Magnetic Resonance Relaxometry for Tumor Cell Density Imaging for Glioma: An Exploratory Study via 11C-Methionine PET and Its Validation via Stereotactic Tissue Sampling
Abstract
Simple Summary
Abstract
1. Introduction
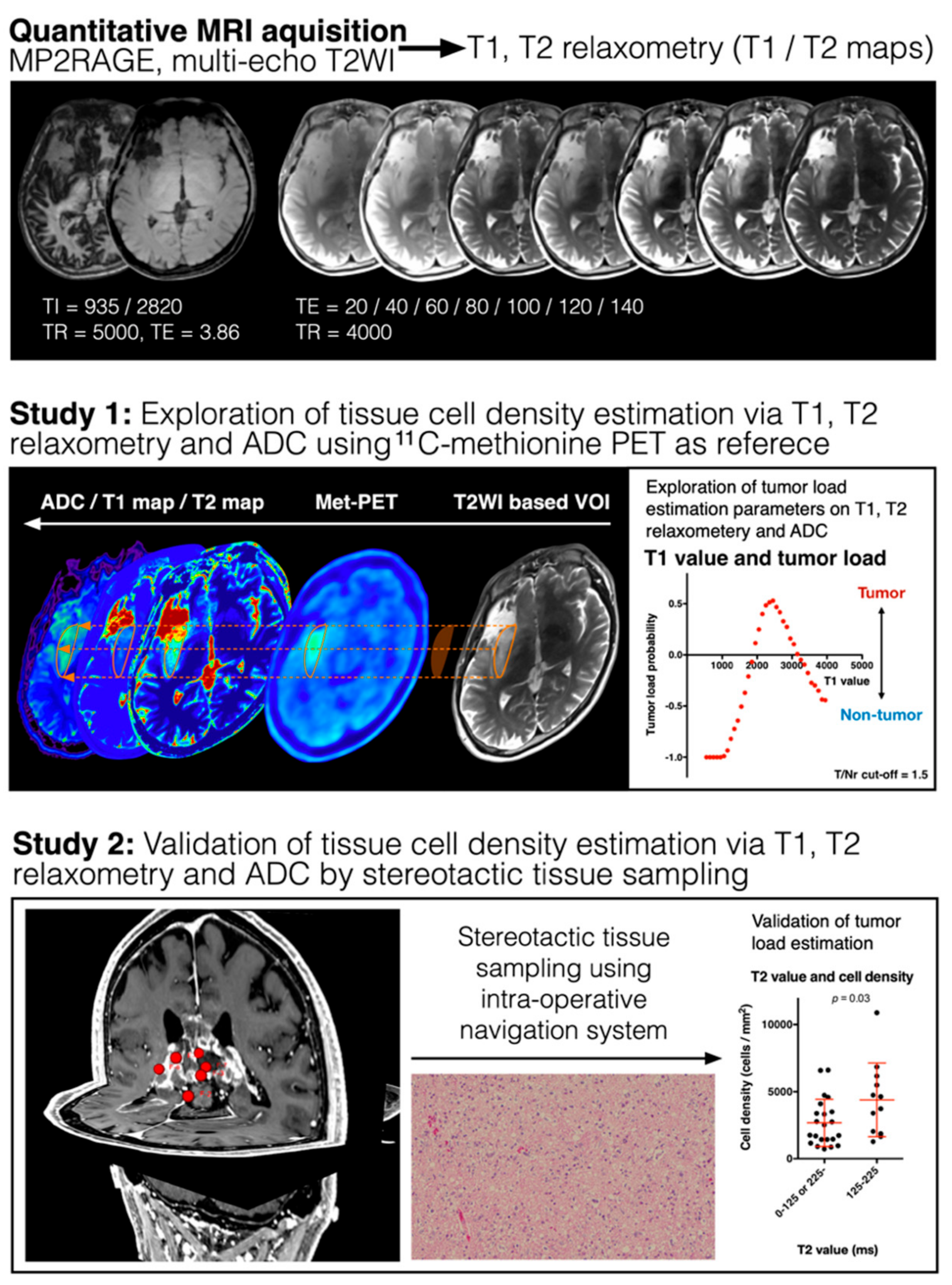
2. Materials and Methods
2.1. Patient Selection
2.2. T1- and T2-Relaxometry
2.3. 11C-Methionine Positron Emission Tomography (MET-PET)
2.4. Diffusion-Weighted Imaging (DWI) and Apparent Diffusion Coefficient (ADC) Measurement
2.5. Image Fusion/Registration and Stereotactic Image-Guided Tissue Sampling
2.6. Histopathological Analysis
2.7. Statistical Analysis
3. Results
3.1. Areas of High and Low MET Uptake Show Different T1- and T2-Relaxation Times
3.2. ADCs within Areas of High and Low MET Uptake
3.3. Histological Validation of T1- and T2-Relaxation Times and ADC as Surrogate Measurements for TCD
3.4. Synthetic Tumor Load Image via T1- and T2-Relaxometry
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Eidel, O.; Burth, S.; Neumann, J.-O.; Kieslich, P.J.; Sahm, F.; Jungk, C.; Kickingereder, P.; Bickelhaupt, S.; Mundiyanapurath, S.; Bäumer, P.; et al. Tumor Infiltration in Enhancing and Non-Enhancing Parts of Glioblastoma: A Correlation with Histopathology. PLoS ONE 2017, 12, e0169292. [Google Scholar] [CrossRef]
- Ellingson, B.M.; Lai, A.; Nguyen, H.N.; Nghiemphu, P.L.; Pope, W.B.; Cloughesy, T.F. Quantification of Nonenhancing Tumor Burden in Gliomas Using Effective T2 Maps Derived from Dual-Echo Turbo Spin-Echo MRI. Clin. Cancer Res. 2015, 21, 4373–4383. [Google Scholar] [CrossRef]
- Kinoshita, M.; Arita, H.; Okita, Y.; Kagawa, N.; Kishima, H.; Hashimoto, N.; Tanaka, H.; Watanabe, Y.; Shimosegawa, E.; Hatazawa, J.; et al. Comparison of Diffusion Tensor Imaging and 11C-Methionine Positron Emission Tomography for Reliable Prediction of Tumor Cell Density in Gliomas. J. Neurosurg. 2016, 125, 1136–1142. [Google Scholar] [CrossRef] [PubMed]
- Hirata, T.; Kinoshita, M.; Tamari, K.; Seo, Y.; Suzuki, O.; Wakai, N.; Achiha, T.; Umehara, T.; Arita, H.; Kagawa, N.; et al. 11C-Methionine-18F-FDG Dual-PET-Tracer–Based Target Delineation of Malignant Glioma: Evaluation of Its Geometrical and Clinical Features for Planning Radiation Therapy. J. Neurosurg. 2018, 131, 676–686. [Google Scholar] [CrossRef]
