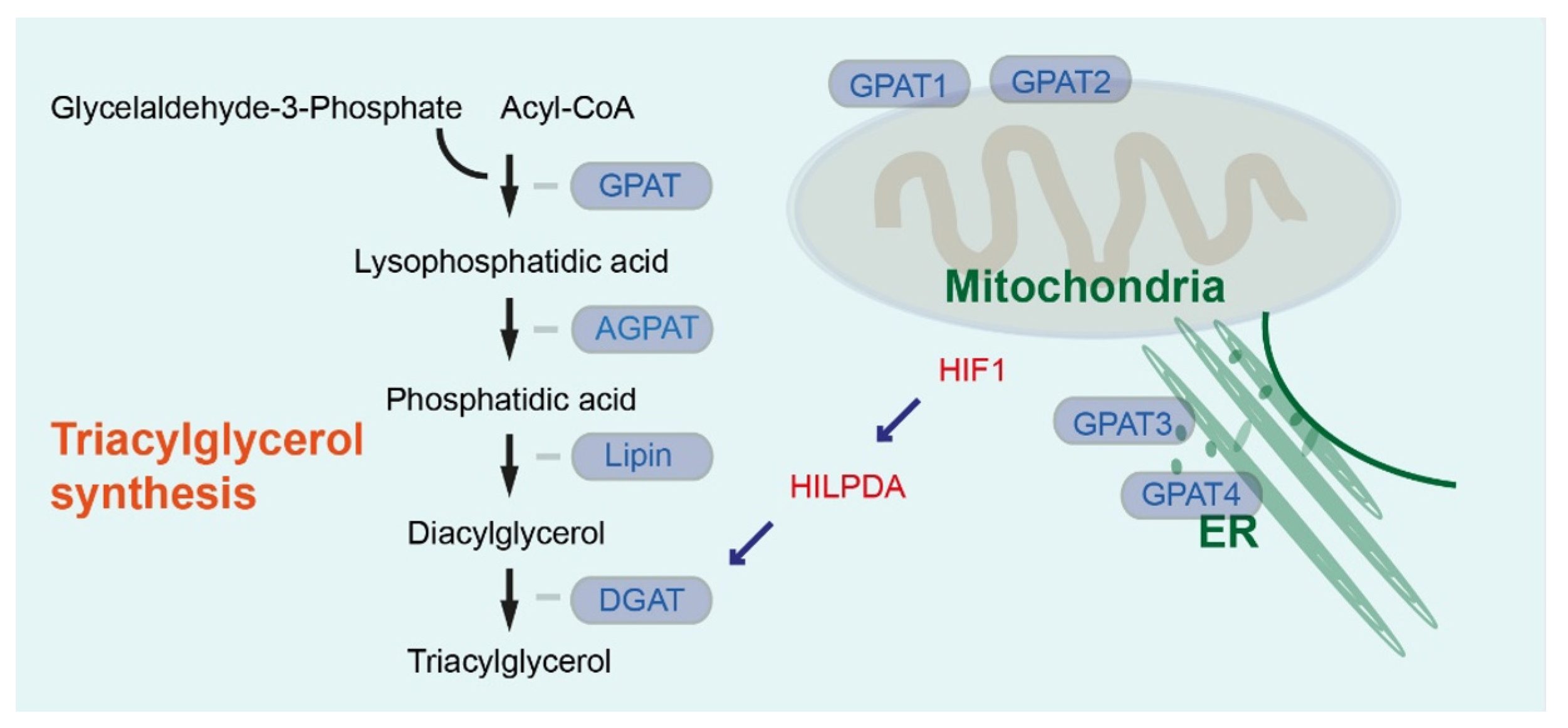
1. Lipid Synthesis and Lipid Droplets in Pancreatic and Hepatic Cancer
2. Role of Lipid Droplets and Lipid Droplet-Associated Factors in Hepatopancreatobiliary Cancer
3. PNPLA3 in Pancreatic and Hepatic Diseases
4. PNPLA3 in Stellate Cells and Cancer-Associated Fibroblasts
5. TM6SF2 Variant in NAFLD, Fibrosis, and Cancer
6. HSD17B11 and HSD17B13 in Pancreatic and Liver Cancer
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting Cancer Incidence and Deaths to 2030: The Unexpected Burden of Thyroid, Liver, and Pancreas Cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, M.; Mihaljevic, A.L.; Probst, P.; Heckler, M.; Klaiber, U.; Heger, U.; Büchler, M.W.; Hackert, T. Meta-analysis of recurrence pattern after resection for pancreatic cancer. Br. J. Surg. 2019, 106, 1590–1601. [Google Scholar] [CrossRef]
- Sunami, Y.; Rebelo, A.; Kleeff, J. Lipid Metabolism and Lipid Droplets in Pancreatic Cancer and Stellate Cells. Cancers 2017, 10, 3. [Google Scholar] [CrossRef] [Green Version]
- Marengo, A.; Rosso, C.; Bugianesi, E. Liver Cancer: Connections with Obesity, Fatty Liver, and Cirrhosis. Annu. Rev. Med. 2016, 67, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Baenke, F.; Peck, B.; Miess, H.; Schulze, A. Hooked on fat: The role of lipid synthesis in cancer metabolism and tumour development. Dis. Model. Mech. 2013, 6, 1353–1363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunami, Y. NASH, Fibrosis and Hepatocellular Carcinoma: Lipid Synthesis and Glutamine/Acetate Signaling. Int. J. Mol. Sci. 2020, 21, 6799. [Google Scholar] [CrossRef]
- Yu, J.; Loh, K.; Song, Z.-Y.; Yang, H.-Q.; Zhang, Y.; Lin, S. Update on glycerol-3-phosphate acyltransferases: The roles in the development of insulin resistance. Nutr. Diabetes 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Karasawa, K.; Tanigawa, K.; Harada, A.; Yamashita, A. Transcriptional Regulation of Acyl-CoA:Glycerol-sn-3-Phosphate Acyltransferases. Int. J. Mol. Sci. 2019, 20, 964. [Google Scholar] [CrossRef] [Green Version]
- Wilfling, F.; Wang, H.; Haas, J.; Krahmer, N.; Gould, T.J.; Uchida, A.; Cheng, J.-X.; Graham, M.; Christiano, R.; Fröhlich, F.; et al. Triacylglycerol Synthesis Enzymes Mediate Lipid Droplet Growth by Relocalizing from the ER to Lipid Droplets. Dev. Cell 2013, 24, 384–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellis, J.M.; Paul, D.S.; DePetrillo, M.A.; Singh, B.P.; Malarkey, D.E.; Coleman, R.A. Mice Deficient in Glycerol-3-Phosphate Acyltransferase-1 Have a Reduced Susceptibility to Liver Cancer. Toxicol. Pathol. 2012, 40, 513–521. [Google Scholar] [CrossRef] [Green Version]
- Pellon-Maison, M.; Montanaro, M.A.; Lacunza, E.; Garcia-Fabiani, M.B.; Soler-Gerino, M.C.; Cattaneo, E.R.; Quiroga, I.Y.; Abba, M.C.; Coleman, R.A.; Gonzalez-Baró, M.R. Glycerol-3-Phosphate Acyltranferase-2 Behaves as a Cancer Testis Gene and Promotes Growth and Tumorigenicity of the Breast Cancer MDA-MB-231 Cell Line. PLoS ONE 2014, 9, e100896. [Google Scholar] [CrossRef]
- Jiang, P.; Sun, W.; Shen, N.; Huang, X.; Fu, S. Identification of a metabolism-related gene expression prognostic model in endometrial carcinoma patients. BMC Cancer 2020, 20, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.K. Lysophospholipid acyltransferases: 1-acylglycerol-3-phosphate O-acyltransferases. From discovery to disease. Curr. Opin. Lipidol. 2012, 23, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Tauber, S.; Jais, A.; Jeitler, M.; Haider, S.; Husa, J.; Lindroos, J.; Knöfler, M.; Mayerhofer, M.; Pehamberger, H.; Wagner, O.; et al. Transcriptome analysis of human cancer reveals a functional role of Heme Oxygenase-1 in tumor cell adhesion. Mol. Cancer 2010, 9, 200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Triantafyllou, E.-A.; Georgatsou, E.; Mylonis, I.; Simos, G.; Paraskeva, E. Expression of AGPAT2, an enzyme involved in the glycerophospholipid/triacylglycerol biosynthesis pathway, is directly regulated by HIF-1 and promotes survival and etoposide resistance of cancer cells under hypoxia. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2018, 1863, 1142–1152. [Google Scholar] [CrossRef]
