Simple Summary
The ligand of numb-protein X1 (LNX1) is reported to be upregulated in various cancers, however the cellular function of LNX1 is not clearly characterized. The aim of the present study was to elucidate the regulation of LNX1 expression and clarify the role of LNX1 in cell-cycle progression and resistance to the cancer therapeutic agent, cisplatin. We found that LNX1 expression is decreased by DNA damage including cisplatin treatment and the levels of S and G2/M populations were correlated with LNX1 expression. We also showed that the upregulation of LNX1 contributes to cell-cycle progression and cisplatin resistance. Our data suggest that LNX1 is the important regulator of the cell cycle, and contributes to tumor progression.
Abstract
The ligand of numb-protein X1 (LNX1) acts as a proto-oncogene by inhibiting p53 stability; however, the regulation of LNX1 expression has not been investigated. In this study, we screened chemicals to identify factors that potentially regulate LNX1 expression. We found that LNX1 expression levels were decreased by DNA damage, including that by cisplatin. Upon treatment with lipopolysaccharide (LPS) and phorbol 12-myristate 13-acetate (PMA), LNX1 expression levels increased. In addition, cell-cycle progression increased upon LNX1 expression; the levels of S and G2/M populations were correlated with LNX1 expression. Moreover, in CRISPR-Cas9-mediated LNX1 knockout cells, we observed a delay in cell-cycle progression and a downregulation of genes encoding the cell-cycle markers cyclin D1 and cyclin E1. Finally, the upregulation of LNX1-activated cell-cycle progression and increased resistance to cisplatin-mediated cell death. Taken together, these results suggest that LNX1 contributes to cell-cycle progression and cisplatin resistance.
1. Introduction
The ligand of numb-protein X1 (LNX1) is a binding partner for the cell-fate-determinant NUMB [1]. There are two isoforms of LNX1: LNX1 p80 and LNX1 p70. LNX1 p80 has been hereafter referred to as LNX1 [2,3]. It is a RING-type E3 ubiquitin ligase that contains a catalytic N-terminal RING domain, an amino acid motif NPAY for binding with phosphotyrosine-binding domains, and four PDZ domains. LNX1, through its RING domain, can ubiquitinate specific isoforms of NUMB and induce their proteasomal degradation [1]. A shorter, alternatively spliced isoform, LNX1 p70, is deficient in the N-terminal RING domain and is distributed in the brain and kidney glomeruli. LNX1 p70 interacts with cell adhesion molecules to reorganize cell junctions and acts as a scaffold to promote the ubiquitination of its ligands by recruiting other E3-ligases [4]. Different studies have predicted and identified the binding partners of LNX1 [5,6]. LNX1 interacts with proto-oncogenes such as erythroblastic leukemia viral oncogene homolog 2 (ERBB2) and c-Src through its PDZ domain [7,8]. Moreover, it can interact with pJAK2 to regulate lung cancer [9]. In glioblastoma, LNX1 regulates Notch1 signaling and induces expansion of the glioma stem cell population [10]. Furthermore, several studies associated with cancer and LNX1 have suggested its role as a tumor regulator in various cancers, including gliomas and colorectal carcinoma [11,12]. LNX1 related protein, LNX2 is also reported to upregulate Wnt/beta-catenin pathway in colorectal cancers [13]. In addition, recent studies also showed the role of LNX1 in the nervous system and pathophysiological cell signaling [14,15].
Previously, we reported that LNX1 acts as a proto-oncogene by inhibiting p53 stability [16]. p53 is a major tumor suppressor that regulates numerous signaling pathways including those involved in cell death, growth, DNA repair, and cellular senescence [17]. In the previous study, we have shown that LNX1 interacts with p53 and is ubiquitinated in an MDM2-dependent manner. In CRISPR-Cas9-mediated LNX1 knockout (KO) cells, tumor growth was reduced because of increased p53 stability. In addition, we demonstrated that LNX1 enhanced tumor growth both in cell culture and xenograft models by inhibiting a p53-dependent signaling pathway in p53 wild-type (WT) cancer cells [16]. However, the regulation of LNX1 remains unclear.
Cisplatin is one of the most widely used anticancer drugs [18]. However, its clinical efficacy is limited owing to frequently emerging cisplatin-resistant cell populations [18,19,20]. Several cellular changes have been implicated in cisplatin resistance, including increased glutathione or metallothionein content, decreased accumulation and/or increased efflux of cisplatin, and increased DNA repair [21]. Moreover, additional oncogenic pathways (such as ras, c-fos/AP1, and bcl-2 pathways) have been characterized, which indicate that the altered expression of oncogenes could subsequently limit cisplatin-mediated DNA damage. Additionally, the activation of antiapoptotic pathways may contribute to cisplatin resistance [22,23].
In this study, we found that DNA-damage inducers decreased LNX1 expression levels; however, LPS and PMA increased these levels. In addition, the cell-cycle progression increased LNX1 expression levels, and S, G2, and M populations correlated with these levels. Moreover, in CRISPR-Cas9-mediated LNX1 KOs, cell-cycle progression was delayed and the expression levels of the genes encoding the cell-cycle markers cyclin D1 and cyclin E1 decreased. Our study suggests that LNX1 expression contributes to cell-cycle progression. Here, we investigated the potential role of LNX1 in cisplatin resistance using overexpression experiments. We found that the overexpression of LNX1 induces cell death upon cisplatin treatment.
2. Materials and Methods
2.1. Cell Culture and Cell Viability Assay
A549 human lung cancer cells, MCF7 human breast cancer cells, H460 human lung cancer cells and HEK293 and HEK293T human embryonic kidney cells were maintained in DMEM (WelGene, Gyeongsan-si, South Korea) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Waltham, MA, USA) and antibiotic-antimycotic solution (WelGene). HEK293 and HEK293T cells were transfected by using Lipofectamine (Thermo Fisher Scientific), according to the manufacturer’s protocol, and calcium phosphate, respectively. Cell proliferation was measured using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Briefly, cells were seeded uniformly into the wells of a 24-well plate. At the indicated time, a final concentration of 1 mg/mL of the MTT solution was added; the mixture was incubated for an additional 3 h. MTT was purchased from Thermo Fisher Scientific. Live cells were counted using a Vi-Cell XR automatic cell counter (Beckman Coulter, Indianapolis, IN, USA), and trypan blue dye was used to differentiate between the live and dead cells. Next, the resuspended cells were collected and delivered to the flow cell for imaging purposes. The counter was set to capture 50 images per sample. Three cycles of both the aspirate and trypan blue were used to maintain single-cell suspension and achieve sufficient mixing.