- Pafundi, D.H.; Laack, N.N.; Youland, R.S.; Parney, I.F.; Lowe, V.J.; Giannini, C.; Kemp, B.J.; Grams, M.P.; Morris, J.M.; Hoover, J.M.; et al. Biopsy Validation of 18F-DOPA PET and Biodistribution in Gliomas for Neurosurgical Planning and Radiotherapy Target Delineation: Results of a Prospective Pilot Study. Neuro-Oncology 2013, 15, 1058–1067. [Google Scholar] [CrossRef]
- Kinoshita, M.; Arita, H.; Goto, T.; Okita, Y.; Isohashi, K.; Watabe, T.; Kagawa, N.; Fujimoto, Y.; Kishima, H.; Shimosegawa, E.; et al. A Novel PET Index, 18F-FDG–11C-Methionine Uptake Decoupling Score, Reflects Glioma Cell Infiltration. J. Nucl. Med. 2012, 53, 1701–1708. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Ono, Y.; Aga, F.; Kawai, N.; Kudomi, N.; Nishiyama, Y. Correlation of 18F-FLT Uptake with Tumor Grade and Ki-67 Immunohistochemistry in Patients with Newly Diagnosed and Recurrent Gliomas. J. Nucl. Med. 2012, 53, 1911–1915. [Google Scholar] [CrossRef]
- Jansen, N.L.; Suchorska, B.; Wenter, V.; Eigenbrod, S.; Schmid-Tannwald, C.; Zwergal, A.; Niyazi, M.; Drexler, M.; Bartenstein, P.; Schnell, O.; et al. Dynamic 18F-FET PET in Newly Diagnosed Astrocytic Low-Grade Glioma Identifies High-Risk Patients. J. Nucl. Med. 2014, 55, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, M.; Goto, T.; Arita, H.; Okita, Y.; Isohashi, K.; Kagawa, N.; Fujimoto, Y.; Kishima, H.; Shimosegawa, E.; Saitoh, Y.; et al. Imaging 18F-Fluorodeoxy Glucose/11C-Methionine Uptake Decoupling for Identification of Tumor Cell Infiltration in Peritumoral Brain Edema. J. Neuro-Oncol. 2012, 106, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Berntsson, S.G.; Falk, A.; Savitcheva, I.; Godau, A.; Zetterling, M.; Hesselager, G.; Alafuzoff, I.; Larsson, E.-M.; Smits, A. Perfusion and Diffusion MRI Combined with 11C-Methionine PET in the Preoperative Evaluation of Suspected Adult Low-Grade Gliomas. J. Neuro-Oncol. 2013, 114, 241–249. [Google Scholar] [CrossRef]
- LaViolette, P.S.; Mickevicius, N.J.; Cochran, E.J.; Rand, S.D.; Connelly, J.; Bovi, J.A.; Malkin, M.G.; Mueller, W.M.; Schmainda, K.M. Precise Ex Vivo Histological Validation of Heightened Cellularity and Diffusion-Restricted Necrosis in Regions of Dark Apparent Diffusion Coefficient in 7 Cases of High-Grade Glioma. Neuro-Oncology 2014, 16, 1599–1606. [Google Scholar] [CrossRef]