- Xu, W.-H.; Xu, Y.; Wang, J.; Wan, F.-N.; Wang, H.-K.; Cao, D.-L.; Shi, G.-H.; Qu, Y.-Y.; Zhang, H.-L.; Ye, D.-W. Prognostic value and immune infiltration of novel signatures in clear cell renal cell carcinoma microenvironment. Aging 2019, 11, 6999–7020. [Google Scholar] [CrossRef]
- Fan, S.-H.; Wang, Y.-Y.; Wu, Z.-Y.; Zhang, Z.-F.; Lu, J.; Li, M.-Q.; Shan, Q.; Wu, D.-M.; Sun, C.-H.; Hu, B.; et al. AGPAT9 suppresses cell growth, invasion and metastasis by counteracting acidic tumor microenvironment through KLF4/LASS2/V-ATPase signaling pathway in breast cancer. Oncotarget 2015, 6, 18406–18417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Rijn, J.M.; Ardy, R.C.; Kuloğlu, Z.; Härter, B.; van Haaften-Visser, D.Y.; van der Doef, H.P.; van Hoesel, M.; Kansu, A.; van Vugt, A.H.; Thian, M.; et al. Intestinal Failure and Aberrant Lipid Metabolism in Patients With DGAT1 Deficiency. Gastroenterology 2018, 155, 130–143.e15. [Google Scholar] [CrossRef] [Green Version]
- Schene, I.F.; Joore, I.P.; Oka, R.; Mokry, M.; van Vugt, A.H.M.; van Boxtel, R.; van der Doef, H.P.J.; van der Laan, L.J.W.; Verstegen, M.M.A.; van Hasselt, P.M.; et al. Prime editing for functional repair in patient-derived disease models. Nat. Commun. 2020, 11, 5352. [Google Scholar] [CrossRef]
- Rodriguez, M.A.D.L.R.; Kersten, S. Regulation of lipid droplet homeostasis by hypoxia inducible lipid droplet associated HILPDA. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2020, 1865, 158738. [Google Scholar] [CrossRef]
- VandeKopple, M.J.; Wu, J.; Auer, E.N.; Giaccia, A.J.; Denko, N.C.; Papandreou, I. HILPDA Regulates Lipid Metabolism, Lipid Droplet Abundance, and Response to Microenvironmental Stress in Solid Tumors. Mol. Cancer Res. 2019, 17, 2089–2101. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, M.A.D.L.R.; Deng, L.; Gemmink, A.; van Weeghel, M.; Aoun, M.L.; Warnecke, C.; Singh, R.; Borst, J.W.; Kersten, S. Hypoxia-inducible lipid droplet-associated induces DGAT1 and promotes lipid storage in hepatocytes. Mol. Metab. 2021, 47, 101168. [Google Scholar] [CrossRef]
- Cheng, X.; Geng, F.; Pan, M.; Wu, X.; Zhong, Y.; Wang, C.; Tian, Z.; Cheng, C.; Zhang, R.; Puduvalli, V.; et al. Targeting DGAT1 Ameliorates Glioblastoma by Increasing Fat Catabolism and Oxidative Stress. Cell Metab. 2020, 32, 229–242.e8. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, T.; Jin, Y.; Shen, J. Dgat2 reduces hepatocellular carcinoma malignancy via downregulation of cell cycle-related gene expression. Biomed. Pharmacother. 2019, 115, 108950. [Google Scholar] [CrossRef]
- Li, S.; Wu, T.; Lu, Y.-X.; Wang, J.-X.; Yu, F.-H.; Yang, M.-Z.; Huang, Y.-J.; Li, Z.-J.; Wang, S.-L.; Huang, L.; et al. Obesity promotes gastric cancer metastasis via diacylglycerol acyltransferase 2-dependent lipid droplets accumulation and redox homeostasis. Redox Biol. 2020, 36, 101596. [Google Scholar] [CrossRef]
- Walther, T.C.; Farese, R.V. Lipid Droplets and Cellular Lipid Metabolism. Annu. Rev. Biochem. 2012, 81, 687–714. [Google Scholar] [CrossRef] [Green Version]
- Walther, T.C.; Chung, J.; Farese, R.V. Lipid Droplet Biogenesis. Annu. Rev. Cell Dev. Biol. 2017, 33, 491–510. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Liu, H.; Luo, X. Lipid droplet and its implication in cancer progression. Am. J. Cancer Res. 2020, 10, 4112–4122. [Google Scholar]
- Rozeveld, C.N.; Johnson, K.M.; Zhang, L.; Razidlo, G.L. KRAS Controls Pancreatic Cancer Cell Lipid Metabolism and Invasive Potential through the Lipase HSL. Cancer Res. 2020, 80, 4932–4945. [Google Scholar] [CrossRef]