2.2. Cell Line Generation
The generation of LNX1 KO cell lines with CRISPR-Cas9 has been described previously [16]. For LNX1 stable cell lines, LNX1 CDSs were cloned into HA-pCDNA3 vectors. HEK293 cells were transfected using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA). Twenty-four hours prior to transfection, cells were seeded at a density of 1.5 × 105 cells/well in 6-well culture plates. Transfection was performed at 60% confluency with 2 μg DNA and 4 μL Lipofectamine 2000 reagent in 200 μL Opti-MEM. Forty-eight hours after transfection, the cells were treated with neomycin to select those with stable expression of the neomycin resistance gene. Thus, the stable cells were selected by detecting neomycin resistance after drug treatment (800 μg/mL) and single-cell clone isolation.
2.3. Virus Production and Transduction
Lentiviral vectors were produced by co-transfection of HEK293T cells with the psPAX2 envelope and pMD2.G packaging plasmids using the calcium phosphate transfection method (2 M CaCl2, 2 × HEPES buffered saline (pH 7.2)) [24]. Cells were plated in a 100 mm culture dish on the day before transfection. The cells were co-transfected with 5 µg of vector plasmids, 1.5 µg of psPAX2, and pMD2.G. The medium was changed 24 h after transfection, and the supernatant medium containing the virus was collected 48 and 72 h post-transfection. The collected medium was filtered through a 0.45 µm filter and mixed with the Lenti-X concentrator (Takara, Kyoto, Japan) in accordance with the manufacturer’s instructions. Viral pellets were stored in Opti-MEM and maintained at −70 °C. The cells in the 6-well tissue culture plates were infected with the medium containing virus and 8 mg/mL polybrene. Twenty-four hours after the infection, the medium was changed, and the cells were selected using puromycin (2 mg/mL).
2.4. Western Blotting
For the Western blot analysis, cells were collected in cell lysis buffer (150 mM NaCl, 50 mM HEPES [pH 7.5], and 1% NP40) containing protease inhibitor cocktail (Roche, Basel, Switzerland). Polypeptides in whole-cell lysates were resolved by SDS-PAGE and transferred onto immune-blot PVDF membrane filters (Bio-Rad, Hercules, CA, USA). Proteins were detected with a 1:1000 or 1:5000 dilution of primary antibodies using an ECL system (Dogen, Seoul, South Korea). Images were acquired using ChemiDoc-it 410 Imaging System (Analytik Jena, Upland, CA, USA) and ImageQuant LAS 4000 System (GE Healthcare, Waukesha, WI, USA). LNX1-specific antibodies were purchased from Lifespan Biosciences (Seattle, WA, USA). Cyclin D1- and cyclin E1-specific antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA), and PARP-specific antibodies were purchased from GeneTex (San Antonio, TX, USA).
2.5. Reporter Plasmid Construction and Reporter Assay
The LNX1 promoter region was amplified from HEK293 genomic DNA using the forward primer 5′-GGCTCGAGGGGATAGCCACACCTACCTTA-3′ and reverse primer 5′-GGAAGCTTGGGAAAGCATTGCTGAGACCT-3′. The amplified fragments were initially cloned into the pTOP TA V2 vector (Enzynomics, Daejeon, Korea, EZ011). The reporter plasmid construct was amplified and subsequently subcloned into the pGL3-promoter vector (Promega, Madison, WI, USA, E1641). For the reporter assay, cells were seeded in 24-well plates in DMEM for 18 h before transfection. Typically, cells in each well were transfected with less than 0.5 μg of the total DNA, and Renilla luciferase was used for normalization in each assay. To examine the cell signaling pathways, HEK293 and A549 cells were treated with cisplatin, topotecan, hydroxyurea, LPS, and PMA and irradiated with UV.
2.6. Quantitative RT-PCR
For quantitative RT-PCR, cells were harvested, and RNA was extracted using TRIzol (Thermo Fisher Scientific). Reverse transcription was conducted with an M-MLV RT kit (Enzynomics, Daejeon, South Korea) according to the manufacturer’s protocol, and PCR was performed using the StepOnePlus Real-Time PCR System (Thermo Fisher Scientific). LNX1 mRNA was amplified using the forward primer 5′-TGAGCCCGGAGGAGTCATAA-3′ and reverse primer 5′-ATTCCAGCCACATGACCCAG-3′. For amplifying p21 mRNA, the following pair of primers was used: forward 5′-CATGTGGACCTGTCACTGTCTTGTA-3′ and reverse 5′-GAAGATCAGCCGGCGTTTG-3′.
2.7. Cell-Cycle Analysis
A double-thymidine block was used to arrest A549 cells at the G1/S transition. A549 cells were synchronized by the addition of 2 mM thymidine from a 100 mM stock solution for 18 h. Next, the cells were washed with phosphate buffered saline (PBS), followed by the addition of regular growth medium (DMED supplemented with 10% FBS). Eight hours after the incubation, the medium was changed to that containing 2 mM thymidine and incubated for another 18 h. The cells were washed twice with PBS and replaced with fresh growth medium. The cells were collected at the indicated time points for the preparation of whole cell extracts. For cell-cycle analysis, the cells were washed and fixed with 70% ethanol. After centrifugation, the cells were resuspended in PBS containing 0.25 mg/mL propidium iodide (PI) and 10 mg/mL RNase A (Sigma, St. Louis, MO, USA). Next, the cell-cycle progression was analyzed using a FACSCalibur flow cytometer (Becton Dickinson, Mountain View, CA, USA).
2.8. Statistical Analysis
The results of the luciferase assay, Western blot, quantitative RT-PCR, cell viability assay, and cell-cycle distribution assay were evaluated using a 2-tailed Student’s t-test using Microsoft Excel 2016 software (Microsoft, Redmond, WA, USA). Statistical significance was set at p < 0.05.