- Marques, J.P.; Kober, T.; Krueger, G.; van der Zwaag, W.; de Moortele, P.-F.V.; Gruetter, R. MP2RAGE, a Self Bias-Field Corrected Sequence for Improved Segmentation and T1-Mapping at High Field. Neuroimage 2010, 49, 1271–1281. [Google Scholar] [CrossRef]
- Hatakeyama, T.; Kawai, N.; Nishiyama, Y.; Yamamoto, Y.; Sasakawa, Y.; Ichikawa, T.; Tamiya, T. 11C-Methionine (MET) and 18F-Fluorothymidine (FLT) PET in Patients with Newly Diagnosed Glioma. Eur. J. Nucl. Med. Mol. I 2008, 35, 2009–2017. [Google Scholar] [CrossRef] [PubMed]
- Okita, Y.; Kinoshita, M.; Goto, T.; Kagawa, N.; Kishima, H.; Shimosegawa, E.; Hatazawa, J.; Hashimoto, N.; Yoshimine, T. 11C-Methionine Uptake Correlates with Tumor Cell Density Rather than with Microvessel Density in Glioma: A Stereotactic Image-Histology Comparison. Neuroimage 2010, 49, 2977–2982. [Google Scholar] [CrossRef]
- Arita, H.; Kinoshita, M.; Kagawa, N.; Fujimoto, Y.; Kishima, H.; Hashimoto, N.; Yoshimine, T. 11C-methionine Uptake and Intraoperative 5-aminolevulinic Acid-induced Fluorescence as Separate Index Markers of Cell Density in Glioma. Cancer 2011, 118, 1619–1627. [Google Scholar] [CrossRef] [PubMed]
- Čížek, J.; Herholz, K.; Vollmar, S.; Schrader, R.; Klein, J.; Heiss, W.-D. Fast and Robust Registration of PET and MR Images of Human Brain. Neuroimage 2004, 22, 434–442. [Google Scholar] [CrossRef]
- Izutsu, N.; Kinoshita, M.; Yanagisawa, T.; Nakanishi, K.; Sakai, M.; Kishima, H. Preservation of Motor Function After Resection of Lower-Grade Glioma at the Precentral Gyrus and Prediction by Presurgical Functional Magnetic Resonance Imaging and Magnetoencephalography. World Neurosurg. 2017, 107, 1045.e5–1045.e8. [Google Scholar] [CrossRef] [PubMed]
- Overton, W.R. Modified Histogram Subtraction Technique for Analysis of Flow Cytometry Data. Cytometry 1988, 9, 619–626. [Google Scholar] [CrossRef]
- Agnès, C.; Jean-Jacques, R.; Hong-Hsu, H.; Daniel, S.; Jean-Jacques, L. Limits of Flow-Cytometry Histogram Analysis Methods to Assess Bladder Tumour Antigen Expression. Anal. Cell Pathol. 1997, 13, 39–47. [Google Scholar] [CrossRef]
- Lasocki, A.; Gaillard, F. Non-Contrast-Enhancing Tumor: A New Frontier in Glioblastoma Research. Am. J. Neuroradiol. 2019, 40, 758–765. [Google Scholar] [CrossRef] [PubMed]