- Grippo, P.J.; Fitchev, P.S.; Bentrem, D.J.; Melstrom, L.G.; Dangi-Garimella, S.; Krantz, S.B.; Heiferman, M.J.; Chung, C.; Adrian, K.; Cornwell, M.L.; et al. Concurrent PEDF deficiency and Kras mutation induce invasive pancreatic cancer and adipose-rich stroma in mice. Gut 2012, 61, 1454–1464. [Google Scholar] [CrossRef] [PubMed]
- Principe, D.R.; Decant, B.; Diaz, A.M.; Mangan, R.J.; Hwang, R.; Lowy, A.; Shetuni, B.B.; Sreekumar, B.K.; Chung, C.; Bentrem, D.J.; et al. PEDF inhibits pancreatic tumorigenesis by attenuating the fibro-inflammatory reaction. Oncotarget 2016, 7, 28218–28234. [Google Scholar] [CrossRef]
- Zechner, R.; Kienesberger, P.C.; Haemmerle, G.; Zimmermann, R.; Lass, A. Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. J. Lipid Res. 2009, 50, 3–21. [Google Scholar] [CrossRef] [Green Version]
- Lass, A.; Zimmermann, R.; Haemmerle, G.; Riederer, M.; Schoiswohl, G.; Schweiger, M.; Kienesberger, P.; Bogner-Strauss, J.; Gorkiewicz, G.; Zechner, R. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab. 2006, 3, 309–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sztalryd, C.; Brasaemle, D.L. The perilipin family of lipid droplet proteins: Gatekeepers of intracellular lipolysis. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2017, 1862, 1221–1232. [Google Scholar] [CrossRef] [PubMed]
- Grace, S.A.; Meeks, M.W.; Chen, Y.; Cornwell, M.; Ding, X.; Hou, P.; Rutgers, J.K.; Crawford, S.E.; Lai, J.-P. Adipose Triglyceride Lipase (ATGL) Expression Is Associated with Adiposity and Tumor Stromal Proliferation in Patients with Pancreatic Ductal Adenocarcinoma. Anticancer. Res. 2017, 37, 699–704. [Google Scholar] [CrossRef] [Green Version]
- Bai, R.; Rebelo, A.; Kleeff, J.; Sunami, Y. Identification of prognostic lipid droplet-associated genes in pancreatic cancer patients via bioinformatics analysis. Lipids Health Dis. 2021, 20, 1–12. [Google Scholar] [CrossRef]
- Kimmel, A.R.; Sztalryd, C. The Perilipins: Major Cytosolic Lipid Droplet–Associated Proteins and Their Roles in Cellular Lipid Storage, Mobilization, and Systemic Homeostasis. Annu. Rev. Nutr. 2016, 36, 471–509. [Google Scholar] [CrossRef]
- Straub, B.K.; Herpel, E.; Singer, S.; Zimbelmann, R.; Breuhahn, K.; Macher-Goeppinger, S.; Warth, A.; Lehmann-Koch, J.; Longerich, T.; Heid, H.; et al. Lipid droplet-associated PAT-proteins show frequent and differential expression in neoplastic steatogenesis. Mod. Pathol. 2010, 23, 480–492. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Cui, S.; He, Q.; Guo, Y.; Pan, X.; Zhang, P.; Huang, N.; Ge, C.; Wang, G.; Gonzalez, F.J.; et al. SUMOylation inhibitors synergize with FXR agonists in combating liver fibrosis. Nat. Commun. 2020, 11, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Halilbasic, E.; Fuchs, C.; Traussnigg, S.; Trauner, M. Farnesoid X Receptor Agonists and Other Bile Acid Signaling Strategies for Treatment of Liver Disease. Dig. Dis. 2016, 34, 580–588. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Ishida, M.; Ryota, H.; Yamamoto, T.; Kosaka, H.; Hirooka, S.; Yamaki, S.; Kotsuka, M.; Matsui, Y.; Yanagimoto, H.; et al. Adipophilin expression is an indicator of poor prognosis in patients with pancreatic ductal adenocarcinoma: An immunohistochemical analysis. Pancreatology 2019, 19, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Qiu, B.; Ackerman, D.; Sanchez, D.J.; Alison, G.; Ochocki, J.D.; Grazioli, A.; Bobrovnikova-Marjon, E.; Diehl, J.A.; Keith, B.; Simon, M.C. HIF2α-Dependent Lipid Storage Promotes Endoplasmic Reticulum Homeostasis in Clear-Cell Renal Cell Carcinoma. Cancer Discov. 2015, 5, 652–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Q.; Ruan, H.; Wang, K.; Song, Z.; Bao, L.; Xu, T.; Xiao, H.; Wang, C.; Cheng, G.; Tong, J.; et al. Overexpression of PLIN2 is a prognostic marker and attenuates tumor progression in clear cell renal cell carcinoma. Int. J. Oncol. 2018, 53, 137–147. [Google Scholar] [CrossRef]