3. Results
3.1. DNA Damage Decreases the Expression Levels of LNX1
To investigate the potential regulation of LNX1 expression, we analyzed the nucleotide sequence of the LNX1 promoter using the Ensemble database [25] and constructed reporter plasmids containing 1.5 kb promoter fragments relative to the transcription start site (TSS) of LNX1. Anticancer drugs such as cisplatin and topotecan, UV, and hydroxyurea induce DNA damage [26]; hence, we transfected A549 cells with reporter plasmids containing the LNX1 promoter regions and treated the cells with DNA damage-inducing reagents. Although the cells with the control reporter (pGL3-promoter) did not exhibit any response to stimuli, a substantial reduction was observed with the reporter constructs containing LNX1 promoter regions (Figure 1A). We tested whether each stimulus could regulate the expression levels of LNX1 and found that the mRNA expression levels of LNX1 decreased in a dose-dependent manner in response to each stimulus (Figure 1B and Figure S1A). Additionally, we found the protein expression level of LNX1 was decreased by DNA damage (Figure 1C,D). These results suggest that DNA damage decreases the expression of LNX1 at both mRNA and protein levels.
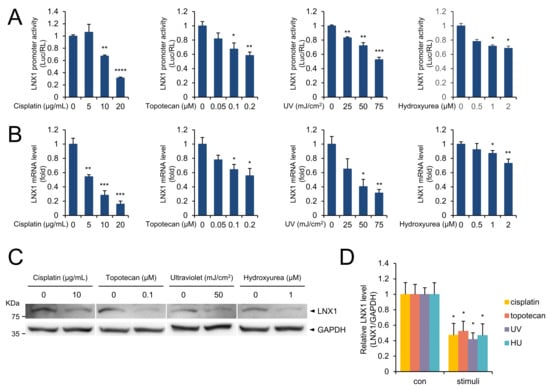
Figure 1.
DNA damage decreased the LNX1 expression levels. (A) A549 cells were transfected with pGL3 plasmids containing LNX1 promoter sequence. After 24 h, cells were treated with the indicated drugs for 24 h. Cells were irradiated with UV and incubated for 4 h, and luciferase activity was measured. Relative luciferase activity was normalized to Renilla luciferase activity and was represented as a fold change compared with control. Experiments were performed in triplicate, and standard deviation is shown as control vs. drug-treated cells, *: p < 0.05, **: p < 0.01, ***: p < 0.005, and ****: p < 0.001. (B) The indicated stimuli decreased LNX1 mRNA levels. Quantitative RT-PCR was used to measure the mRNA expression levels of LNX1. Control vs. drug-treated cells, *: p < 0.05, **: p < 0.01, and ***: p < 0.005. (C,D) LNX1 protein levels decreased in response to the indicated stimuli. Each cell lysate was probed with the indicated antibodies. The uncropped Western Blot images can be found in Figure S3.
3.2. LPS, PMA, and Serum Addition Increase the Expression Levels of LNX1
Lipopolysaccharide (LPS) treatment leads to the early activation of the NF-κB, IRF3, and MAPK kinase signaling pathways [27,28,29]. Moreover, PMA is involved in the activation of the PKC-mediated NF-κB and MAPK signaling pathways [30,31]. Therefore, we examined whether LPS and PMA could regulate the expression of LNX1. Both LPS and PMA increased the promoter activity of LNX1 (Figure 2A). Growth medium containing FBS provides many factors that are essential for cell growth. LNX1 expression levels in cells grown in serum-free medium were lower than those in cells grown in growth medium. However, replacing the growth medium reactivated the expression of LNX1 (Figure 2B and Figure S1B). Furthermore, these factors increased the protein expression levels of LNX1 (Figure 2C,D). Therefore, signaling pathways associated with cell growth influence the expression of LNX1.

Figure 2.
LPS, PMA, and serum addition increase the LNX1 expression levels. (A) A549 cells were transfected with pGL3-LNX1 plasmids. After 24 h, cells were incubated with the indicated concentrations of LPS, PMA or serum-free medium. Serum-free medium was replaced with growth medium (GM) and cells were incubated for the indicated hours. Relative luciferase activity was measured and represented as a fold change compared with control. Experiments were performed in triplicate, and standard deviation is shown as control vs. drug-treated cells, *: p < 0.05, **: p < 0.01, ***: p < 0.005 (B) The indicated stimuli increased LNX1 mRNA levels. Expression levels were calculated by quantitative real-time PCR analysis. The results are represented as relative mRNA levels. (C,D) LNX1 protein levels increased in cells grown in LPS, PMA, and FBS-containing medium. Each cell lysate was probed with the indicated antibodies and quantified. The uncropped Western Blot images can be found in Figure S3.
In addition, we conducted a bioinformatic analysis (AliBaba2.1, [32]) for potential binding sites for oncogenic transcription factors. The LNX1 promoter region (−500 bp) contains several oncogenic transcription factors, including NF-kB, c-myc and AP-1 (Figure S2A). We also analyzed different publicly available gene expression datasets (GSE13333, GSE17511, GSE37219, and GSE6077) (Figure S2B). These data included gene expression profiles in mouse serum response factor (SRF)-deficient mice (GSE13333) [33], the back skin from transgenic mice with the inhibitor of NF-κB kinase beta (IKK-β) overexpression (GSE17511) [34], osteoclasts lacking the transcription factor NFATc1 (GSE37219) [35], and the lungs of transgenic embryos overexpressing N-myc in the lung epithelia (GSE6077) [36]. LNX1 expression levels significantly decreased in SRF-deficient cells, NFATc1-deficient cells, and IKK-β-overexpressing cells. The expression levels of LNX1 increased in proto-oncogene N-myc transgenic models. These data suggest that LNX1 mRNA expression levels are affected by the growth signaling pathways containing AP-1, NFATc1, N-myc, and NF-kB.
3.3. LNX1 Expression Correlates with Cell-Cycle Progression
Since the expression levels of LNX1 increased in growth medium, we examined whether this was related to cell-cycle progression. First, A549 cells were synchronized by using a double-thymidine block, which obstructs DNA replication and synchronizes cells at the G1-S border [37,38]. According to the DNA content of cells determined by PI, the cells progressed into S phase 4 h after the release of the block; after 6 h, the cells reached the G2 phase. (Figure 3A). We measured the mRNA expression levels of LNX1 under the same conditions. LNX1 expression levels decreased in G1/S arrest (0 h); however, these levels increased during cell-cycle progression (2–6 h) (Figure 3C). LNX1 expression levels were correlated with the S/G2/M population of the cells (Figure 3D). These data suggest a relationship between LNX1 and cell-cycle dynamics.
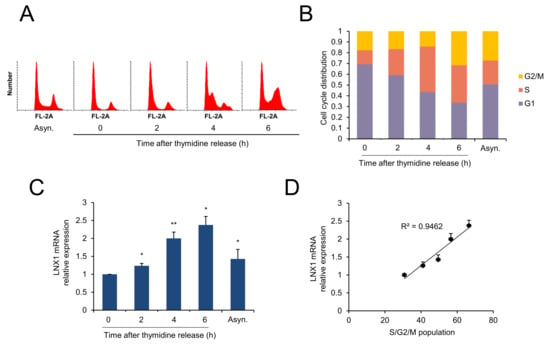
Figure 3.