- Verburg, N.; Hoefnagels, F.W.A.; Barkhof, F.; Boellaard, R.; Goldman, S.; Guo, J.; Heimans, J.J.; Hoekstra, O.S.; Jain, R.; Kinoshita, M.; et al. Diagnostic Accuracy of Neuroimaging to Delineate Diffuse Gliomas within the Brain: A Meta-Analysis. Am. J. Neuroradiol. 2017, 38, 1884–1891. [Google Scholar] [CrossRef]
- Kinoshita, M.; Hashimoto, N.; Goto, T.; Yanagisawa, T.; Okita, Y.; Kagawa, N.; Kishima, H.; Tanaka, H.; Fujita, N.; Shimosegawa, E.; et al. Use of Fractional Anisotropy for Determination of the Cut-off Value in 11C-Methionine Positron Emission Tomography for Glioma. Neuroimage 2009, 45, 312–318. [Google Scholar] [CrossRef]
- Kinoshita, M.; Goto, T.; Okita, Y.; Kagawa, N.; Kishima, H.; Hashimoto, N.; Yoshimine, T. Diffusion Tensor-Based Tumor Infiltration Index Cannot Discriminate Vasogenic Edema from Tumor-Infiltrated Edema. J. Neuro-Oncol. 2009, 96, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Ali, T.S.; Bjarnason, T.A.; Senger, D.L.; Dunn, J.F.; Joseph, J.T.; Mitchell, J.R. QuantitativeT2: Interactive Quantitative T2 MRI Witnessed in Mouse Glioblastoma. J. Med. Imaging 2015, 2, 036002. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ellingson, B.M.; Cloughesy, T.F.; Lai, A.; Nghiemphu, P.L.; Lalezari, S.; Zaw, T.; Motevalibashinaeini, K.; Mischel, P.S.; Pope, W.B. Quantification of Edema Reduction Using Differential Quantitative T2 (DQT2) Relaxometry Mapping in Recurrent Glioblastoma Treated with Bevacizumab. J. Neuro-Oncol. 2011, 106, 111–119. [Google Scholar] [CrossRef]
- Chang, P.D.; Malone, H.R.; Bowden, S.G.; Chow, D.S.; Gill, B.J.A.; Ung, T.H.; Samanamud, J.; Englander, Z.K.; Sonabend, A.M.; Sheth, S.A.; et al. A Multiparametric Model for Mapping Cellularity in Glioblastoma Using Radiographically Localized Biopsies. Am. J. Neuroradiol. 2017, 38, 890–898. [Google Scholar] [CrossRef]
- Prager, A.J.; Martinez, N.; Beal, K.; Omuro, A.; Zhang, Z.; Young, R.J. Diffusion and Perfusion MRI to Differentiate Treatment-Related Changes Including Pseudoprogression from Recurrent Tumors in High-Grade Gliomas with Histopathologic Evidence. Am. J. Neuroradiol. 2015, 36, 877–885. [Google Scholar] [CrossRef]
- Lee, J.; Choi, S.H.; Kim, J.; Sohn, C.; Lee, S.; Jeong, J. Glioma Grading Using Apparent Diffusion Coefficient Map: Application of Histogram Analysis Based on Automatic Segmentation. Nmr. Biomed. 2014, 27, 1046–1052. [Google Scholar] [CrossRef] [PubMed]