- Wang, K.; Ruan, H.; Song, Z.; Cao, Q.; Bao, L.; Liu, D.; Xu, T.; Xiao, H.; Wang, C.; Cheng, G.; et al. PLIN3 is up-regulated and correlates with poor prognosis in clear cell renal cell carcinoma. Urol. Oncol. Semin. Orig. Investig. 2018, 36, 343.e9–343.e19. [Google Scholar] [CrossRef]
- Yang, A.; Mottillo, E.P.; Mladenovic-Lucas, L.; Zhou, L.; Granneman, J.G. Dynamic interactions of ABHD5 with PNPLA3 regulate triacylglycerol metabolism in brown adipocytes. Nat. Metab. 2019, 1, 560–569. [Google Scholar] [CrossRef]
- Huang, Y.; He, S.; Li, J.Z.; Seo, Y.-K.; Osborne, T.F.; Cohen, J.C.; Hobbs, H.H. A feed-forward loop amplifies nutritional regulation of PNPLA3. Proc. Natl. Acad. Sci. USA 2010, 107, 7892–7897. [Google Scholar] [CrossRef] [Green Version]
- Qiao, A.; Liang, J.; Ke, Y.; Li, C.; Cui, Y.; Shen, L.; Zhang, H.; Cui, A.; Liu, X.; Liu, C.; et al. Mouse patatin-like phospholipase domain-containing 3 influences systemic lipid and glucose homeostasis. Hepatology 2011, 54, 509–521. [Google Scholar] [CrossRef]
- Li, J.Z.; Huang, Y.; Karaman, R.; Ivanova, P.T.; Brown, H.A.; Roddy, T.; Castro-Perez, J.; Cohen, J.C.; Hobbs, H.H. Chronic overexpression of PNPLA3I148M in mouse liver causes hepatic steatosis. J. Clin. Investig. 2012, 122, 4130–4144. [Google Scholar] [CrossRef] [Green Version]
- Pingitore, P.; Romeo, S. The role of PNPLA3 in health and disease. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2019, 1864, 900–906. [Google Scholar] [CrossRef]
- Wang, Y.; Kory, N.; Basuray, S.; Cohen, J.C.; Hobbs, H.H. PNPLA3, CGI-58, and Inhibition of Hepatic Triglyceride Hydrolysis in Mice. Hepatology 2019, 69, 2427–2441. [Google Scholar] [CrossRef] [Green Version]
- Basuray, S.; Smagris, E.; Cohen, J.C.; Hobbs, H.H. The PNPLA3 variant associated with fatty liver disease (I148M) accumulates on lipid droplets by evading ubiquitylation. Hepatology 2017, 66, 1111–1124. [Google Scholar] [CrossRef] [Green Version]
- Smagris, E.; Basuray, S.; Li, J.Z.; Huang, Y.; Lai, K.V.; Gromada, J.; Cohen, J.C.; Hobbs, H.H. Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology 2014, 61, 108–118. [Google Scholar] [CrossRef] [Green Version]
- Banini, B.A.; Kumar, D.P.; Cazanave, S.; Seneshaw, M.; Mirshahi, F.; Santhekadur, P.K.; Wang, L.; Guan, H.P.; Oseini, A.M.; Alonso, C.; et al. Identification of a Metabolic, Transcriptomic, and Molecular Signature of Patatin-Like Phospholipase Domain Containing 3–Mediated Acceleration of Steatohepatitis. Hepatology 2020, 73, 1290–1306. [Google Scholar] [CrossRef]
- Kovarova, M.; Königsrainer, I.; Königsrainer, A.; Machicao, F.; Häring, H.-U.; Schleicher, E.; Peter, A. The Genetic Variant I148M inPNPLA3Is Associated With Increased Hepatic Retinyl-Palmitate Storage in Humans. J. Clin. Endocrinol. Metab. 2015, 100, E1568–E1574. [Google Scholar] [CrossRef] [Green Version]
- Romeo, S.; Kozlitina, J.; Xing, C.; Pertsemlidis, A.; Cox, D.; Pennacchio, L.; Boerwinkle, E.; Cohen, J.C.; Hobbs, H.H. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2008, 40, 1461–1465. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.-L.; Patman, G.; Leathart, J.; Piguet, A.-C.; Burt, A.; Dufour, J.-F.; Day, C.; Daly, A.; Reeves, H.; Anstee, Q.M. Carriage of the PNPLA3 rs738409 C >G polymorphism confers an increased risk of non-alcoholic fatty liver disease associated hepatocellular carcinoma. J. Hepatol. 2014, 61, 75–81. [Google Scholar] [CrossRef]
- Burza, M.A.; Pirazzi, C.; Maglio, C.; Sjöholm, K.; Mancina, R.M.; Svensson, P.-A.; Jacobson, P.; Adiels, M.; Baroni, M.G.; Borén, J.; et al. PNPLA3 I148M (rs738409) genetic variant is associated with hepatocellular carcinoma in obese individuals. Dig. Liver Dis. 2012, 44, 1037–1041. [Google Scholar] [CrossRef]
- Unalp-Arida, A.; Ruhl, C.E. Patatin-Like Phospholipase Domain-Containing Protein 3 I148M and Liver Fat and Fibrosis Scores Predict Liver Disease Mortality in the U.S. Population. Hepatology 2019, 71, 820–834. [Google Scholar] [CrossRef]