LNX1 expression correlates with cell-cycle progression. A549 cells were synchronized using a double-thymidine block. Once released, cells were collected at the indicated time points. (A,B) Cell-cycle distribution was obtained by flow cytometry analysis of the DNA of PI-stained cells. (C) Expression levels were calculated by quantitative real-time PCR analysis. Synchronous vs. thymidine release or asynchronous, *: p < 0.05 and **: p < 0.01, (D) LNX1 expression is correlated with S, G2, and M populations. The R2 values were calculated and have been shown in the graphs.
3.4. LNX1 KO Delays Cell-Cycle Progression
To examine the potential role of LNX1 in the cell cycle, we generated LNX1 KO A549 cells using the CRISPR-CRISPR associated protein 9 (CRISPR-Cas9) genome-editing system. Western blot analysis was conducted using anti-LNX1 antibodies and revealed that LNX1 expression levels decreased in A549 cells (Figure 4A). The expression levels of genes encoding cyclin D1 and cyclin E1 decreased in LNX1 KO cells; however, those encoding the cyclin-dependent kinase inhibitors p16INK4A and p21WAF1/Cip1 increased (Figure 4A). These results suggest that LNX1 KO cells exhibit aberrant cell-cycle dynamics.
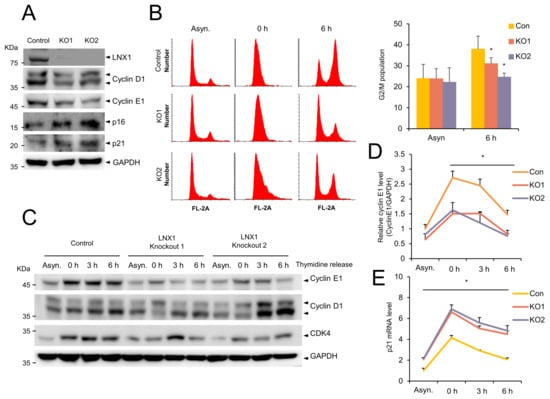
Figure 4.
Knocking out LNX1 delays cell-cycle progression. (A) Expression levels of genes encoding the cell-cycle markers cyclin D1 and cyclin E1 decreased in LNX1 knockout (KO) cells. LNX1 KO cell lines were generated using the lentiviral-mediated CRISPR-Cas9 genome editing. A549 cells were infected with the lentivirus encoding LNX1 gRNA. Equal amounts of cell lysates were probed with the indicated antibodies. (B) G2 and M populations decreased in LNX1 KO cells. Cells were synchronized by a double-thymidine block. Once released, cells were collected at indicated time points. Cell-cycle distributions were obtained following flow cytometry analysis of DNA in PI-stained cells. Control vs. LNX1 KO, *: p < 0.05 (n = 3). (C) Knocking out LNX1 disrupts the cell-cycle dynamics and expression of genes encoding cyclin D1 and cyclin E1. Control and LNX1 KO cells were synchronized by a double thymidine block. Once released, cell lysates were collected at indicated time points. Equal amounts of cell lysates were probed with the indicated antibodies. (D) The bands of gene encoding cyclin E1 were quantified, and the relative expression levels are shown in the graph. Control cells vs. LNX1 KO cells, *: p < 0.05. (E) mRNA levels of p21 were measured based on quantitative real-time PCR analysis. Control cells vs. LNX1 KO cells, *: p < 0.05. The uncropped Western Blot images can be found in Figure S3.
To investigate whether LNX1 KO contributes to cell-cycle progression, we examined the cell-cycle profiles of WT and LNX1 KO cells. The WT and LNX1 KO cells were synchronized by using a double-thymidine block, and the cell-cycle progression was determined by flow cytometry analysis (Figure 4B). The cell-cycle profiles were similar in WT and LNX1 KO cells during G1/S arrest (0 h). However, the entry into the G2/M-phase (6 h after release) was delayed in LNX1 KO cells. In WT cells, the progression through the G2/M-phase (47%) was completed after 6 h, and a significant fraction of LNX1 KO1 cells (34%) and KO2 cells (27%) did not reach the G2/M-phase even after 6 h of release (Figure 4B). Consistent with this data, genes encoding cyclin D1 and cyclin E1 were found to be differentially expressed in LNX1 KO cells compared with those in WT cells. In particular, expression levels of gene encoding cyclin E1 decreased and cell-cycle progression was delayed in LNX1 KO cells at the release time points (Figure 4C,D). In addition, increased expression levels of the CDK inhibitor p21 were observed in LNX1 KO cells (Figure 4E).
3.5. LNX1 Overexpression Leads to Cell-Cycle Progression and Facilitates Cisplatin Resistance
Once we had found that the cell growth conditions increase expression of LNX1 and disruption of LNX1 delays cell-cycle progression, we wondered what effect increased expression of LNX1 would have on the cell cycle. We found that the expression of genes encoding cyclin D1 and cyclin E1 was induced in cells with an increased dosage of transfected plasmid encoding LNX1 (Figure 5A–C). The endogenous expression levels of genes encoding cyclin D1 and cyclin E1 increased depending on the amount of transfected plasmid encoding LNX1, thus indicating that the upregulation of these genes was dependent on the dose of LNX1. We constructed HEK293 cell lines that stably expressed LNX1 for subsequent analysis (Figure 5D). LNX1-expressing HEK293 cells were then incubated with thymidine to synchronize cells and were harvested at the indicated time points after the release of thymidine. The overexpression of LNX1 increased the proportion of cycling cells (S/G2/M phase) (Figure 5E,F). In addition, the proliferation of cells stably expressing LNX1 increased, in comparison with that of the control cells (Figure 5G). These data suggest that overexpression of LNX1 leads to cell-cycle progression.
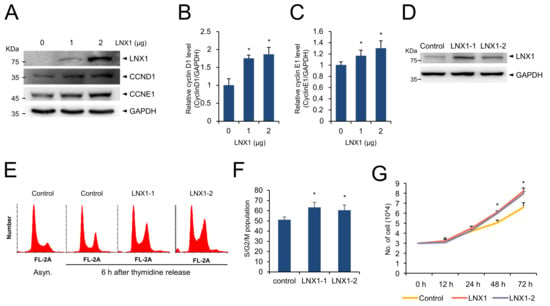
Figure 5.