- Ryu, Y.J.; Choi, S.H.; Park, S.J.; Yun, T.J.; Kim, J.-H.; Sohn, C.-H. Glioma: Application of Whole-Tumor Texture Analysis of Diffusion-Weighted Imaging for the Evaluation of Tumor Heterogeneity. PLoS ONE 2014, 9, e108335. [Google Scholar] [CrossRef]
- Ellingson, B.M.; Cloughesy, T.F.; Lai, A.; Mischel, P.S.; Nghiemphu, P.L.; Lalezari, S.; Schmainda, K.M.; Pope, W.B. Graded Functional Diffusion Map–Defined Characteristics of Apparent Diffusion Coefficients Predict Overall Survival in Recurrent Glioblastoma Treated with Bevacizumab. Neuro-Oncology 2011, 13, 1151–1161. [Google Scholar] [CrossRef] [PubMed]
- Verburg, N.; Koopman, T.; Yaqub, M.M.; Hoekstra, O.S.; Lammertsma, A.A.; Barkhof, F.; Pouwels, P.J.W.; Reijneveld, J.C.; Heimans, J.J.; Rozemuller, A.J.M.; et al. Improved Detection of Diffuse Glioma Infiltration with Imaging Combinations: A Diagnostic Accuracy Study. Neuro-Oncology 2019, 22, 412–422. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, M.; Hashimoto, N.; Goto, T.; Kagawa, N.; Kishima, H.; Izumoto, S.; Tanaka, H.; Fujita, N.; Yoshimine, T. Fractional Anisotropy and Tumor Cell Density of the Tumor Core Show Positive Correlation in Diffusion Tensor Magnetic Resonance Imaging of Malignant Brain Tumors. Neuroimage 2008, 43, 29–35. [Google Scholar] [CrossRef] [PubMed]



| # | Sex | Age | Pathology | Molecular Characteristics | MET-PET | ADC | Number of Stereotactic Tissues (Enclosed: Contrast-Enhanced Lesions) | ||
|---|---|---|---|---|---|---|---|---|---|
| T1/T2 map | MET-PET | ADC | |||||||
| 1 | M | 68 | AA | IDH-wt | Performed | Performed | 4 | 4 | 4 |
| 2 | M | 68 | AA | IDH-wt | - | Performed | 5 | - | 5 |
| 3 | F | 81 | DA | IDH-wt | Performed | Performed | 5 | 5 | 5 |
| 4 | M | 72 | GBM | IDH-wt | - | Performed | 3 (3) | - | 3 (3) |
| 5 | M | 19 | GBM | IDH-wt | - | - | 4 (2) | - | - |
| 6 | M | 70 | GBM | IDH-wt | - | - | 6 (3) | - | - |
| 7 | F | 72 | GBM | IDH-wt | - | Performed | 1 (1) | - | 1 (1) |
| 8 | F | 68 | GBM | IDH-wt | Performed | Performed | 2 (2) | 2 (2) | 2 (2) |
| 9 | M | 52 | GBM | IDH-wt | - | - | 3 (2) | - | - |
| 10 | M | 76 | GBM | IDH-wt | - | Performed | 3 (2) | - | 3 (2) |
| 11 | F | 34 | GBM | IDH-wt | Performed | Performed | 7 (6) | 7 (6) | 7 (6) |
| 12 | M | 30 | rec. AA | IDH-wt | - | - | 3 | - | - |
| 13 | F | 45 | rec. GBM | IDH-wt | - | Performed | 3 (2) | - | 3 (2) |