- Valenti, L.; Al-Serri, A.; Daly, A.; Galmozzi, E.; Rametta, R.; Dongiovanni, P.; Nobili, V.; Mozzi, E.; Roviaro, G.; Vanni, E.; et al. Homozygosity for the patatin-like phospholipase-3/adiponutrin I148M polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology 2010, 51, 1209–1217. [Google Scholar] [CrossRef]
- Pennisi, G.; Pipitone, R.M.; Cammà, C.; Di Marco, V.; Di Martino, V.; Spatola, F.; Zito, R.; Craxì, A.; Grimaudo, S.; Petta, S. PNPLA3 rs738409 C>G Variant Predicts Fibrosis Progression by Noninvasive Tools in Nonalcoholic Fatty Liver Disease. Clin. Gastroenterol. Hepatol. 2020, 19, 1979–1981. [Google Scholar] [CrossRef]
- Rosendahl, J.; Tonjes, A.; Schleinitz, R.; Kovács, P.; Wiegand, J.; Ruffert, C.; Jesinghaus, M.; Schober, R.; Herms, M.; Grützmann, R.; et al. A Common Variant of PNPLA3 (p.I148M) Is Not Associated with Alcoholic Chronic Pancreatitis. PLoS ONE 2012, 7, e29433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruschi, F.V.; Claudel, T.; Tardelli, M.; Starlinger, P.; Marra, F.; Trauner, M. PNPLA3 I148M Variant Impairs Liver X Receptor Signaling and Cholesterol Homeostasis in Human Hepatic Stellate Cells. Hepatol. Commun. 2019, 3, 1191–1204. [Google Scholar] [CrossRef] [Green Version]
- Pirazzi, C.; Valenti, L.; Motta, B.M.; Pingitore, P.; Hedfalk, K.; Mancina, R.M.; Burza, M.A.; Indiveri, C.; Ferro, Y.; Montalcini, T.; et al. PNPLA3 has retinyl-palmitate lipase activity in human hepatic stellate cells. Hum. Mol. Genet. 2014, 23, 4077–4085. [Google Scholar] [CrossRef] [Green Version]
- Kozlitina, J.; Smagris, E.; Stender, S.; Nordestgaard, B.G.; Zhou, H.H.; Tybjærg-Hansen, A.; Vogt, T.F.; Hobbs, H.H.; Cohen, J.C. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2014, 46, 352–356. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Llauradó, G.; Oresic, M.; Hyötyläinen, T.; Orho-Melander, M.; Yki-Järvinen, H. Circulating triacylglycerol signatures and insulin sensitivity in NAFLD associated with the E167K variant in TM6SF2. J. Hepatol. 2015, 62, 657–663. [Google Scholar] [CrossRef] [Green Version]
- Sookoian, S.; Castaño, G.O.; Scian, R.; Mallardi, P.; Gianotti, T.F.; Burgueño, A.L.; Martino, J.S.; Pirola, C.J. Genetic variation in transmembrane 6 superfamily member 2 and the risk of nonalcoholic fatty liver disease and histological disease severity. Hepatology 2015, 61, 515–525. [Google Scholar] [CrossRef]
- Liu, Y.-L.; Reeves, H.L.; Burt, A.; Tiniakos, D.G.; McPherson, S.; Leathart, J.B.S.; Allison, M.; Alexander, G.J.; Piguet, A.-C.; Anty, R.; et al. TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non-alcoholic fatty liver disease. Nat. Commun. 2014, 5, 4309. [Google Scholar] [CrossRef] [Green Version]
- Schwantes-An, T.; Darlay, R.; Mathurin, P.; Masson, S.; Liangpunsakul, S.; Mueller, S.; Aithal, G.P.; Eyer, F.; Gleeson, D.; Thompson, A.; et al. Genome-wide Association Study and Meta-analysis on Alcohol-Associated Liver Cirrhosis Identifies Genetic Risk Factors. Hepatology 2020, 73, 1920–1931. [Google Scholar] [CrossRef]
- Ma, Y.; Belyaeva, O.V.; Brown, P.; Fujita, K.; Valles, K.; Karki, S.; de Boer, Y.; Koh, C.; Chen, Y.; Du, X.; et al. 17-Beta Hydroxysteroid Dehydrogenase 13 Is a Hepatic Retinol Dehydrogenase Associated with Histological Features of Nonalcoholic Fatty Liver Disease. Hepatology 2018, 69, 1504–1519. [Google Scholar] [CrossRef]
- Abul-Husn, N.S.; Cheng, X.; Li, A.H.; Xin, Y.; Schurmann, C.; Stevis, P.; Liu, Y.; Kozlitina, J.; Stender, S.; Wood, G.C.; et al. A Protein-TruncatingHSD17B13Variant and Protection from Chronic Liver Disease. N. Engl. J. Med. 2018, 378, 1096–1106. [Google Scholar] [CrossRef]