Overexpression of LNX1 promotes cell-cycle progression. (A) The expression levels of genes encoding cyclin D1 and cyclin E1 increased in a dose-dependent manner in HEK293 cells that were transfected with plasmids encoding LNX1. Cell lysates were probed with indicated antibodies. (B,C) The bands of genes encoding cyclin D1 and cyclin E1 were quantified, and the relative expression levels are shown in the graph. Control cells vs. LNX1-overexpressing cells, *: p < 0.05. (D) HEK293 cells with stable expression of LNX1 were developed, and (E,F) cell-cycle distribution was analyzed. Cells were synchronized by using double-thymidine block and harvested at 6 h after thymidine release. The cell-cycle profiles were obtained following flow cytometry analysis of DNA in PI-stained cells. (G) Equal numbers of control and LNX1 (stable expression) cells were seeded and incubated for the indicated hours. Each cell was counted via Vi-Cell XR counter (Beckman Coulter, Brea, CA, USA), which is a trypan blue-based automatic cell counter. The uncropped Western Blot images can be found in Figure S3.
As LNX1 expression induced the cell cycle, we sought to determine its role in cancer therapy using cisplatin. We treated the control and LNX1-expressing cells with cisplatin and measured cell viability using the MTT assay. Trypan blue-stained cells were counted using the cell counter. We found that the expression of LNX1 reduced cisplatin-mediated cell death (Figure 6A–C). Transient LNX1 overexpression resisted cisplatin-induced apoptosis (Figure 6A). In addition, LNX1-overexpressing cells showed decreased cell death following cisplatin treatment (Figure 6B,C). Further, in cells stably expressing LNX1, the subG1 population and aberrant distribution of cell cycles induced by cisplatin were reduced (Figure 6D). Additionally, upon cisplatin treatment, the cleavage of PARP1 (an apoptosis marker protein) decreased in cells stably expressing LNX1 compared with that in control cells (Figure 6E).
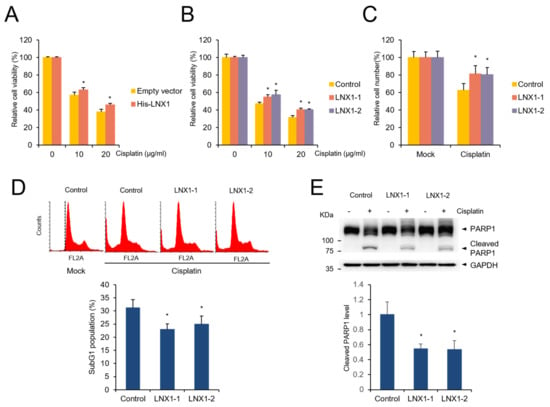
Figure 6.
Overexpression of LNX1 increases resistance to cisplatin. (A) HEK293 cells were transfected with plasmids encoding LNX1 and treated with the indicated concentrations of cisplatin for 24 h. (B) Control and LNX1 (stable expression) cells were treated with cisplatin. Cell viability was measured using MTT assay. Control cells vs. LNX1-overexpressing cells or cells stably expressing LNX1, *: p < 0.05 (C) HEK293 cells stably expressing LNX1 were treated with 10 µg/mL of cisplatin for 24 h. Live cells were counted using Vi-Cell XR counter, which is a trypan blue-based automatic cell counter. (D) Upon cisplatin treatment, subG1 population was reduced in cells stably expressing LNX1. Cells that were treated with 10 µg/mL of cisplatin for 24 h were stained with PI, and the subG1 population was analyzed using flow cytometry analysis. Control cells vs. cells stably expressing LNX1, *: p < 0.05. (E) Cisplatin-treated control and cells stably expressing LNX1 were harvested 24 h after treatment. Cell lysates were probed with PARP1 antibodies. Control cells vs. cells stably expressing LNX1, *: p < 0.05. The uncropped Western Blot images can be found in Figure S3.
As the overexpression of LNX1 decreased cisplatin-induced cell death in HEK293 cells, we examined whether LNX1-overexpressing cancer cells were resistant to cisplatin treatment. LNX1-overexpressing cells were generated with A549, MCF7 and H460 cells and the expression of cyclin D1 and cyclin E1 were examined. We found cyclin D1 expression is increased in LNX1-overexpressing cells (Figure 7A). Next, we treated control and LNX1-overexpressing cells with cisplatin, and found that LNX1 expression resulted in enhanced cell viability upon cisplatin treatment (Figure 7B). These data suggest that the overexpression of LNX1 contributes to cisplatin resistance.

Figure 7.
Overexpression of LNX1 increases resistance to cisplatin in cancer cells. (A) LNX1-overexpression cells were generated using A549, MCF7 and H460 cells, and the cell lysates were subject to Western blots with indicated antibodies. (B) Control and LNX1 (stable expression) cells were treated with cisplatin. Cell viability was measured using MTT assay. Control cells vs. LNX1-overexpressing cells or cells stably expressing LNX1, *: p < 0.05. The uncropped Western Blot images can be found in Figure S3.
4. Discussion
In a previous study, we reported that LNX1 acts as an oncogene and found that the expression levels of LNX1 increased in cancers using the TCGA database [16]. However, the factors that regulate LNX1 expression in cells was unknown. In this study, we found that several stimuli regulate LNX1 expression (Figure 1 and Figure 2). DNA damaging agents, including the anticancer drugs cisplatin and topotecan, UV, and hydroxyurea, decreased the expression levels of LNX1. Moreover, the promoter activity and mRNA and protein levels of LNX1 were decreased by these stimuli. We analyzed the promoter region of LNX1 based on the Ensemble database [25] and constructed a reporter plasmid containing the LNX1 promoter region (−1.5 kb upstream from TSS).
LPS, PMA, and replacement of serum-free medium with FBS-containing medium increased LNX1 expression levels, including promoter activity and levels of mRNA and protein (Figure 2). LPS leads to the early activation of the NF-κB, IRF3, and MAPK kinase signaling pathways [27,28,29]. Additionally, PMA is involved in the PKC-mediated NF-κB and MAPK signaling pathways [30,31]. Therefore, we expected that signaling associated with the regulation of cell growth, such as MAPK and NF-kB, was involved in the regulation of LNX1 expression. Other studies have shown that cell-growth-associated signaling pathways, including N-myc, NF-Kb, and NFATc1, regulate the expression of LNX1. Using publicly available gene expression datasets (GSE13333, GSE17511, GSE37219, GSE6077), several studies have implicated that the mRNA expression of LNX1 is affected by the growth signaling pathway containing NFATc1, N-myc, and NF-kB [33,34,35,36]. In particular, LNX1 expression significantly decreased in SRF-deficient mice. In our results, the expression of LNX1 decreased in cells grown in serum-free medium compared with that in cells grown in growth medium. Replacement of the growth medium with serum-free medium induced the expression of LNX1. These data suggest that the expression of LNX1 is associated with oncogenic responses, such as growth factors and other oncogenes.