| 14 | F | 29 | OL | IDH-mt, 1p/19q-codeleted | Performed | Performed | 6 | 6 | 6 |
| 15 | F | 48 | OL | IDH-mt, 1p/19q-codeleted | Performed | Performed | 4 | 4 | 4 |
| 16 | M | 37 | OL | IDH-mt, 1p/19q-codeleted | Performed | Performed | - | - | - |
| 17 | M | 30 | OL | IDH-mt, 1p/19q-codeleted | Performed | Performed | 4 | 4 | 4 |
| 18 | F | 46 | rec. AO | IDH-mt, 1p/19q-codeleted | - | - | 2 (2) | - | - |
| 19 | M | 48 | rec. AO | IDH-mt, 1p/19q-codeleted | Performed | Performed | 3 | 3 | 3 |
| 20 | F | 35 | AA | IDH-mt | - | Performed | 5 | - | 6 |
| 21 | M | 34 | DA | IDH-mt | Performed | Performed | 4 | 4 | 4 |
| 22 | F | 48 | DMG | H3 K27M-mt | - | Performed | 1 | - | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kinoshita, M.; Uchikoshi, M.; Tateishi, S.; Miyazaki, S.; Sakai, M.; Ozaki, T.; Asai, K.; Fujita, Y.; Matsuhashi, T.; Kanemura, Y.; et al. Magnetic Resonance Relaxometry for Tumor Cell Density Imaging for Glioma: An Exploratory Study via 11C-Methionine PET and Its Validation via Stereotactic Tissue Sampling. Cancers 2021, 13, 4067. https://doi.org/10.3390/cancers13164067
Kinoshita M, Uchikoshi M, Tateishi S, Miyazaki S, Sakai M, Ozaki T, Asai K, Fujita Y, Matsuhashi T, Kanemura Y, et al. Magnetic Resonance Relaxometry for Tumor Cell Density Imaging for Glioma: An Exploratory Study via 11C-Methionine PET and Its Validation via Stereotactic Tissue Sampling. Cancers. 2021; 13(16):4067. https://doi.org/10.3390/cancers13164067
Chicago/Turabian StyleKinoshita, Manabu, Masato Uchikoshi, Souichiro Tateishi, Shohei Miyazaki, Mio Sakai, Tomohiko Ozaki, Katsunori Asai, Yuya Fujita, Takahiro Matsuhashi, Yonehiro Kanemura, and et al. 2021. "Magnetic Resonance Relaxometry for Tumor Cell Density Imaging for Glioma: An Exploratory Study via 11C-Methionine PET and Its Validation via Stereotactic Tissue Sampling" Cancers 13, no. 16: 4067. https://doi.org/10.3390/cancers13164067
APA StyleKinoshita, M., Uchikoshi, M., Tateishi, S., Miyazaki, S., Sakai, M., Ozaki, T., Asai, K., Fujita, Y., Matsuhashi, T., Kanemura, Y., Shimosegawa, E., Hatazawa, J., Nakatsuka, S.-i., Kishima, H., & Nakanishi, K. (2021). Magnetic Resonance Relaxometry for Tumor Cell Density Imaging for Glioma: An Exploratory Study via 11C-Methionine PET and Its Validation via Stereotactic Tissue Sampling. Cancers, 13(16), 4067. https://doi.org/10.3390/cancers13164067