- Luukkonen, P.K.; Tukiainen, T.; Juuti, A.; Sammalkorpi, H.; Haridas, P.N.; Niemelä, O.; Arola, J.; Orho-Melander, M.; Hakkarainen, A.; Kovanen, P.T.; et al. Hydroxysteroid 17-β dehydrogenase 13 variant increases phospholipids and protects against fibrosis in nonalcoholic fatty liver disease. JCI Insight 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Trépo, E.; Nahon, P.; Cao, Q.; Moreno, C.; Letouzé, E.; Imbeaud, S.; Bayard, Q.; Gustot, T.; Deviere, J.; et al. A 17-Beta-Hydroxysteroid Dehydrogenase 13 Variant Protects From Hepatocellular Carcinoma Development in Alcoholic Liver Disease. Hepatology 2019, 70, 231–240. [Google Scholar] [CrossRef]
- Stickel, F.; Lutz, P.; Buch, S.; Nischalke, H.D.; Silva, I.; Rausch, V.; Fischer, J.; Weiss, K.H.; Gotthardt, D.; Rosendahl, J.; et al. Genetic Variation in HSD17B13 Reduces the Risk of Developing Cirrhosis and Hepatocellular Carcinoma in Alcohol Misusers. Hepatology 2019, 72, 88–102. [Google Scholar] [CrossRef] [PubMed]
- Gellert-Kristensen, H.; Nordestgaard, B.G.; Tybjaerg-Hansen, A.; Stender, S. High Risk of Fatty Liver Disease Amplifies the Alanine Transaminase–Lowering Effect of a HSD17B13 Variant. Hepatology 2019, 71, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Bruschi, F.V.; Claudel, T.; Tardelli, M.; Caligiuri, A.; Stulnig, T.; Marra, F.; Trauner, M. The PNPLA3 I148M variant modulates the fibrogenic phenotype of human hepatic stellate cells. Hepatology 2017, 65, 1875–1890. [Google Scholar] [CrossRef] [Green Version]
- Pingitore, P.; Dongiovanni, P.; Motta, B.M.; Meroni, M.; Lepore, S.M.; Mancina, R.M.; Pelusi, S.; Russo, C.; Caddeo, A.; Rossi, G.; et al. PNPLA3 overexpression results in reduction of proteins predisposing to fibrosis. Hum. Mol. Genet. 2016, 25, 5212–5222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blaner, W.S. Retinol-Binding Protein: The Serum Transport Protein for Vitamin A. Endocr. Rev. 1989, 10, 308–316. [Google Scholar] [CrossRef]
- Saeed, A.; Bartuzi, P.; Heegsma, J.; Dekker, D.; Kloosterhuis, N.; de Bruin, A.; Jonker, J.W.; van de Sluis, B.; Faber, K.N. Impaired Hepatic Vitamin A Metabolism in NAFLD Mice Leading to Vitamin A Accumulation in Hepatocytes. Cell. Mol. Gastroenterol. Hepatol. 2020, 11, 309–325.e3. [Google Scholar] [CrossRef] [PubMed]
- Dobrotkova, V.; Chlapek, P.; Mazanek, P.; Sterba, J.; Veselska, R. Traffic lights for retinoids in oncology: Molecular markers of retinoid resistance and sensitivity and their use in the management of cancer differentiation therapy. BMC Cancer 2018, 18, 1–13. [Google Scholar] [CrossRef]
- McCarroll, J.; Phillips, P.; Santucci, N.; Pirola, R.C.; Wilson, J.; Apte, M.V. Vitamin A inhibits pancreatic stellate cell activation: Implications for treatment of pancreatic fibrosis. Gut 2006, 55, 79–89. [Google Scholar] [CrossRef]
- Froeling, F.E.; Feig, C.; Chelala, C.; Dobson, R.; Mein, C.E.; Tuveson, D.A.; Clevers, H.; Hart, I.R.; Kocher, H. Retinoic Acid–Induced Pancreatic Stellate Cell Quiescence Reduces Paracrine Wnt–β-Catenin Signaling to Slow Tumor Progression. Gastroenterology 2011, 141, 1486–1497.e14. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Tang, S. WNT/β-catenin signaling in the development of liver cancers. Biomed. Pharmacother. 2020, 132, 110851. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, K.Y.; Dawson, D.W. WNT Ligand Dependencies in Pancreatic Cancer. Front. Cell Dev. Biol. 2021, 9. [Google Scholar] [CrossRef]
- Hingorani, S.R.; Wang, L.; Multani, A.S.; Combs, C.; Deramaudt, T.B.; Hruban, R.H.; Rustgi, A.K.; Chang, S.; Tuveson, D.A. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal ade-nocarcinoma in mice. Cancer Cell 2005, 7, 469–483. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.-W.; Tao, L.-Y.; Yang, J.-Y.; Jiang, Y.-S.; Fu, X.-L.; Liu, W.; Huo, Y.-M.; Li, J.; Zhang, J.-F.; Hua, R.; et al. SFRP4 is a prognostic marker and correlated with Treg cell infiltration in pancreatic ductal adenocarcinoma. Am. J. Cancer Res. 2019, 9, 363–377. [Google Scholar] [PubMed]