As we anticipated that cell growth could be affected by LNX1, we analyzed its relationship with the cell cycle. We applied the widely used double-thymidine-block method to analyze the cell cycle [38,39]. The double-thymidine block causes G1/S arrest, and replacement with fresh medium aids progression to the next stage of the cell cycle. The expression levels of LNX1 decreased in synchronous cells compared with those in asynchronous cells. However, LNX1 expression levels increased after thymidine release. Therefore, we found a correlation between LNX1 expression and S/G2/M populations (Figure 3). When we used hydroxyurea to block the G1 to S transition, we observed that the expression pattern of LNX1 and its correlation with the cell-cycle distribution were similar to those when thymidine was used. Moreover, CRISPR-Cas9-mediated LNX1 KO cells delayed the progression of the cell cycle. The population of G2/M cells after 6 h of thymidine release was decreased in LNX1 KO (Figure 4B). This result is consistent with the previous report that downregulation of LNX1 by siRNA induces cell-cycle arrest in HEK293 cells [40]. The expression levels of genes encoding cyclin D1 and cyclin E1 decreased in LNX1 KO cells, and the cyclin kinetics collapsed in the thymidine-mediated cell-cycle analysis. In particular, the expression level of gene-encoding cyclin E1 decreased and cell-cycle progression was consequently delayed in LNX1 KO cells (Figure 4D). This could be because gene encoding cyclin E1 is a crucial regulator and is dynamically expressed during the G1 to S transition [41,42]. In LNX1 overexpression experiments, the expression level of the gene-encoding cyclin D1 was found to be higher than that of gene-encoding cyclin E1 (Figure 5A). As most cells remained in the G1 phase, gene-encoding cyclin D1 was more dominant than that encoding cyclin E1 in this stage of the cell cycle [43,44].
In a previous study, we demonstrated that LNX1 acts as a proto-oncogene by inhibiting p53 stability [16]. p53 plays a crucial role in DNA repair and is a determinant of the development of cisplatin resistance [45]. The main mechanism underlying the anticancer property of cisplatin is DNA damage caused by binding platinum to DNA, thereby forming DNA adducts. The DNA damage by cisplatin induces cell-cycle arrest and initiates apoptosis in fast-cycling cells [21,46]. However, cancer stem cells remain in a quiescent, slow-proliferating state and escape from chemotherapy, including cisplatin treatment that targets fast-cycling cells [46].
In this report, we studied the regulation of LNX1 expression and its consequences. Cisplatin treatment resulted in the downregulation of LNX1, and LNX1 overexpression contributes to enhanced cell viability upon cisplatin treatment. These results support the notion that LNX1 expression regulates cisplatin resistance or cisplatin sensitivity. We found that LNX1 contributes to cell-cycle progression and the stability of p53 [16]; therefore, we examined whether it is related to cisplatin resistance or sensitivity. The overexpression of LNX1 reduced cell death following cisplatin treatment. The subG1 population in cisplatin-treated WT cells was higher than that in LNX1-stable cells, and the extent of the collapse of the cell-cycle distribution of WT cells was greater than that of cisplatin-treated LNX1-stable cells (Figure 5). Although LNX1 overexpression accelerated the cell cycle, the sensitivity of cisplatin did not increase in LNX1-expressing cells. It is possible that cisplatin has various mechanisms that induce cell death and compensate for the absence of LNX1 [47,48,49]. Moreover, apoptosis signaling involving p53 is directly associated with cell death induced by cisplatin. Knocking out LNX1 did not influence the viability of cisplatin. Both WT and LNX1 KO cells might have efficient p53 activity that can induce cell death by cisplatin treatment. However, because the overexpression of LNX1 can lead to the degradation of p53, cell death is reduced by cisplatin treatment [16,50]. Further studies are required to elucidate the relationship between p53 and LNX1 expression in cisplatin resistance and the role of LNX1 in DNA damage response. In addition, we need to confirm the role of LNX1 in various other cancer cell lines and in vivo.
5. Conclusions
Here, we demonstrate that LNX1 expression is decreased by DNA damage, and increased by cell cycle progression. We also showed that LNX1 knockout results in a delay in cell cycle progression and LNX1 overexpression activates cell cycle progression. Finally, LNX1 overexpression increased resistance to cisplatin treatment. These results indicate that elevated level of LNX1 in cancer cells contributes to tumor growth and enhanced drug resistance.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers13164066/s1, Figure S1. DNA damage and cell growth were associated with expression of LNX1 in MCF7 and H460 cells, Figure S2. Several oncogenic transcription factors are involved with the expression of LNX1, Figure S3. Full length blots cropped for representative figures.
Author Contributions
Conceptualization, M.J., R.P. and J.P.; methodology, M.J. and R.P.; investigation, M.J., R.P. and Y.-I.P.; materials, E.-J.L.; writing—original draft preparation, M.J.; writing—review and editing, M.J., Y.P., S.N., J.I.L. and J.P. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (2019R1A2C1006511).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
Authors have no conflict of interest to declare.
References
- Nie, J.; McGill, M.A.; Dermer, M.; Dho, S.E.; Wolting, C.D.; McGlade, C.J. LNX functions as a RING type E3 ubiquitin ligase that targets the cell fate determinant Numb for ubiquitin-dependent degradation. EMBO J. 2002, 21, 93–102. [Google Scholar] [CrossRef]
- Lenihan, J.A.; Saha, O.; Mansfield, L.M.; Young, P.W. Tight, cell type-specific control of LNX expression in the nervous system, at the level of transcription, translation and protein stability. Gene 2014, 552, 39–50. [Google Scholar] [CrossRef]
- Dho, S.E.; Jacob, S.; Wolting, C.D.; French, M.B.; Rohrschneider, L.R.; McGlade, C.J. The mammalian numb phosphotyrosine-binding domain: Characterization of binding specificity and identification of a novel PDZ domain-containing numb binding protein, LNX. J. Biol. Chem. 1998, 273, 9179–9187. [Google Scholar] [CrossRef] [PubMed]
- Kansaku, A.; Hirabayashi, S.; Mori, H.; Fujiwara, N.; Kawata, A.; Ikeda, M.; Rokukawa, C.; Kurihara, H.; Hata, Y. Ligand-of-Numb protein X is an endocytic scaffold for junctional adhesion molecule 4. Oncogene 2006, 25, 5071–5084. [Google Scholar] [CrossRef] [PubMed]
- Wolting, C.D.; Griffiths, E.K.; Sarao, R.; Prevost, B.C.; Wybenga-Groot, L.E.; McGlade, C.J. Biochemical and computational analysis of LNX1 interacting proteins. PLoS ONE 2011, 6, e26248. [Google Scholar] [CrossRef] [PubMed]
- Lenihan, J.A.; Saha, O.; Young, P.W. Proteomic analysis reveals novel ligands and substrates for LNX1 E3 ubiquitin ligase. PLoS ONE 2017, 12, e0187352. [Google Scholar] [CrossRef]
- Weiss, A.; Baumgartner, M.; Radziwill, G.; Dennler, J.; Moelling, K. c-Src is a PDZ interaction partner and substrate of the E3 ubiquitin ligase Ligand-of-Numb protein X1. FEBS Lett. 2007, 581, 5131–5136. [Google Scholar] [CrossRef] [PubMed]
- Young, P.; Nie, J.; Wang, X.; McGlade, C.J.; Rich, M.M.; Feng, G. LNX1 is a perisynaptic Schwann cell specific E3 ubiquitin ligase that interacts with ErbB2. Mol. Cell. Neurosci. 2005, 30, 238–248. [Google Scholar] [CrossRef]
- Lee, C.-H.; Yang, J.-R.; Chen, C.-Y.; Tsai, M.-H.; Hung, P.-F.; Chen, S.-J.; Chiang, S.-L.; Chang, H.; Lin, P. Novel STAT3 Inhibitor LDOC1 Targets Phospho-JAK2 for Degradation by Interacting with LNX1 and Regulates the Aggressiveness of Lung Cancer. Cancers 2019, 11, 63. [Google Scholar] [CrossRef]
- Baisiwala, S.; Hall, R.R.; Saathoff, M.R.; Shireman, J.M.; Park, C.; Budhiraja, S.; Goel, C.; Warnke, L.; Hardiman, C.; Wang, J.Y.; et al. LNX1 Modulates Notch1 Signaling to Promote Expansion of the Glioma Stem Cell Population during Temozolomide Therapy in Glioblastoma. Cancers 2020, 12, 3505. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Wang, L.; Shan, Y.; Nafees, M.; Ihab, E.; Zhang, R.; Wang, F.; Yin, W. Suppression of cancer stemness by upregulating Ligand-of-Numb protein X1 in colorectal carcinoma. PLoS ONE 2017, 12, e0188665. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, J.; Xu, J.; Zhao, W.; Hu, G.; Cheng, H.; Kang, Y.; Xie, Y.; Lu, Y. Characterization of human LNX, a novel ligand of Numb protein X that is downregulated in human gliomas. Int. J. Biochem. Cell Biol. 2005, 37, 2273–2283. [Google Scholar] [CrossRef] [PubMed]
- Camps, J.; Pitt, J.J.; Emons, G.; Hummon, A.B.; Case, C.M.; Grade, M.; Jones, T.L.; Nguyen, Q.T.; Ghadimi, B.M.; Beissbarth, T.; et al. Genetic amplification of the NOTCH modulator LNX2 upregulates the WNT/beta-catenin pathway in colorectal cancer. Cancer Res. 2013, 73, 2003–2013. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Won, M.; Ro, H. The Molecular and Pathophysiological Functions of Members of the LNX/PDZRN E3 Ubiquitin Ligase Family. Molecules 2020, 25, 5938. [Google Scholar] [CrossRef]
- Young, P.W. LNX1/LNX2 proteins: Functions in neuronal signalling and beyond. Neuronal Signal. 2018, 2, NS20170191. [Google Scholar] [CrossRef]
- Park, R.; Kim, H.; Jang, M.; Jo, D.; Park, Y.I.; Namkoong, S.; Lee, J.I.; Jang, I.S.; Park, J. LNX1 contributes to tumor growth by down-regulating p53 stability. FASEB J. 2019, 33, 13216–13227. [Google Scholar] [CrossRef]
- Vogelstein, B.; Lane, D.; Levine, A.J. Surfing the p53 network. Nature 2000, 408, 307–310. [Google Scholar] [CrossRef]
- Siddik, Z.H. Cisplatin: Mode of cytotoxic action and molecular basis of resistance. Oncogene 2003, 22, 7265–7279. [Google Scholar] [CrossRef]
- Kelland, L. The molecular basis of cisplatin sensitivity/resistance. Eur. J. Cancer 1994, 30, 725–727. [Google Scholar] [CrossRef]
- Weiss, R.B.; Christian, M.C. New cisplatin analogues in development. Drugs 1993, 46, 360–377. [Google Scholar] [CrossRef]
- Andrews, P.A. Mechanisms of acquired resistance to cisplatin. Anticancer. Drug Resist. 1994, 73, 217–248. [Google Scholar]
- Moorehead, R.A.; Singh, G. Influence of the proto-oncogene c-fos on cisplatin sensitivity. Biochem. Pharmacol. 2000, 59, 337–345. [Google Scholar] [CrossRef]
- Dempke, W.; Voigt, W.; Grothey, A.; Hill, B.T.; Schmoll, H.-J. Cisplatin resistance and oncogenes-a review. Anti-Cancer Drugs 2000, 11, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Kingston, R.E.; Chen, C.A.; Rose, J.K. Calcium phosphate transfection. Curr. Protoc. Mol. Biol. 2003, 63, 9.1.1–9.1.11. [Google Scholar] [CrossRef] [PubMed]
- Ensemble Database. Available online: http://www.ensembl.org/ (accessed on 12 July 2021).