- Anstee, Q.M.; Darlay, R.; Cockell, S.; Meroni, M.; Govaere, O.; Tiniakos, D.; Burt, A.D.; Bedossa, P.; Palmer, J.; Liu, Y.-L.; et al. Genome-wide association study of non-alcoholic fatty liver and steatohepatitis in a histologically characterised cohort. J. Hepatol. 2020, 73, 505–515. [Google Scholar] [CrossRef]
- Waghray, M.; Yalamanchili, M.; Dziubinski, M.; Zeinali, M.; Erkkinen, M.; Yang, H.; Schradle, K.A.; Urs, S.; di Magliano, M.P.; Welling, T.H.; et al. GM-CSF Mediates Mesenchymal–Epithelial Cross-talk in Pancreatic Cancer. Cancer Discov. 2016, 6, 886–899. [Google Scholar] [CrossRef] [Green Version]
- Sunami, Y.; Häußler, J.; Kleeff, J. Cellular Heterogeneity of Pancreatic Stellate Cells, Mesenchymal Stem Cells, and Cancer-Associated Fibroblasts in Pancreatic Cancer. Cancers 2020, 12, 3770. [Google Scholar] [CrossRef]
- Van Name, M.; Savoye, M.; Chick, J.M.; Galuppo, B.T.; Feldstein, A.; Pierpont, B.; Johnson, C.; Shabanova, V.; Ekong, U.; Valentino, P.L.; et al. A Low ω-6 to ω-3 PUFA Ratio (n–6:n–3 PUFA) Diet to Treat Fatty Liver Disease in Obese Youth. J. Nutr. 2020, 150, 2314–2321. [Google Scholar] [CrossRef]
- Newberry, E.P.; Hall, Z.; Xie, Y.; Molitor, E.A.; Bayguinov, P.O.; Strout, G.W.; Fitzpatrick, J.A.; Brunt, E.M.; Griffin, J.L.; Davidson, N.O. Liver-Specific Deletion of Mouse Tm6sf2 Promotes Steatosis, Fibrosis, and Hepatocellular Cancer. Hepatology 2021. [Google Scholar] [CrossRef]
- Bucher, S.S.; Stickel, F.; Trépo, E.; Way, M.; Herrmann, A.; Nischalke, H.D.; Brosch, M.; Rosendahl, J.J.; Berg, T.; Ridinger, M.M.; et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat. Genet. 2015, 47, 1443–1448. [Google Scholar] [CrossRef]
- Liu, S.; Murakami, E.; Nakahara, T.; Ohya, K.; Teraoka, Y.; Makokha, G.N.; Uchida, T.; Morio, K.; Fujino, H.; Ono, A.; et al. In vitro analysis of hepatic stellate cell activation influenced by transmembrane 6 superfamily 2 polymorphism. Mol. Med. Rep. 2020, 23. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Mao, Z.; Liu, Y.; Zhang, X.; Zhang, W.; Gustafsson, J.-A.; Guan, Y. Role of HSD17B13 in the liver physiology and pathophysiology. Mol. Cell. Endocrinol. 2018, 489, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Horiguchi, Y.; Araki, M.; Motojima, K. 17β-Hydroxysteroid dehydrogenase type 13 is a liver-specific lipid droplet-associated protein. Biochem. Biophys. Res. Commun. 2008, 370, 235–238. [Google Scholar] [CrossRef]
- Su, W.; Wang, Y.; Jia, X.; Wu, W.; Li, L.; Tian, X.; Li, S.; Wang, C.; Xu, H.; Cao, J.; et al. Comparative proteomic study reveals 17 -HSD13 as a pathogenic protein in nonalcoholic fatty liver disease. Proc. Natl. Acad. Sci. USA 2014, 111, 11437–11442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adam, M.; Heikelä, H.; Sobolewski, C.; Portius, D.; Mäki-Jouppila, J.; Mehmood, A.; Adhikari, P.; Esposito, I.; Elo, L.L.; Zhang, F.-P.; et al. Hydroxysteroid (17β) dehydrogenase 13 deficiency triggers hepatic steatosis and inflammation in mice. FASEB J. 2018, 32, 3434–3447. [Google Scholar] [CrossRef] [Green Version]
- Su, W.; Peng, J.; Li, S.; Dai, Y.-B.; Wang, C.-J.; Xu, H.; Gao, M.; Ruan, X.-Z.; Gustafsson, J.; Guan, Y.-F.; et al. Liver X receptor α induces 17β-hydroxysteroid dehydrogenase-13 expression through SREBP-1c. Am. J. Physiol. Metab. 2017, 312, E357–E367. [Google Scholar] [CrossRef]
- Liu, S.; Huang, C.; Li, D.; Ren, W.; Zhang, H.; Qi, M.; Li, X.; Yu, L. Molecular cloning and expression analysis of a new gene for short-chain dehydrogenase/reductase 9. Acta Biochim. Pol. 2007, 54, 213–218. [Google Scholar] [CrossRef]
- Brereton, P.; Suzuki, T.; Sasano, H.; Li, K.; Duarte, C.; Obeyesekere, V.; Haeseleer, F.; Palczewski, K.; Smith, I.; Komesaroff, P.; et al. Pan1b (17βHSD11)-enzymatic activity and distribution in the lung. Mol. Cell. Endocrinol. 2001, 171, 111–117. [Google Scholar] [CrossRef]