- Chang, L.-C.; Sheu, H.-M.; Huang, Y.-S.; Tsai, T.-R.; Kuo, K.-W. A novel function of emodin: Enhancement of the nucleotide excision repair of UV-and cisplatin-induced DNA damage in human cells. Biochem. Pharmacol. 1999, 58, 49–57. [Google Scholar] [CrossRef]
- Fitzgerald, K.A.; Rowe, D.C.; Barnes, B.J.; Caffrey, D.R.; Visintin, A.; Latz, E.; Monks, B.; Pitha, P.M.; Golenbock, D.T. LPS-TLR4 signaling to IRF-3/7 and NF-κB involves the toll adapters TRAM and TRIF. J. Exp. Med. 2003, 198, 1043–1055. [Google Scholar] [CrossRef]
- He, W.; Wang, Z.; Luo, Z.; Yu, Q.; Jiang, Y.; Zhang, Y.; Zhou, Z.; Smith, A.J.; Cooper, P.R. LPS promote the odontoblastic differentiation of human dental pulp stem cells via MAPK signaling pathway. J. Cell. Physiol. 2015, 230, 554–561. [Google Scholar] [CrossRef]
- Lu, Y.-C.; Yeh, W.-C.; Ohashi, P.S. LPS/TLR4 signal transduction pathway. Cytokine 2008, 42, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Shelly, C.; Petruzzelli, L.; Herrera, R. PMA-induced phenotypic changes in K562 cells: MAPK-dependent and-independent events. Leukemia 1998, 12, 1951–1961. [Google Scholar] [CrossRef] [PubMed]
- Ueberla, K.; Lu, Y.; Chung, E.; Haseltine, W.A. The NF-kB p65 promoter. J. Acquir. Immune Defic. Syndr. 1998, 6, 227–230. [Google Scholar]
- AliBaba2.1. Available online: http://gene-regulation.com/pub/programs/alibaba2/ (accessed on 12 July 2021).
- Sun, K.; Battle, M.A.; Misra, R.P.; Duncan, S.A. Hepatocyte expression of serum response factor is essential for liver function, hepatocyte proliferation and survival, and postnatal body growth in mice. Hepatology 2009, 49, 1645–1654. [Google Scholar] [CrossRef] [PubMed]
- Page, A.; Navarro, M.; Garín, M.; Pérez, P.; Casanova, M.L.; Moreno, R.; Jorcano, J.L.; Cascallana, J.L.; Bravo, A.; Ramírez, A. IKKβ leads to an inflammatory skin disease resembling interface dermatitis. J. Investig. Dermatol. 2010, 130, 1598–1610. [Google Scholar] [CrossRef] [PubMed]
- Aliprantis, A.O.; Ueki, Y.; Sulyanto, R.; Park, A.; Sigrist, K.S.; Sharma, S.M.; Ostrowski, M.C.; Olsen, B.R.; Glimcher, L.H. NFATc1 in mice represses osteoprotegerin during osteoclastogenesis and dissociates systemic osteopenia from inflammation in cherubism. J. Clin. Investig. 2008, 118, 3775–3789. [Google Scholar] [CrossRef]
- Cox, B.; Kislinger, T.; Wigle, D.A.; Kannan, A.; Brown, K.; Okubo, T.; Hogan, B.; Jurisica, I.; Frey, B.; Rossant, J. Integrated proteomic and transcriptomic profiling of mouse lung development and Nmyc target genes. Mol. Syst. Biol. 2007, 3, 109. [Google Scholar] [CrossRef]
- Apraiz, A.; Mitxelena, J.; Zubiaga, A. Studying cell cycle-regulated gene expression by two complementary cell synchronization protocols. J. Vis. Exp. JoVE 2017, 124, 55745. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.-Y.; Ma, W.-L.; Liang, S.; Zeng, Y.; Shi, R.; Yu, H.-L.; Xiao, W.-W.; Zheng, W.-L. Analysis of microRNA expression profiles during the cell cycle in synchronized HeLa cells. BMB Rep. 2009, 42, 593–598. [Google Scholar] [CrossRef]
- Sadasivam, S.; Duan, S.; DeCaprio, J.A. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 2012, 26, 474–489. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Gu, S.; Li, Y.; Ji, C.; Xie, Y.; Mao, Y. A global genomic view on LNX siRNA-mediated cell cycle arrest. Mol. Biol. Rep. 2011, 38, 2771–2783. [Google Scholar] [CrossRef]
- Wu, W.; Sato, K.; Koike, A.; Nishikawa, H.; Koizumi, H.; Venkitaraman, A.R.; Ohta, T. HERC2 is an E3 ligase that targets BRCA1 for degradation. Cancer Res. 2010, 70, 6384–6392. [Google Scholar] [CrossRef]
- Hayami, R.; Sato, K.; Wu, W.; Nishikawa, T.; Hiroi, J.; Ohtani-Kaneko, R.; Fukuda, M.; Ohta, T. Down-regulation of BRCA1-BARD1 ubiquitin ligase by CDK2. Cancer Res. 2005, 65, 6–10. [Google Scholar]
- Santra, M.K.; Wajapeyee, N.; Green, M.R. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature 2009, 459, 722–725. [Google Scholar] [CrossRef]
- Peurala, E.; Koivunen, P.; Haapasaari, K.-M.; Bloigu, R.; Jukkola-Vuorinen, A. The prognostic significance and value of cyclin D1, CDK4 and p16 in human breast cancer. Breast Cancer Res. 2013, 15, R5. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Howell, S.B. DNA mismatch repair and p53 function are major determinants of the rate of development of cisplatin resistance. Mol. Cancer Ther. 2006, 5, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
- Raudenska, M.; Balvan, J.; Fojtu, M.; Gumulec, J.; Masarik, M. Unexpected therapeutic effects of cisplatin. Metallomics 2019, 11, 1182–1199. [Google Scholar] [CrossRef]
- Ravi, R.; Somani, S.M.; Rybak, L.P. Mechanism of cisplatin ototoxicity: Antioxidant system. Pharmacol. Toxicol. 1995, 76, 386–394. [Google Scholar] [CrossRef]
- Reedijk, J.; Lohman, P. Cisplatin: Synthesis, antitumour activity and mechanism of action. Pharm. Weekbl. 1985, 7, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Shen, D.-W.; Pouliot, L.M.; Hall, M.D.; Gottesman, M.M. Cisplatin resistance: A cellular self-defense mechanism resulting from multiple epigenetic and genetic changes. Pharmacol. Rev. 2012, 64, 706–721. [Google Scholar] [CrossRef]
- Yang, X.; Fraser, M.; Moll, U.M.; Basak, A.; Tsang, B.K. Akt-mediated cisplatin resistance in ovarian cancer: Modulation of p53 action on caspase-dependent mitochondrial death pathway. Cancer Res. 2006, 66, 3126–3136. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).