- Zillikens, M.C.; Demissie, S.; Hsu, Y.-H.; Yerges-Armstrong, L.M.; Chou, W.-C.; Stolk, L.; Livshits, G.; Broer, L.; Johnson, T.; Koller, D.L.; et al. Large meta-analysis of genome-wide association studies identifies five loci for lean body mass. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, S.; Zhang, C.; Zhu, X.; Hammad, M.A.; Zhang, X.; Christian, M.; Zhang, H.; Liu, P. Hydroxysteroid dehydrogenase family proteins on lipid droplets through bacteria, C. elegans, and mammals. Biochim. et Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2018, 1863, 881–894. [Google Scholar] [CrossRef] [PubMed]

| Gene | Variant | Amino Acid Change | Functional Relevance/Phenotypical Change | Reference |
|---|---|---|---|---|
| PNPLA3 | rs738409 C > G | I148M | Inhibits PNPLA2 | [51] |
| PNPLA3 | rs738409 C > G | I148M | Increased hepatic TAG content | [55] |
| PNPLA3 | rs738409 C > G | I148M | Confers susceptibility to NAFLD, associated with hepatic fat content | [56] |
| PNPLA3 | rs738409 C > G | I148M | Increased risk of NAFLD-associated HCC | [57] |
| PNPLA3 | rs738409 C > G | I148M | Associated with HCC in obese patients | [58] |
| PNPLA3 | rs738409 C > G | I148M | Associated with increased liver disease mortality | [59] |
| PNPLA3 | rs738409 C > G | I148M | Associated with the severity of liver fibrosis and fibrosis progression in patients with NAFLD | [60,61] |
| PNPLA3 | rs738409 C > G | I148M | Not associated with alcoholic chronic pancreatitis | [62] |
| PNPLA3 | rs738409 C > G | I148M | Reduced LXRα expression and transcriptional activity in HSCs | [63] |
| PNPLA3 | rs738409 C > G | I148M | Homozygotes have lower circulating levels of RBP4 | [64] |
| TM6SF2 | rs58542926 C > T | E167K | Impaired function contributes to NAFLD | [65] |
| TM6SF2 | rs58542926 C > T | E167K | Associated with increased circulating TAGs in patients with NAFLD | [66] |
| TM6SF2 | rs58542926 C > T | E167K | Associated with increased hepatic TAG content | [67] |
| TM6SF2 | rs58542926 C > T | E167K | Associated with hepatic fibrosis and cirrhosis, increased risk of NAFLD-HCC | [68] |
| Near HSD17B13 | rs4607179 A > C | Associated with lower risk of alcohol-associated liver cirrhosis | [69] | |
| Near HSD17B13 | rs6834314 A > G | Associated with increased steatosis and NAFLD histology | [70] | |
| HSD17B13 | rs72613567 A insertion | Associated with reduced levels of ALT, AST, reduced risk of chronic liver disease and of progression from steatosis to steatohepatitis | [71] | |
| HSD17B13 | rs72613567 A insertion | Increases phospholipids and protects against fibrosis in NAFLD | [72] | |
| HSD17B13 | rs72613567 A insertion | Protects from HCC development in alcohol liver disease | [73] | |
| HSD17B13 | rs72613567 A insertion | Reduces the risk of developing cirrhosis and HCC in alcohol misusers | [74] | |
| HSD17B13 | rs72613567 A insertion | Reduced risk of cirrhosis and HCC | [75] | |
| HSD17B13 | rs62305723 G > A | P260S | Retains LD localization but lacks RDH activity, decreased ballooning and inflammation | [70] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sunami, Y.; Rebelo, A.; Kleeff, J. Lipid Droplet-Associated Factors, PNPLA3, TM6SF2, and HSD17B Proteins in Hepatopancreatobiliary Cancer. Cancers 2021, 13, 4391. https://doi.org/10.3390/cancers13174391
Sunami Y, Rebelo A, Kleeff J. Lipid Droplet-Associated Factors, PNPLA3, TM6SF2, and HSD17B Proteins in Hepatopancreatobiliary Cancer. Cancers. 2021; 13(17):4391. https://doi.org/10.3390/cancers13174391
Chicago/Turabian StyleSunami, Yoshiaki, Artur Rebelo, and Jörg Kleeff. 2021. "Lipid Droplet-Associated Factors, PNPLA3, TM6SF2, and HSD17B Proteins in Hepatopancreatobiliary Cancer" Cancers 13, no. 17: 4391. https://doi.org/10.3390/cancers13174391
APA StyleSunami, Y., Rebelo, A., & Kleeff, J. (2021). Lipid Droplet-Associated Factors, PNPLA3, TM6SF2, and HSD17B Proteins in Hepatopancreatobiliary Cancer. Cancers, 13(17), 4391. https://doi.org/10.3390/cancers13174391








